Deep Learning for the Classification of Non-Hodgkin Lymphoma on Histopathological Images
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort, Tissue Microarray Construction, and Scanning of Tissue Slides
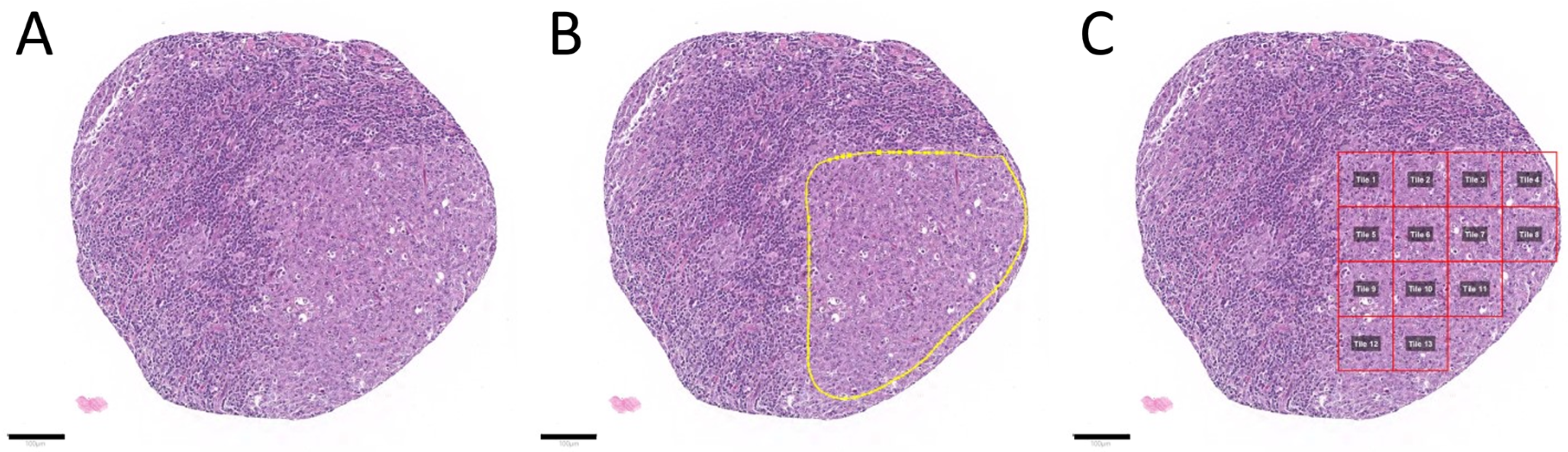
2.2. Tumor Annotation and Image Patch Extraction
2.3. Hardware and Software
2.4. Analytical Subsets
2.5. Model Training and Optimization
3. Results
3.1. Patient Cohort, Annotation, Image Patch Extraction, and Subset Analysis
3.2. Convolutional Neuronal Network Selection and Hyperparameter Optimization
3.3. Evaluation of the Test Set and Quality Control
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- National Cancer Institute. Cancer Stat Facts: Non-Hodgkin Lymphoma. Available online: https://seer.cancer.gov/statfacts/html/nhl.html (accessed on 21 January 2021).
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Di Napoli, A.; Remotti, D.; Agostinelli, C.; Ambrosio, M.R.; Ascani, S.; Carbone, A.; Facchetti, F.; Lazzi, S.; Leoncini, L.; Lucioni, M.; et al. A practical algorithmic approach to mature aggressive B cell lymphoma diagnosis in the double/triple hit era: Selecting cases, matching clinical benefit. Virchows Archiv 2019, 475, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Rosai, J. The Continuing Role of Morphology in the Molecular Age. Mod. Pathol. 2001, 14, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Märkl, B.; Füzesi, L.; Huss, R.; Bauer, S.; Schaller, T. Number of pathologists in Germany: Comparison with European countries, USA, and Canada. Virchows Archiv 2021, 478, 335–341. [Google Scholar] [CrossRef]
- A Adeyi, O. Pathology services in developing countries-the West African experience. Arch. Pathol. Lab. Med. 2011, 135, 183–186. [Google Scholar] [CrossRef]
- Adesina, A.; Chumba, D.; Nelson, A.M.; Orem, J.; Roberts, D.J.; Wabinga, H.; Wilson, M.; Rebbeck, T.R. Improvement of pathology in sub-Saharan Africa. Lancet Oncol. 2013, 14, e152–e157. [Google Scholar] [CrossRef]
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef]
- Wang, S.; Wang, T.; Yang, L.; Yang, D.M.; Fujimoto, J.; Yi, F.; Luo, X.; Yang, Y.; Yao, B.; Lin, S.; et al. ConvPath: A software tool for lung adenocarcinoma digital pathological image analysis aided by a convolutional neural network. EBioMedicine 2019, 50, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.W.; Tafe, L.J.; Linnik, Y.A.; Vaickus, L.J.; Tomita, N.; Hassanpour, S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Landhuis, E. Deep learning takes on tumours. Nat. Cell Biol. 2020, 580, 551–553. [Google Scholar] [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- El Achi, H.; Belousova, T.; Chen, L.; Wahed, A.; Wang, I.; Hu, Z.; Kanaan, Z.; Rios, A.; Nguyen, A.N.D. Automated Diagnosis of Lymphoma with Digital Pathology Images Using Deep Learning. Ann. Clin. Lab. Sci. 2019, 49, 153–160. [Google Scholar]
- Syrykh, C.; Abreu, A.; Amara, N.; Siegfried, A.; Maisongrosse, V.; Frenois, F.X.; Martin, L.; Rossi, C.; Laurent, C.; Brousset, P. Accurate diagnosis of lymphoma on whole-slide histopathology images using deep learning. npj Digit. Med. 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Li, D.; Bledsoe, J.R.; Zeng, Y.; Liu, W.; Hu, Y.; Bi, K.; Liang, A.; Li, S. A deep learning diagnostic platform for diffuse large B-cell lymphoma with high accuracy across multiple hospitals. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Swiderska-Chadaj, Z.; Hebeda, K.M.; Brand, M.V.D.; Litjens, G. Artificial intelligence to detect MYC translocation in slides of diffuse large B-cell lymphoma. Virchows Archiv 2020, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Sato, K.; Kabeya, Y.; Yonezawa, S.; Nakano, H.; Takeuchi, Y.; Ozawa, I.; Higo, S.; Yanagida, E.; Yamada, K.; et al. Deep learning shows the capability of high-level computer-aided diagnosis in malignant lymphoma. Lab. Investig. 2020, 100, 1300–1310. [Google Scholar] [CrossRef]
- Kriegsmann, M.; Haag, C.; Weis, C.-A.; Steinbuss, G.; Warth, A.; Zgorzelski, C.; Muley, T.; Winter, H.; Eichhorn, M.E.; Eichhorn, F.; et al. Deep Learning for the Classification of Small-Cell and Non-Small-Cell Lung Cancer. Cancers 2020, 12, 1604. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- University, H. BwForCluster MLS&WISO Production Hardware. Available online: https://wiki.bwhpc.de/e/BwForCluster_MLS%26WISO_Production_Hardware (accessed on 20 January 2021).
- Mohlman, J.S.; Leventhal, S.D.; Hansen, T.; Kohan, J.; Pascucci, V.; E Salama, M. Improving Augmented Human Intelligence to Distinguish Burkitt Lymphoma From Diffuse Large B-Cell Lymphoma Cases. Am. J. Clin. Pathol. 2020, 153, 743–759. [Google Scholar] [CrossRef]
- Grandini, M.; Bagli, E.; Visani, G. Metrics for Multi-Class Classification: An Overview. arXiv 2017, arXiv:2008.05756. [Google Scholar]
- Smilkov, D.; Thorat, N.; Kim, B.; Viégas, F.; Wattenberg, M. SmoothGrad: Removing Noise by Adding Noise. Available online: https://arxiv.org/abs/1706.03825 (accessed on 22 January 2021).
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Yamashita, R.; Long, J.; Longacre, T.; Peng, L.; Berry, G.; Martin, B.; Higgins, J.; Rubin, D.L.; Shen, J. Deep learning model for the prediction of microsatellite instability in colorectal cancer: A diagnostic study. Lancet Oncol. 2021, 22, 132–141. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, M.; Wang, S.; Li, X.; Sun, Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. 2020, 40, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Aeffner, F.; Zarella, M.D.; Buchbinder, N.; Bui, M.M.; Goodman, M.R.; Hartman, D.J.; Lujan, G.M.; Molani, M.A.; Parwani, A.V.; Lillard, K.; et al. Introduction to digital image analysis in whole-slide imaging: A white paper from the digital pathology association. J. Pathol. Informatics 2019, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Steiner, D.F.; Macdonald, R.; Liu, Y.; Truszkowski, P.; Hipp, J.D.; Gammage, C.; Thng, F.; Peng, L.; Stumpe, M.C. Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. Am. J. Surg. Pathol. 2018, 42, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Chen, H.; Graham, S.; Dou, Q.; Rajpoot, N.; Heng, P.-A. Fast ScanNet: Fast and Dense Analysis of Multi-Gigapixel Whole-Slide Images for Cancer Metastasis Detection. IEEE Trans. Med. Imaging 2019, 38, 1948–1958. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.W.M.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Informatics 2016, 7, 29. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, W.; Guo, X.; Wang, B.; Wang, Z. Classification of digital pathological images of non-Hodgkin’s lymphoma subtypes based on the fusion of transfer learning and principal component analysis. Med. Phys. 2020, 47, 4241–4253. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Wang, J.; Liu, Q.; Xie, H.; Yang, Z.; Zhou, H. Boosted EfficientNet: Detection of Lymph Node Metastases in Breast Cancer Using Convolutional Neural Networks. Cancers 2021, 13, 661. [Google Scholar] [CrossRef] [PubMed]
- Bhinder, P.; Chahin, M.; Zuberi, L. Concurrent Squamous Cell Carcinoma and Chronic Lymphocytic Leukemia Presenting as an Enlarging Neck Mass. J. Investig. Med. High. Impact Case Rep. 2019, 7, 2324709619842904. [Google Scholar] [CrossRef] [PubMed]
- Lyapichev, K.A.; Kurt, H.; Sukswai, N.; Konoplev, S.; Bueso-Ramos, C.E.; Khoury, J.D.; Huh, Y.O. Chronic lymphocytic leukemia with plasmacytic differentiation. Ann. Hematol. 2019, 98, 2437–2438. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Spina, V.; Gaidano, G. Biology and treatment of Richter syndrome. Blood 2018, 131, 2761–2772. [Google Scholar] [CrossRef] [PubMed]
- Crombie, J.; Davids, M.S. IGHVmutational status testing in chronic lymphocytic leukemia. Am. J. Hematol. 2017, 92, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef] [PubMed]




| Group | SLL/CLL | DLBCL | LN Lung | LN Colon | LN Pancreas |
|---|---|---|---|---|---|
| Total cases, n | 129 | 119 | 64 | 230 | 87 |
| Training set, ~60% of cases | |||||
| Cases, n | 78 | 80 | 34 | 134 | 52 |
| Image patches, n | |||||
| Total | 11,404 | 8625 | 5064 | 18,488 | 7004 |
| Minimum | 4 | 7 | 33 | 4 | 2 |
| Maximum | 231 | 278 | 238 | 222 | 245 |
| Mean | 146 | 108 | 149 | 138 | 135 |
| Median | 149 | 99.5 | 150 | 142 | 139 |
| Validation set, ~20% of cases | |||||
| Cases, n | 22 | 18 | 15 | 57 | 14 |
| Image patches, n | |||||
| Total | 3086 | 1815 | 2436 | 7870 | 1387 |
| Minimum | 3 | 13 | 115 | 24 | 18 |
| Maximum | 214 | 329 | 251 | 255 | 242 |
| Mean | 140 | 101 | 162 | 138 | 99 |
| Median | 146.5 | 88 | 156 | 140 | 80.5 |
| Test set, ~20% of cases | |||||
| Cases, n | 29 | 21 | 15 | 39 | 21 |
| Image patches, n | |||||
| Total | 4631 | 2236 | 2393 | 4966 | 2734 |
| Minimum | 18 | 22 | 105 | 17 | 48 |
| Maximum | 226 | 225 | 265 | 184 | 237 |
| Mean | 160 | 106 | 160 | 127 | 130 |
| Median | 189 | 103 | 162 | 132 | 131 |
| CQC Threshold | 50% | 60% | 70% | 80% | 90% | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PQC Threshold | PQC not met (%) | BACC (%) | CQC not met (%) | BACC (%) | CQC not met (%) | BACC (%) | CQC not met (%) | BACC (%) | CQC not met (%) | BACC (%) | CQC not met (%) |
| 50% | 0.54 | 84.48 | 0.8 | 88.61 | 5.6 | 89.55 | 8 | 94.12 | 17.6 | 95.56 | 24.8 |
| 60% | 3.44 | 83.74 | 0 | 87.67 | 4.8 | 89.55 | 8 | 94.12 | 16.8 | 93.75 | 22.4 |
| 70% | 6.43 | 85.25 | 0.8 | 85.99 | 3.2 | 89.64 | 7.2 | 94.74 | 15.2 | 93.75 | 21.6 |
| 80% | 10.05 | 85.32 | 0 | 86.79 | 4 | 89.64 | 7.2 | 92.42 | 11.2 | 93.75 | 20.8 |
| 90% | 15.74 | 85.25 | 0.8 | 85.24 | 2.4 | 89.64 | 6.4 | 91.3 | 9.6 | 93.75 | 18.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steinbuss, G.; Kriegsmann, M.; Zgorzelski, C.; Brobeil, A.; Goeppert, B.; Dietrich, S.; Mechtersheimer, G.; Kriegsmann, K. Deep Learning for the Classification of Non-Hodgkin Lymphoma on Histopathological Images. Cancers 2021, 13, 2419. https://doi.org/10.3390/cancers13102419
Steinbuss G, Kriegsmann M, Zgorzelski C, Brobeil A, Goeppert B, Dietrich S, Mechtersheimer G, Kriegsmann K. Deep Learning for the Classification of Non-Hodgkin Lymphoma on Histopathological Images. Cancers. 2021; 13(10):2419. https://doi.org/10.3390/cancers13102419
Chicago/Turabian StyleSteinbuss, Georg, Mark Kriegsmann, Christiane Zgorzelski, Alexander Brobeil, Benjamin Goeppert, Sascha Dietrich, Gunhild Mechtersheimer, and Katharina Kriegsmann. 2021. "Deep Learning for the Classification of Non-Hodgkin Lymphoma on Histopathological Images" Cancers 13, no. 10: 2419. https://doi.org/10.3390/cancers13102419
APA StyleSteinbuss, G., Kriegsmann, M., Zgorzelski, C., Brobeil, A., Goeppert, B., Dietrich, S., Mechtersheimer, G., & Kriegsmann, K. (2021). Deep Learning for the Classification of Non-Hodgkin Lymphoma on Histopathological Images. Cancers, 13(10), 2419. https://doi.org/10.3390/cancers13102419







