Interception of Signaling Circuits of Esophageal Adenocarcinoma Cells by Resveratrol Reveals Molecular and Immunomodulatory Signatures
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Line and Materials
2.2. Assessment of the Impact of Resveratrol on the Viability of OE33, OE19 and FLO-1 Cells
2.3. Muse Flow Cytometric Analysis
2.4. Proteomic Analysis
2.5. Protein Lysate Preparation
2.6. Minimal CyDye Labeling
2.7. Statistical Analysis
3. Results
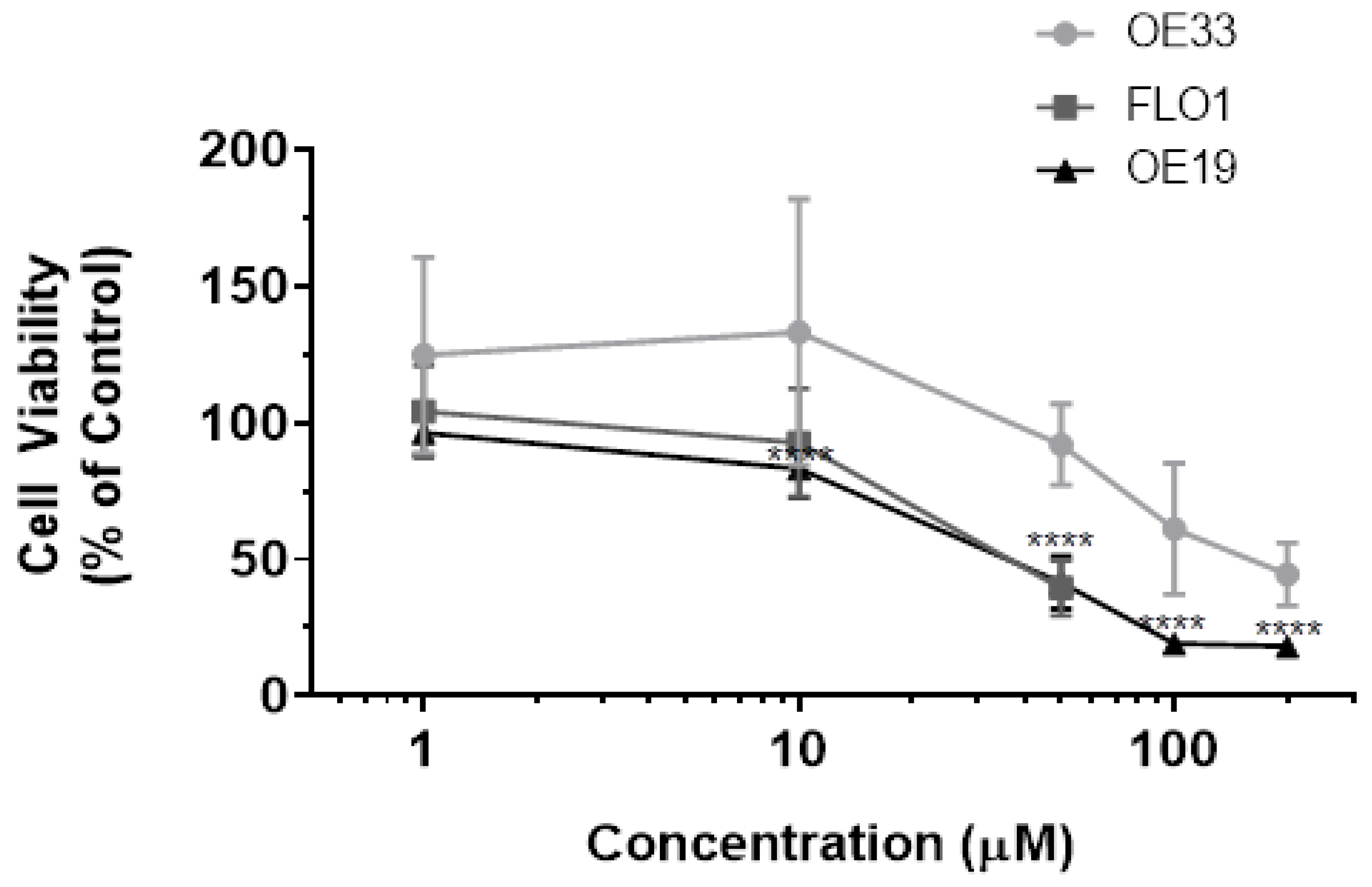
3.1. Resveratrol Impacts Cell Viability of OE33, OE19 and FLO-1 Cells
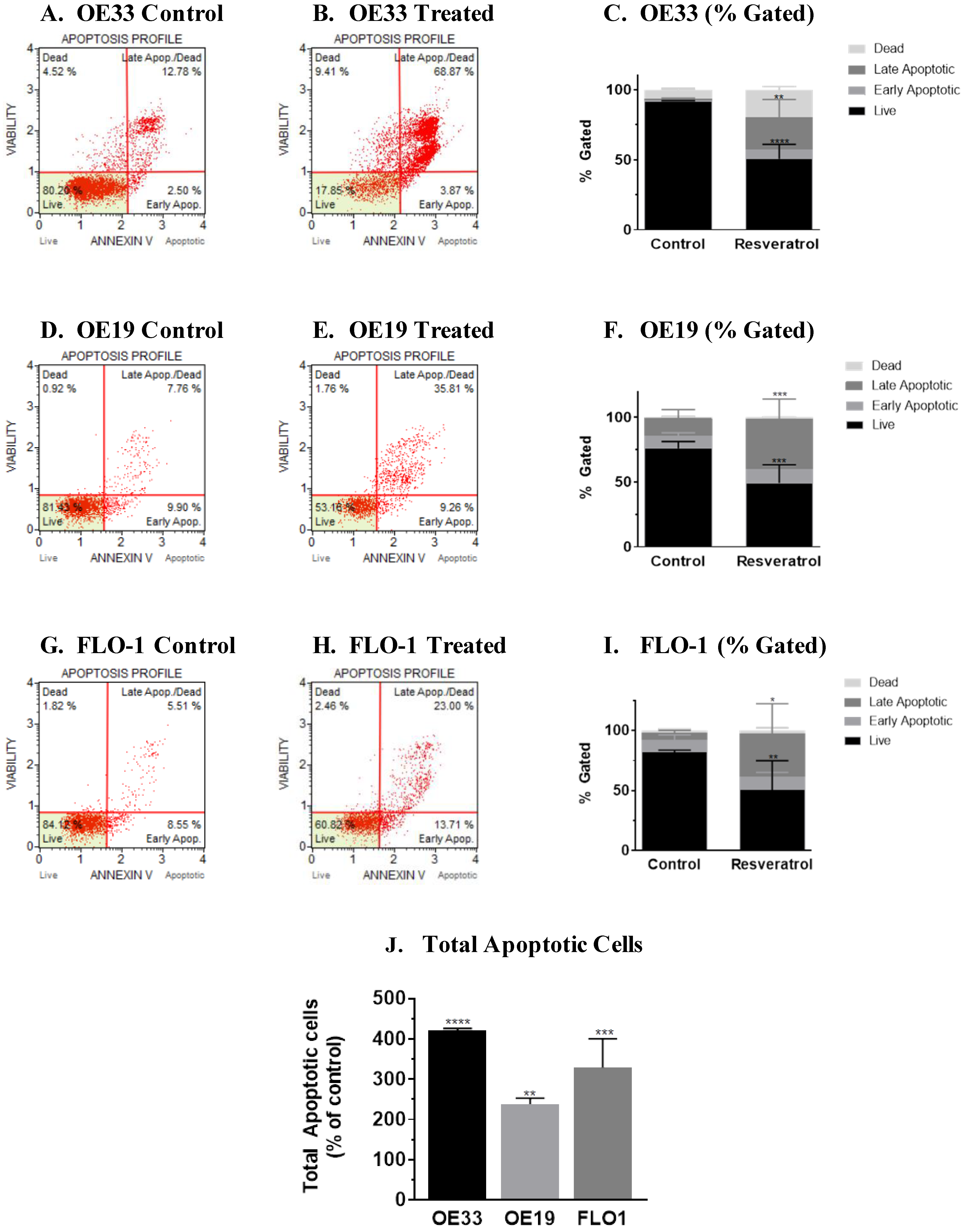
3.2. Molecular Analysis of Resveratrol’s Action Reveals the Impact on the Programmed Cell Death Pathway in the Three Esophageal Adenocarcinoma Cell Systems (OE33, OE19 and FLO-1) Studied
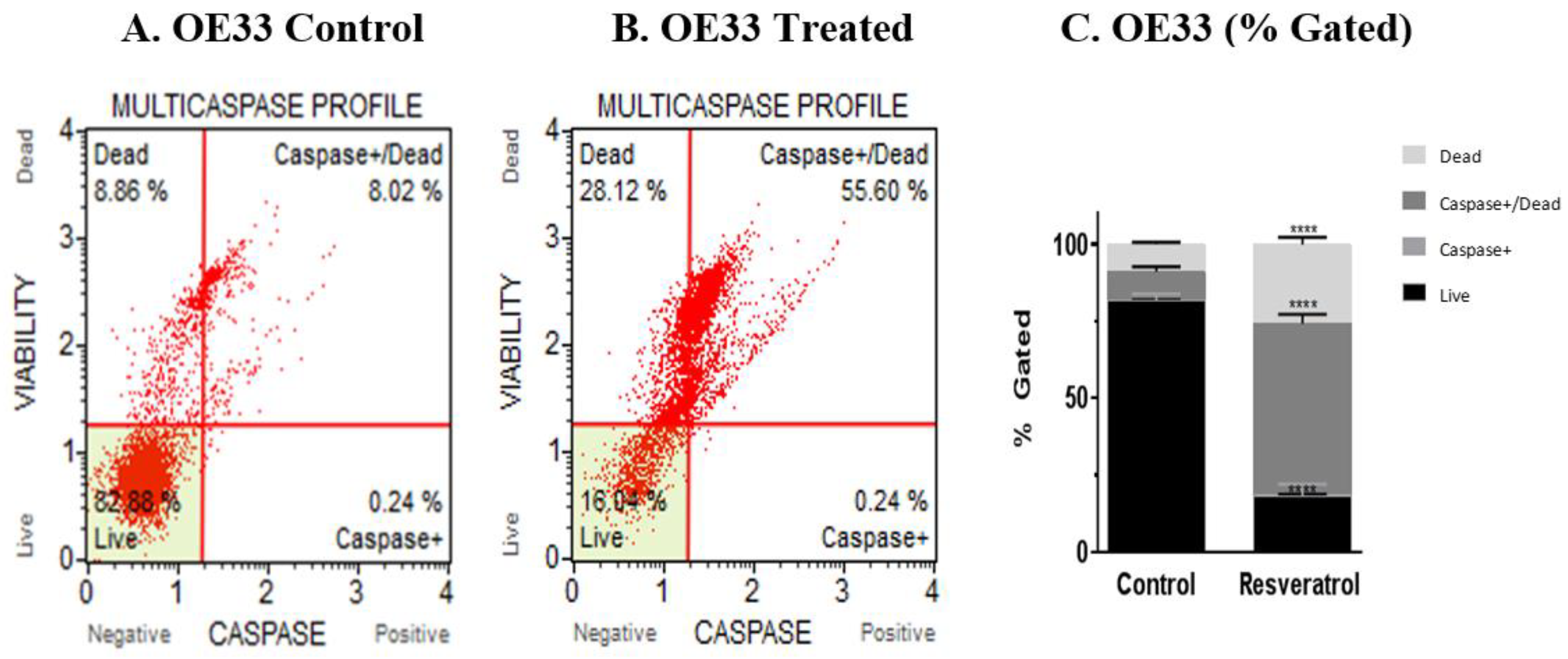
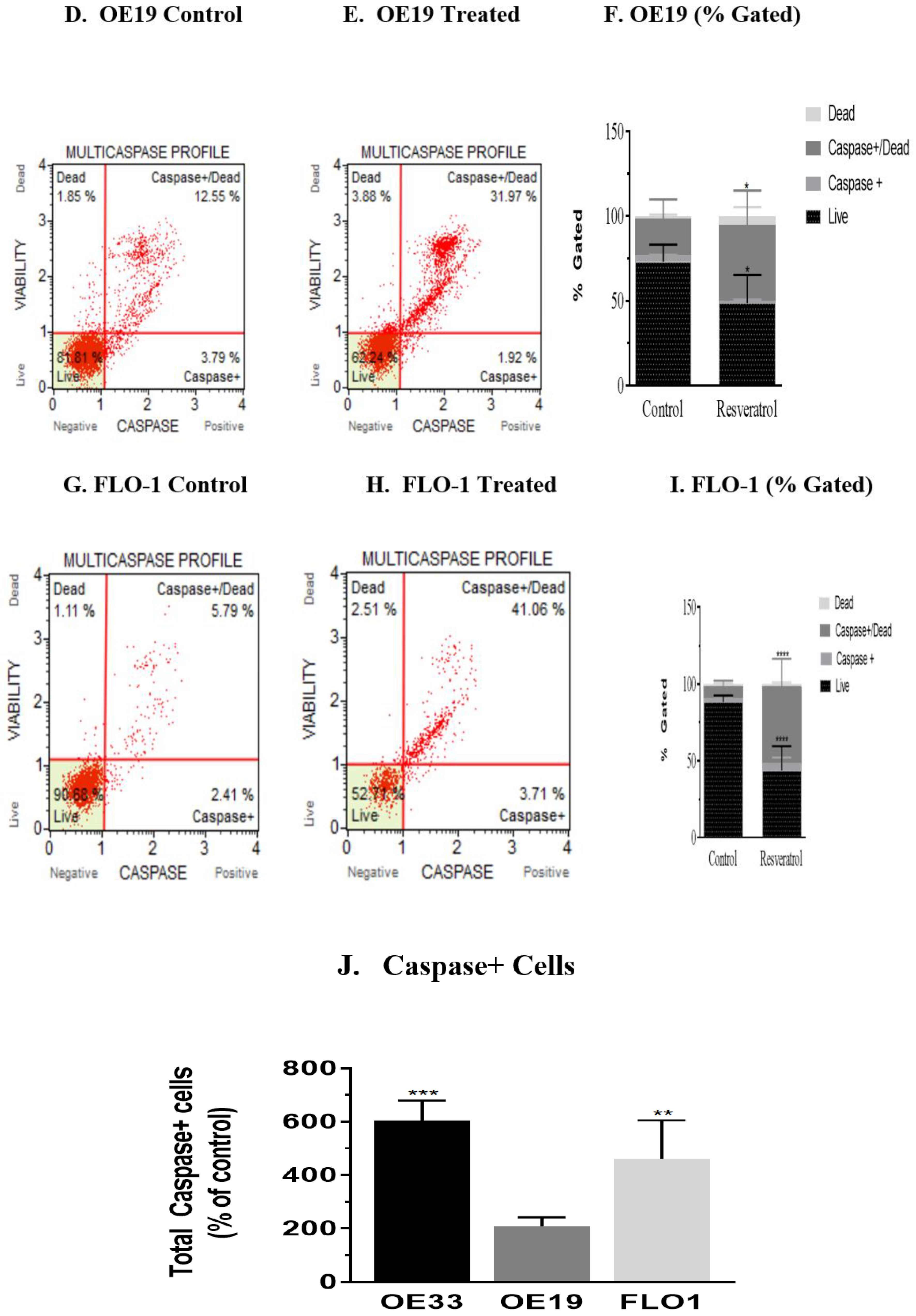
3.3. Activation of Caspases by Resveratrol in Esophageal Adenocarcinoma Cells
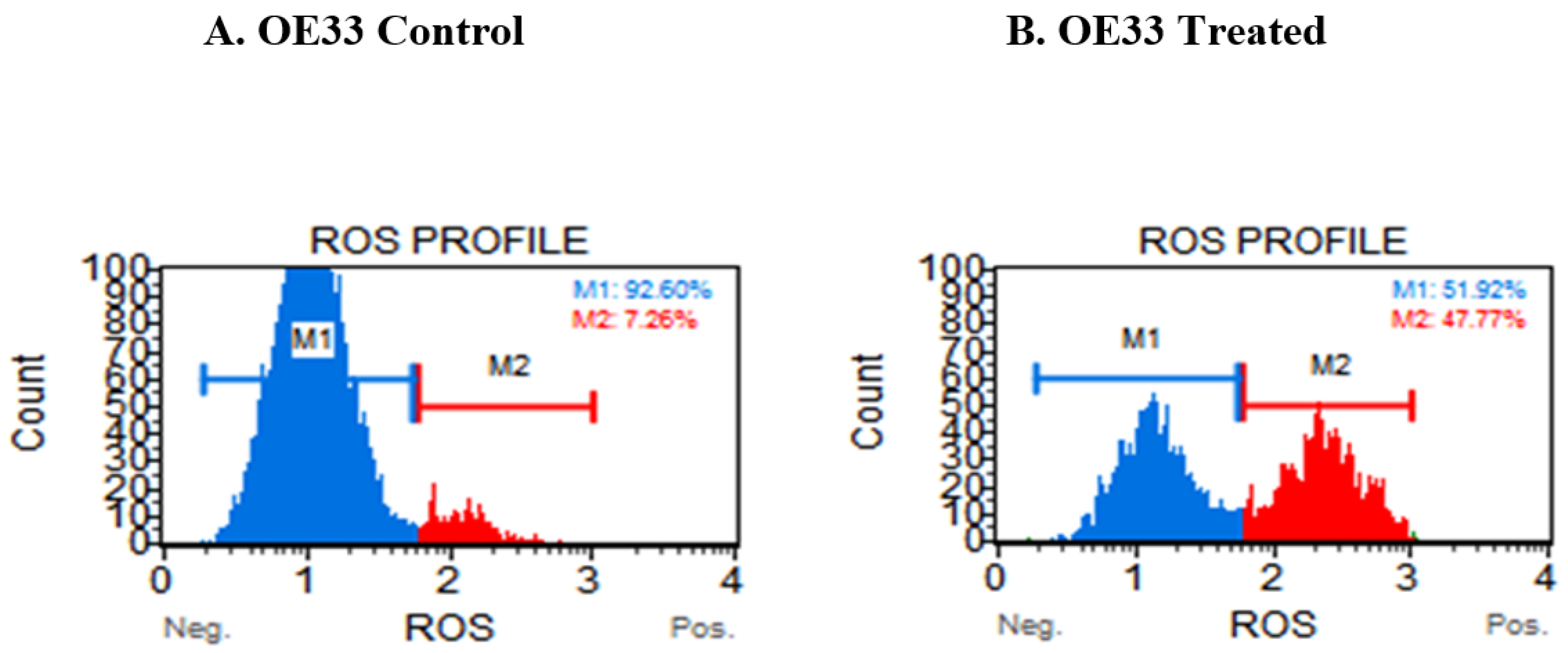
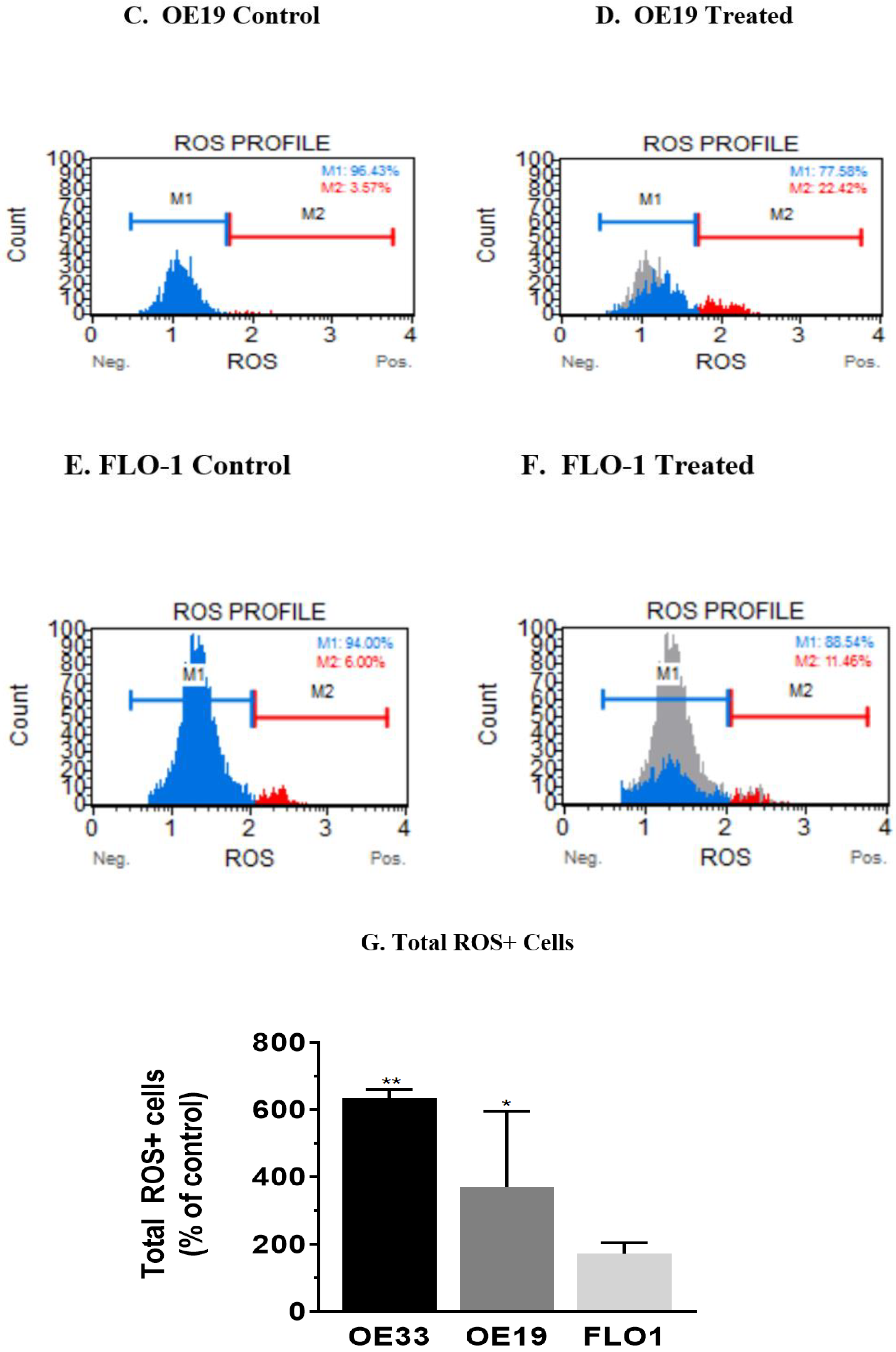
3.4. Resveratrol Impacts the Levels of Reactive Oxygen Species (ROS) in OE33, OE19 and FLO-1 Cells
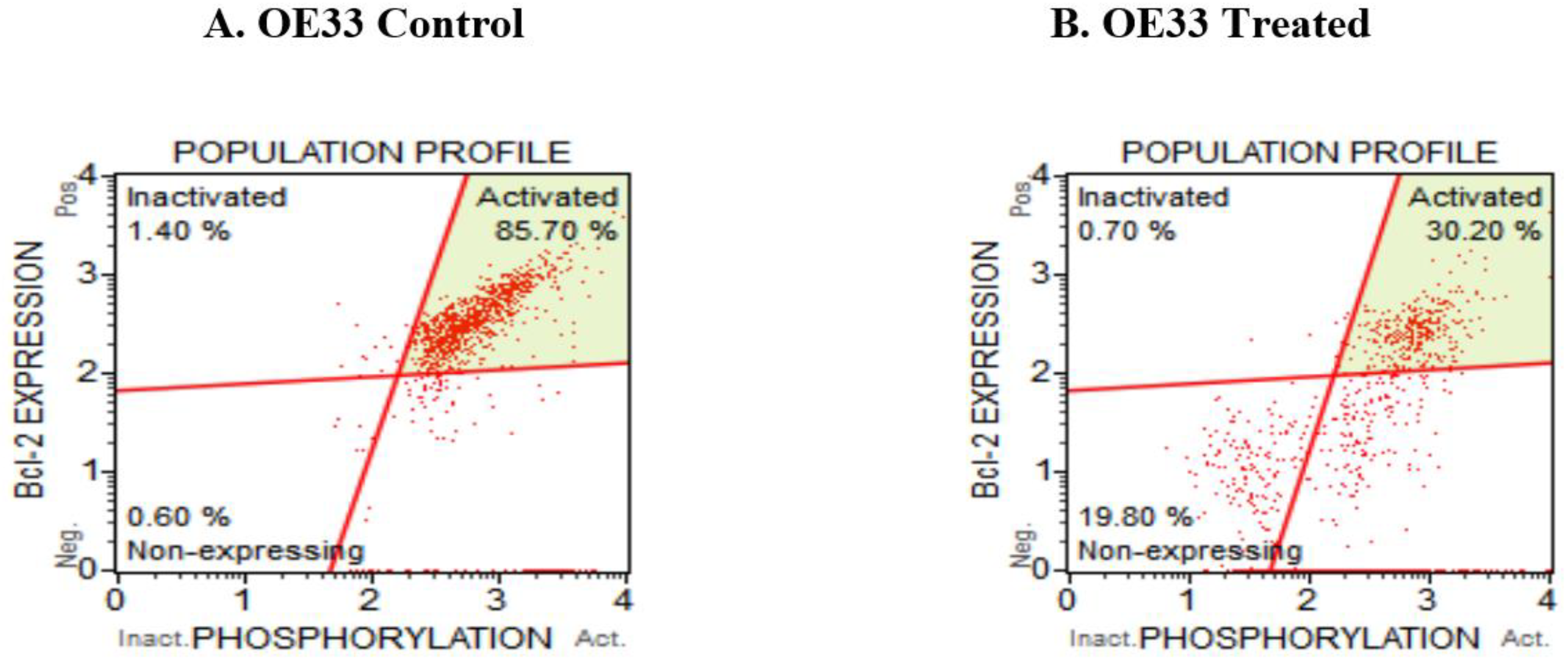
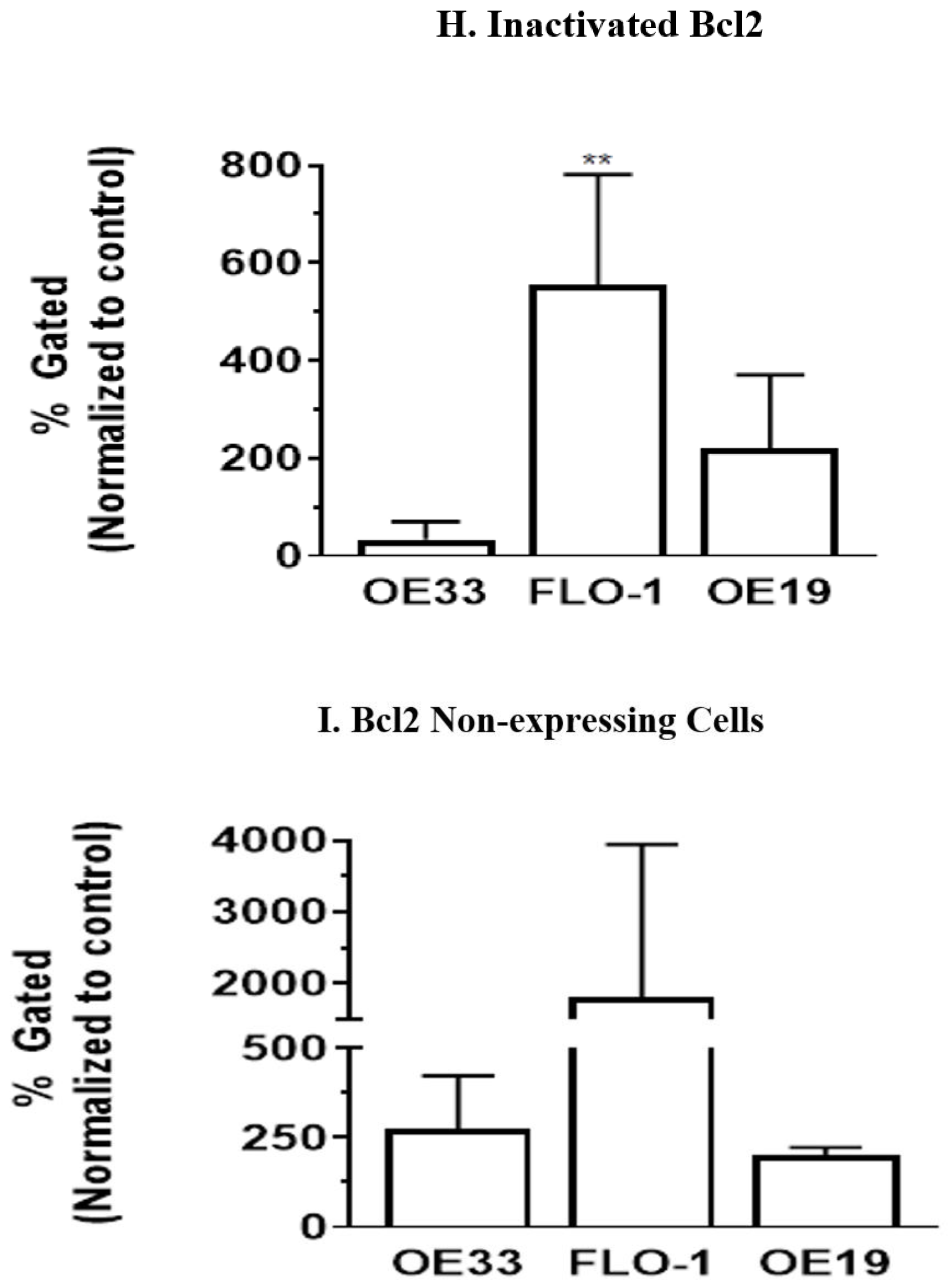
3.5. Resveratrol Affects the Bcl2 Levels in Esophageal Adenocarcinoma Cells
3.6. Proteomic Analysis Reveals Entities of Pivotal Signaling Pathways That Were Differentially Regulated on the Administration of Resveratrol in the Esophageal Adenocarcinoma Cell Lines (OE33, OE19 and FLO-1)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martincorena, I.; Fowler, J.C.; Wabik, A.; Lawson, A.R.J.; Abascal, F.; Hall, M.W.J.; Cagan, A.; Murai, K.; Mahbubani, K.; Stratton, M.R.; et al. Somatic mutant clones colonize the human esophagus with age. Science 2018, 362, 911–917. [Google Scholar] [CrossRef] [Green Version]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Estimating the world cancer burden: Globocan 2000. Int. J. Cancer 2001, 94, 153–156. [Google Scholar] [CrossRef]
- Koshy, M.; Esiashvilli, N.; Landry, J.C.; Thomas, C.R., Jr.; Matthews, R.H. Multiple management modalities in esophageal cancer: Combined modality management approaches. Oncologist 2004, 9, 147–159. [Google Scholar] [CrossRef] [Green Version]
- Kuwano, H.; Sadanaga, N.; Watanabe, M.; Yasuda, M.; Nozoe, T.; Sugimachi, K. Oesophageal cancer composed of mixed histological types. Eur. J. Surg. Oncol. 1996, 22, 225–231. [Google Scholar] [CrossRef]
- Klingelhofer, D.; Zhu, Y.; Braun, M.; Bruggmann, D.; Schoffel, N.; Groneberg, D.A. A world map of esophagus cancer research: A critical accounting. J. Transl. Med. 2019, 17, 150. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Qiu, R.L.; Lin, Y.; Cai, Y.; Bian, Y.; Fan, Y.; Gao, X.J. Resveratrol suppresses human cervical carcinoma cell proliferation and elevates apoptosis via the mitochondrial and p53 signaling pathways. Oncol. Lett. 2018, 15, 9845–9851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Ginkel, P.R.; Sareen, D.; Subramanian, L.; Walker, Q.; Darjatmoko, S.R.; Lindstrom, M.J.; Kulkarni, A.; Albert, D.M.; Polans, A.S. Resveratrol inhibits tumor growth of human neuroblastoma and mediates apoptosis by directly targeting mitochondria. Clin. Cancer Res. 2007, 13, 5162–5169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, L.; Yan, B.; Chen, K.; Jiang, Z.; Zhou, C.; Cao, J.; Qian, W.; Li, J.; Sun, L.; Ma, J.; et al. Resveratrol-Induced Downregulation of NAF-1 Enhances the Sensitivity of Pancreatic Cancer Cells to Gemcitabine via the ROS/Nrf2 Signaling Pathways. Oxid. Med. Cell. Longev. 2018, 2018, 9482018. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.G.; Hong, T.; Shimada, Y.; Komoto, I.; Kawabe, A.; Ding, Y.; Kaganoi, J.; Hashimoto, Y.; Imamura, M. Suppression of N-nitrosomethylbenzylamine (NMBA)-induced esophageal tumorigenesis in F344 rats by resveratrol. Carcinogenesis 2002, 23, 1531–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.W.; Choi, Y.J.; Jang, M.A.; Lee, Y.S.; Jun, D.Y.; Suh, S.I.; Baek, W.K.; Suh, M.H.; Jin, I.N.; Kwon, T.K. Chemopreventive agent resveratrol, a natural product derived from grapes, reversibly inhibits progression through S and G2 phases of the cell cycle in U937 cells. Cancer Lett. 2001, 163, 43–49. [Google Scholar] [CrossRef]
- Woodall, C.E.; Li, Y.; Liu, Q.H.; Wo, J.; Martin, R.C. Chemoprevention of metaplasia initiation and carcinogenic progression to esophageal adenocarcinoma by resveratrol supplementation. Anticancer Drugs 2009, 20, 437–443. [Google Scholar] [CrossRef]
- Whitlock, N.C.; Baek, S.J. The anticancer effects of resveratrol: Modulation of transcription factors. Nutr. Cancer 2012, 64, 493–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locatelli, G.A.; Savio, M.; Forti, L.; Shevelev, I.; Ramadan, K.; Stivala, L.A.; Vannini, V.; Hubscher, U.; Spadari, S.; Maga, G. Inhibition of mammalian DNA polymerases by resveratrol: Mechanism and structural determinants. Biochem. J. 2005, 389, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Park, J.H.; Woo, J.S. Resveratrol induces cell death through ROSdependent downregulation of Notch1/PTEN/Akt signaling in ovarian cancer cells. Mol. Med. Rep. 2019, 19, 3353–3360. [Google Scholar] [CrossRef]
- Bergman, M.; Levin, G.S.; Bessler, H.; Djaldetti, M.; Salman, H. Resveratrol affects the cross talk between immune and colon cancer cells. Biomed. Pharmacother. 2013, 67, 43–47. [Google Scholar] [CrossRef]
- Schwager, J.; Richard, N.; Widmer, F.; Raederstorff, D. Resveratrol distinctively modulates the inflammatory profiles of immune and endothelial cells. BMC Complementary Altern. Med. 2017, 17, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, E.; Hayden, A.; Birts, C.; Britton, E.; Cowie, A.; Pickard, K.; Mellone, M.; Choh, C.; Derouet, M.; Duriez, P.; et al. Authentication and characterisation of a new oesophageal adenocarcinoma cell line: MFD-1. Sci. Rep. 2016, 6, 32417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Patil, K.; Vashi, A.; Wang, Y.; Strickland, E.; Pai, S.B. Cellular molecular and proteomic profiling deciphers the SIRT1 controlled cell death pathways in esophageal adenocarcinoma cells. Cancer Treat. Res. Commun. 2021, 26, 100271. [Google Scholar] [CrossRef]
- Lodise, O.; Patil, K.; Karshenboym, I.; Prombo, S.; Chukwueke, C.; Pai, S.B. Inhibition of Prostate Cancer Cells by 4,5-Dicaffeoylquinic Acid through Cell Cycle Arrest. Prostate Cancer 2019, 2019, 4520645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Y.; Olsen, J.R.; Yuan, X.; Cheng, P.F.; Levesque, M.P.; Brokstad, K.A.; Hoffman, P.S.; Oyan, A.M.; Zhang, W.; Kalland, K.H.; et al. Small molecule promotes beta-catenin citrullination and inhibits Wnt signaling in cancer. Nat. Chem. Biol. 2018, 14, 94–101. [Google Scholar] [CrossRef]
- Tonge, R.; Shaw, J.; Middleton, B.; Rowlinson, R.; Rayner, S.; Young, J.; Pognan, F.; Hawkins, E.; Currie, I.; Davison, M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 2001, 1, 377–396. [Google Scholar] [CrossRef]
- Campos-Silva, C.; Suarez, H.; Jara-Acevedo, R.; Linares-Espinos, E.; Martinez-Pineiro, L.; Yanez-Mo, M.; Vales-Gomez, M. High sensitivity detection of extracellular vesicles immune-captured from urine by conventional flow cytometry. Sci. Rep. 2019, 9, 2042. [Google Scholar] [CrossRef] [Green Version]
- Jayasena, T.; Poljak, A.; Braidy, N.; Zhong, L.; Rowlands, B.; Muenchhoff, J.; Grant, R.; Smythe, G.; Teo, C.; Raftery, M.; et al. Application of Targeted Mass Spectrometry for the Quantification of Sirtuins in the Central Nervous System. Sci. Rep. 2016, 6, 35391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, N.; Chen, X.; Liu, R.; Wang, D.; Su, M.; Wang, Q.; He, A.; Gu, H. Eukaryotic elongation factors 2 promotes tumor cell proliferation and correlates with poor prognosis in ovarian cancer. Tissue Cell 2018, 53, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Song, H.; Chen, G.; Yang, X.; Liu, J.; Ge, Y.; Lu, J.; Qin, Q.; Zhang, C.; Xu, L.; et al. eEF2K promotes progression and radioresistance of esophageal squamous cell carcinoma. Radiother. Oncol. 2017, 124, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Galat, A. Peptidylproline cis-trans-isomerases: Immunophilins. Eur. J. Biochem. 1993, 216, 689–707. [Google Scholar] [CrossRef]
- Khoei, S.G.; Mohammadi, C.; Mohammadi, Y.; Sameri, S.; Najafi, R. Prognostic value of peptidyl-prolyl cis-trans isomerase 1 (PIN1) in human malignant tumors. Clin. Transl. Oncol. 2020, 22, 1067–1077. [Google Scholar] [CrossRef]
- Jin, H.; Jiang, J.; Sun, L.; Zheng, F.; Wu, C.; Peng, L.; Zhao, Y.; Wu, X. The prolyl isomerase Pin1 is overexpressed in human esophageal cancer. Oncol. Lett. 2011, 2, 1191–1196. [Google Scholar] [CrossRef] [Green Version]
- Tuteja, R.; Tuteja, N. Ku autoantigen: A multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 1–33. [Google Scholar] [CrossRef]
- Liu, Z.H.; Wang, N.; Wang, F.Q.; Dong, Q.; Ding, J. High expression of XRCC5 is associated with metastasis through Wnt signaling pathway and predicts poor prognosis in patients with hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7835–7847. [Google Scholar] [CrossRef]
- Wang, S.; Xi, J.; Lin, Z.; Hao, J.; Yao, C.; Zhan, C.; Jiang, W.; Shi, Y.; Wang, Q. Clinical values of Ku80 upregulation in superficial esophageal squamous cell carcinoma. Cancer Med. 2018, 7, 1006–1018. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J.; Ciccone, S.L.; Freie, B.; Kurimasa, A.; Chen, D.J.; Li, G.C.; Clapp, D.W.; Lee, S.H. A positive role for the Ku complex in DNA replication following strand break damage in mammals. J. Biol. Chem. 2004, 279, 6046–6055. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.W.; Kim, S.Y.; Yi, S.L.; Son, S.H.; Song, D.Y.; Moon, S.Y.; Kim, J.H.; Choi, E.K.; Ahn, S.D.; Shin, S.S.; et al. Expression of Ku80 correlates with sensitivities to radiation in cancer cell lines of the head and neck. Oral Oncol. 2006, 42, 979–986. [Google Scholar] [CrossRef]
- Zahid, S.; Seif El Dahan, M.; Iehl, F.; Fernandez-Varela, P.; Le Du, M.H.; Ropars, V.; Charbonnier, J.B. The Multifaceted Roles of Ku70/80. Int. J. Mol. Sci. 2021, 22, 4134. [Google Scholar] [CrossRef]
- Karantza, V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene 2011, 30, 127–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Kapur, S.; Chattopadhyay, I.; Purkayastha, J.; Sharma, J.; Mishra, A.; Hewitt, S.M.; Saxena, S. Cytokeratin immunoexpression in esophageal squamous cell carcinoma of high-risk population in Northeast India. Appl. Immunohistochem. Mol. Morphol. 2009, 17, 419–424. [Google Scholar] [CrossRef] [Green Version]
- Boukakis, G.; Patrinou-Georgoula, M.; Lekarakou, M.; Valavanis, C.; Guialis, A. Deregulated expression of hnRNP A/B proteins in human non-small cell lung cancer: Parallel assessment of protein and mRNA levels in paired tumour/non-tumour tissues. BMC Cancer 2010, 10, 434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuyama, S.; Goto, Y.; Sueoka, N.; Ohkura, Y.; Tanaka, Y.; Nakachi, K.; Sueoka, E. Heterogeneous nuclear ribonucleoprotein B1 expressed in esophageal squamous cell carcinomas as a new biomarker for diagnosis. Jpn. J. Cancer Res. 2000, 91, 658–663. [Google Scholar] [CrossRef]
- Deng, J.; Chen, S.; Wang, F.; Zhao, H.; Xie, Z.; Xu, Z.; Zhang, Q.; Liang, P.; Zhai, X.; Cheng, Y. Effects of hnRNP A2/B1 Knockdown on Inhibition of Glioblastoma Cell Invasion, Growth and Survival. Mol. Neurobiol. 2016, 53, 1132–1144. [Google Scholar] [CrossRef]
- Krajewska, M.; Zapata, J.M.; Meinhold-Heerlein, I.; Hedayat, H.; Monks, A.; Bettendorf, H.; Shabaik, A.; Bubendorf, L.; Kallioniemi, O.P.; Kim, H.; et al. Expression of Bcl-2 family member Bid in normal and malignant tissues. Neoplasia 2002, 4, 129–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusumawidjaja, G.; Kayed, H.; Giese, N.; Bauer, A.; Erkan, M.; Giese, T.; Hoheise, J.D.; Friess, H.; Kleeff, J. Basic transcription factor 3 (BTF3) regulates transcription of tumor-associated genes in pancreatic cancer cells. Cancer Biol. Ther. 2007, 6, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Sui, J.; Xing, J.; Cao, F.; Wang, H.; Fu, C.; Wang, H. Basic transcription factor 3 expression silencing attenuates colon cancer cell proliferation and migration in vitro. Oncol. Lett. 2019, 17, 113–118. [Google Scholar] [CrossRef] [Green Version]
- Nieto-Miguel, T.; Gajate, C.; Gonzalez-Camacho, F.; Mollinedo, F. Proapoptotic role of Hsp90 by its interaction with c-Jun N-terminal kinase in lipid rafts in edelfosine-mediated antileukemic therapy. Oncogene 2008, 27, 1779–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delmas, D.; Rebe, C.; Lacour, S.; Filomenko, R.; Athias, A.; Gambert, P.; Cherkaoui-Malki, M.; Jannin, B.; Dubrez-Daloz, L.; Latruffe, N.; et al. Resveratrol-induced apoptosis is associated with Fas redistribution in the rafts and the formation of a death-inducing signaling complex in colon cancer cells. J. Biol. Chem. 2003, 278, 41482–41490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajate, C.; Mollinedo, F. Lipid raft-mediated Fas/CD95 apoptotic signaling in leukemic cells and normal leukocytes and therapeutic implications. J. Leukoc. Biol. 2015, 98, 739–759. [Google Scholar] [CrossRef]
- Medema, J.P.; Schuurhuis, D.H.; Rea, D.; van Tongeren, J.; de Jong, J.; Bres, S.A.; Laban, S.; Toes, R.E.; Toebes, M.; Schumacher, T.N.; et al. Expression of the serpin serine protease inhibitor 6 protects dendritic cells from cytotoxic T lymphocyte-induced apoptosis: Differential modulation by T helper type 1 and type 2 cells. J. Exp. Med. 2001, 194, 657–667. [Google Scholar] [CrossRef]
- Kim, T.W.; Hung, C.F.; Boyd, D.A.; He, L.; Lin, C.T.; Kaiserman, D.; Bird, P.I.; Wu, T.C. Enhancement of DNA vaccine potency by coadministration of a tumor antigen gene and DNA encoding serine protease inhibitor-6. Cancer Res. 2004, 64, 400–405. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, N.; Kinoshita, T.; Nohata, N.; Yoshino, H.; Itesako, T.; Fujimura, L.; Mitsuhashi, A.; Usui, H.; Enokida, H.; Nakagawa, M.; et al. Tumor-suppressive microRNA-29a inhibits cancer cell migration and invasion via targeting HSP47 in cervical squamous cell carcinoma. Int. J. Oncol. 2013, 43, 1855–1863. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhou, Y.; Lou, Y.; Zhong, H. Knockdown of HNRNPA1 inhibits lung adenocarcinoma cell proliferation through cell cycle arrest at G0/G1 phase. Gene 2016, 576, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Ran, L.; Liu, Y.; Zhong, S.H.; Zhou, P.P.; Liao, M.X.; Fang, W. Knockdown of hnRNP A2/B1 inhibits cell proliferation, invasion and cell cycle triggering apoptosis in cervical cancer via PI3K/AKT signaling pathway. Oncol. Rep. 2018, 39, 939–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevet, E.; Fessart, D.; Delom, F.; Mulot, A.; Vojtesek, B.; Hrstka, R.; Murray, E.; Gray, T.; Hupp, T. Emerging roles for the pro-oncogenic anterior gradient-2 in cancer development. Oncogene 2013, 32, 2499–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, F.M.; He, J.Z.; Wang, S.H.; Liu, D.K.; Xu, X.E.; Wu, J.Y.; Li, E.M.; Xu, L.Y. A Novel Three-Gene Model Predicts Prognosis and Therapeutic Sensitivity in Esophageal Squamous Cell Carcinoma. Biomed. Res. Int. 2019, 2019, 9828637. [Google Scholar] [CrossRef] [Green Version]
- Shimoda, T.; Wada, R.; Kure, S.; Ishino, K.; Kudo, M.; Ohashi, R.; Fujita, I.; Uchida, E.; Yoshida, H.; Naito, Z. Expression of protein disulfide isomerase A3 and its clinicopathological association in gastric cancer. Oncol. Rep. 2019, 41, 2265–2272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, G.; Lu, H.; Li, C. Proapoptotic activities of protein disulfide isomerase (PDI) and PDIA3 protein, a role of the Bcl-2 protein Bak. J. Biol. Chem. 2015, 290, 8949–8963. [Google Scholar] [CrossRef] [Green Version]
- Caorsi, C.; Niccolai, E.; Capello, M.; Vallone, R.; Chattaragada, M.S.; Alushi, B.; Castiglione, A.; Ciccone, G.; Mautino, A.; Cassoni, P.; et al. Protein disulfide isomerase A3-specific Th1 effector cells infiltrate colon cancer tissue of patients with circulating anti-protein disulfide isomerase A3 autoantibodies. Transl. Res. 2016, 171, 17–28 e2. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, H.; Chen, R.; Zhang, Y.; Sun, X.X.; Huang, W.; Bian, H.; Chen, Z.N. Combination of CALR and PDIA3 is a potential prognostic biomarker for non-small cell lung cancer. Oncotarget 2017, 8, 96945–96957. [Google Scholar] [CrossRef] [Green Version]
- Gunnersen, J.M.; Spirkoska, V.; Smith, P.E.; Danks, R.A.; Tan, S.S. Growth and migration markers of rat C6 glioma cells identified by serial analysis of gene expression. Glia 2000, 32, 146–154. [Google Scholar] [CrossRef]
- Smith-Beckerman, D.M.; Fung, K.W.; Williams, K.E.; Auersperg, N.; Godwin, A.K.; Burlingame, A.L. Proteome changes in ovarian epithelial cells derived from women with BRCA1 mutations and family histories of cancer. Mol. Cell. Proteom. 2005, 4, 156–168. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wyckoff, J.B.; Goswami, S.; Wang, Y.; Sidani, M.; Segall, J.E.; Condeelis, J.S. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007, 67, 3505–3511. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Huang, Y.; Zhao, J.; Wu, L.; Qi, Q.; Liu, Y.; Li, G.; Li, J.; Liu, H.; Wu, H. Cofilin: A Promising Protein Implicated in Cancer Metastasis and Apoptosis. Front. Cell Dev. Biol. 2021, 9, 599065. [Google Scholar] [CrossRef]
- Mourikis, T.P.; Benedetti, L.; Foxall, E.; Temelkovski, D.; Nulsen, J.; Perner, J.; Cereda, M.; Lagergren, J.; Howell, M.; Yau, C.; et al. Patient-specific cancer genes contribute to recurrently perturbed pathways and establish therapeutic vulnerabilities in esophageal adenocarcinoma. Nat. Commun. 2019, 10, 3101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dar, A.A.; Goff, L.W.; Majid, S.; Berlin, J.; El-Rifai, W. Aurora kinase inhibitors--rising stars in cancer therapeutics? Mol. Cancer Ther. 2010, 9, 268–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sehdev, V.; Peng, D.; Soutto, M.; Washington, M.K.; Revetta, F.; Ecsedy, J.; Zaika, A.; Rau, T.T.; Schneider-Stock, R.; Belkhiri, A.; et al. The aurora kinase A inhibitor MLN8237 enhances cisplatin-induced cell death in esophageal adenocarcinoma cells. Mol. Cancer Ther. 2012, 11, 763–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]

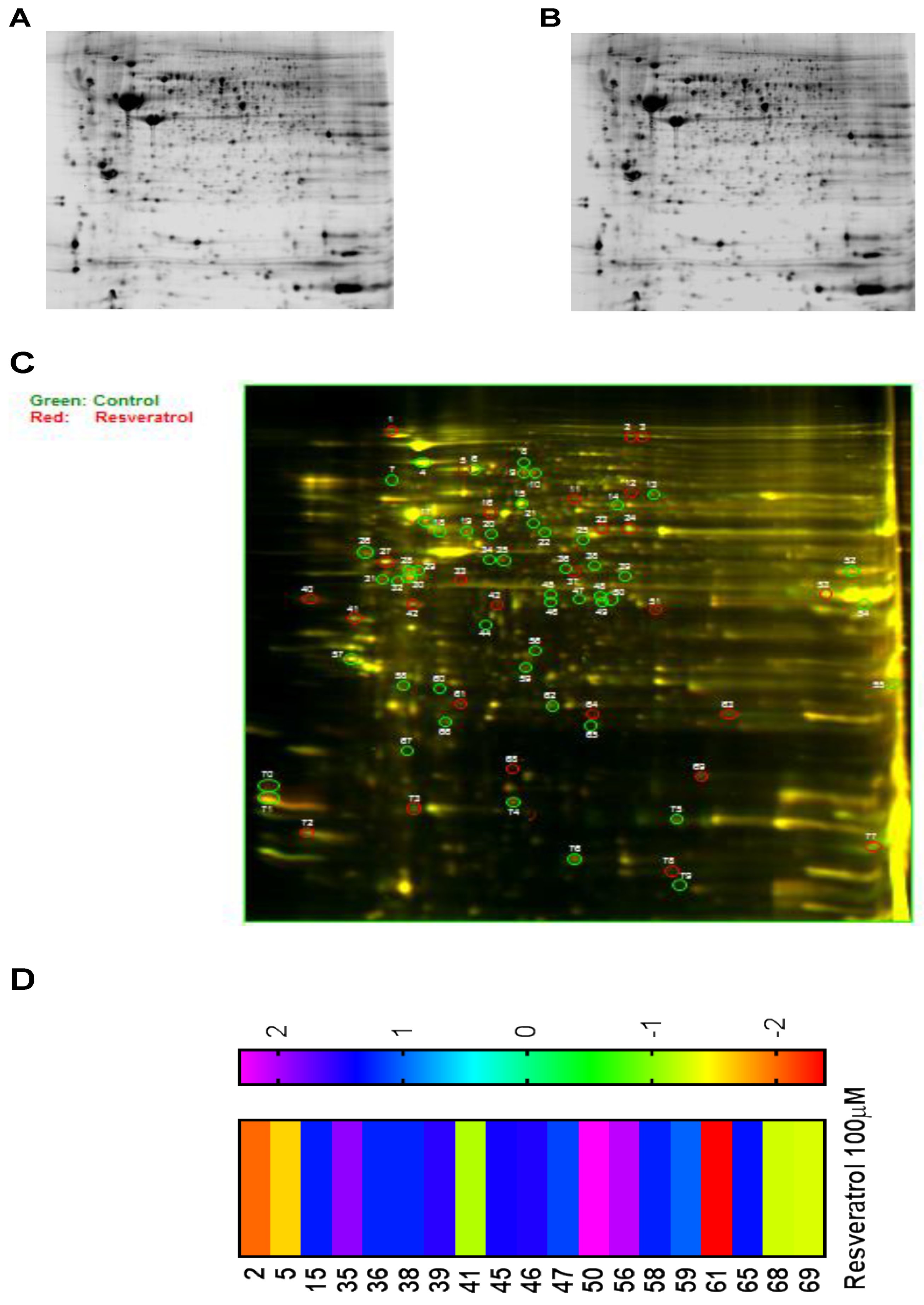
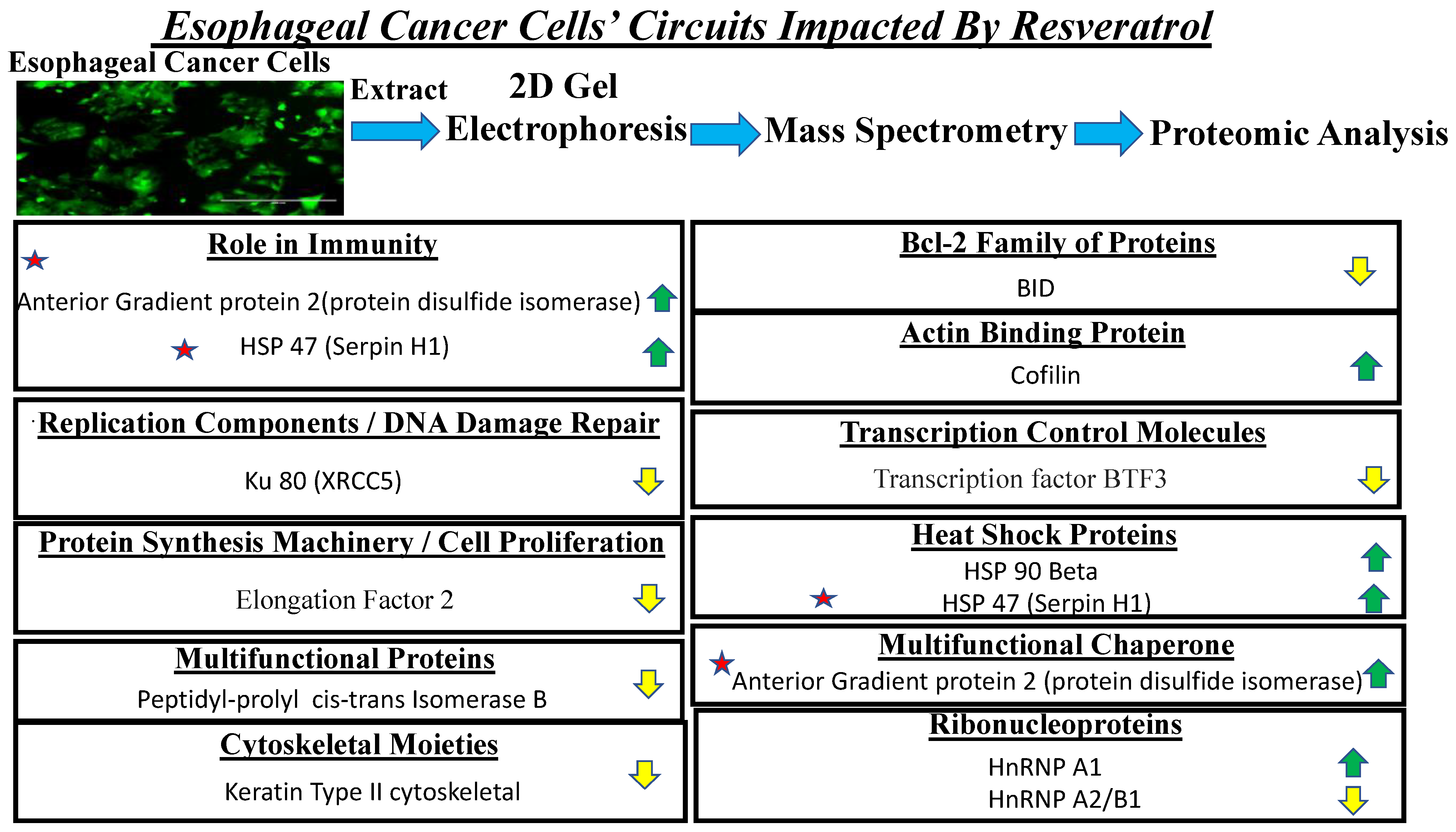
| SI# | Protein | Fold Change in OE33 | Fold Change in OE19 | Fold Change in FLO-1 |
|---|---|---|---|---|
| 1 | Elongation Factor 2 | −1.83 | −1.97 | −1.94 |
| 2 | X-ray repair cross-complementing protein 5 | −2.05 | −2.25 | −1.76 |
| 3 | Keratin type II cytoskeletal 8 | −6.86 | −7.48 | −2.22 |
| 4 | Heterogenous nuclear ribonucleoproteins A2/B1 | −1.9 | −3.8 | −1.3 |
| 5 | BH3-interacting domain death agonist | −1.66 | −2.03 | −1.6 |
| 6 | Transcription factor BTF3 | −2.31 | −4.82 | −1.29 |
| 7 | Peptidyl prolyl cis-trans isomerase B | −1.55 | −1.92 | −5.27 |
| 8 | Heat shock protein HSP 90-beta | 2.00 | 1.74 | 1.98 |
| 9 | Serpin H1 | 3.06 | 1.35 | 1.75 |
| 10 | Heterogenous nuclear ribonucleoprotein A1 | 3.25 | 2.21 | 2.51 |
| 11 | Cofilin-1 | 2.13 | 3.25 | 2.33 |
| 12 | Anterior gradient protein 2 homolog | 2.18 | 2.97 | 2.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhir, H.; Choudhury, M.; Patil, K.; Cheung, C.; Bodlak, A.; Pardo, D.; Adams, A.; Travaglino, S.; Rojas, J.A.; Pai, S.B. Interception of Signaling Circuits of Esophageal Adenocarcinoma Cells by Resveratrol Reveals Molecular and Immunomodulatory Signatures. Cancers 2021, 13, 5811. https://doi.org/10.3390/cancers13225811
Dhir H, Choudhury M, Patil K, Cheung C, Bodlak A, Pardo D, Adams A, Travaglino S, Rojas JA, Pai SB. Interception of Signaling Circuits of Esophageal Adenocarcinoma Cells by Resveratrol Reveals Molecular and Immunomodulatory Signatures. Cancers. 2021; 13(22):5811. https://doi.org/10.3390/cancers13225811
Chicago/Turabian StyleDhir, Hardika, Monica Choudhury, Ketki Patil, Candice Cheung, Adriana Bodlak, Danny Pardo, Asana Adams, Stefano Travaglino, Jose Araque Rojas, and S. Balakrishna Pai. 2021. "Interception of Signaling Circuits of Esophageal Adenocarcinoma Cells by Resveratrol Reveals Molecular and Immunomodulatory Signatures" Cancers 13, no. 22: 5811. https://doi.org/10.3390/cancers13225811
APA StyleDhir, H., Choudhury, M., Patil, K., Cheung, C., Bodlak, A., Pardo, D., Adams, A., Travaglino, S., Rojas, J. A., & Pai, S. B. (2021). Interception of Signaling Circuits of Esophageal Adenocarcinoma Cells by Resveratrol Reveals Molecular and Immunomodulatory Signatures. Cancers, 13(22), 5811. https://doi.org/10.3390/cancers13225811






