Simple Summary
Rectal cancer is the 8th most common cancer globally. Most patients with locally advanced rectal cancer receive neoadjuvant therapy based on 5-fluorouracil and radiotherapy showing variable responses. About 70–90% of patients present partial response, while 20% show treatment resistance. Repositioning drugs approved by regulatory agencies or drugs currently in clinical trials is a strategy to accelerate the development of drug-based cancer therapies. We compared rectal cancer gene expression signatures with reverse drug-induced gene-expression profiles of cancer cell lines to identify potential drugs for repositioning. Our analyses revealed that approved topoisomerase II inhibitors are candidate drugs for rectal cancer treatment. We also verified TOP2A copy number gains and increased expression in rectal tumors. These TOP2A alterations were independent predictive markers of topoisomerase inhibitor efficacy in colorectal cancer cells that closely represent rectal cancer signatures. Topoisomerase inhibitors are potentially helpful to treat rectal cancer patients with TOP2A imbalances.
Abstract
Rectal cancer is a common disease with high mortality rates and limited therapeutic options. Here we combined the gene expression signatures of rectal cancer patients with the reverse drug-induced gene-expression profiles to identify drug repositioning candidates for cancer therapy. Among the predicted repurposable drugs, topoisomerase II inhibitors (doxorubicin, teniposide, idarubicin, mitoxantrone, and epirubicin) presented a high potential to reverse rectal cancer gene expression signatures. We showed that these drugs effectively reduced the growth of colorectal cancer cell lines closely representing rectal cancer signatures. We also found a clear correlation between topoisomerase 2A (TOP2A) gene copy number or expression levels with the sensitivity to topoisomerase II inhibitors. Furthermore, CRISPR-Cas9 and shRNA screenings confirmed that loss-of-function of the TOP2A has the highest efficacy in reducing cellular proliferation. Finally, we observed significant TOP2A copy number gains and increased expression in independent cohorts of rectal cancer patients. These findings can be translated into clinical practice to evaluate TOP2A status for targeted and personalized therapies based on topoisomerase II inhibitors in rectal cancer patients.
1. Introduction
Cancer is one of the leading causes of death worldwide [1]. Unfortunately, the discovery and translation of new drugs for cancer treatment is costly [2], requires years of research and development [3,4], and only a few drugs entering clinical trials are eventually approved for clinical use [5,6]. Drug repositioning has provided emerging opportunities for cancer therapeutic discoveries [7,8]. The definition of drug repositioning (also called drug repositioning, reprofiling, or re-tasking) has been explored since the publication of Ashburn and Thor in 2004 [9]. According to these authors, drug repositioning is the process of identifying and developing new uses for approved drugs or those under investigation outside the original indication [9]. The major advantages of repurposing old drugs are the use of de-risked compounds, with potentially shorter development time and reduced costs [8]. Therefore, repositioning drugs approved by regulatory agencies or currently in clinical trials are valuable alternatives for finding new cancer therapies that can be rapidly translated to the clinic [10]. This approach can benefit cancer patients by increasing the number of new therapeutic indications to non-oncology drugs or improving the effectiveness of personalized and targeted therapies [11].
Rectal cancer (ReCa) is the 8th most common cancer globally, with more than 300,000 deaths annually [1], and its incidence has risen exponentially in individuals aged between 18 and 50 years [12,13,14]. Most patients with locally advanced ReCa receive a complex multimodal therapy based on 5-fluorouracil and radiotherapy before surgery [15,16,17]. This treatment strategy was implemented in the mid-1990s [18] and has yielded an effective reduction in recurrence rates and improved survival [15]. Despite these improved outcomes, patients still suffer from high morbidity of the surgery [19,20], diverse side effects, and poor functional results of the treatment [21,22]. A complete pathological response is observed in only ~15% of patients after neoadjuvant chemoradiotherapy [23] a positive prognostic marker [24]. In the postoperative setting, medical treatment aims to decrease the risk of relapse and death [16,17]. However, only a minor fraction of the patients will benefit [25], culminating in the metastatic spread of ReCa, which is the leading cause of death [26]. More effective systemic treatments may increase the resectability of metastases and increase overall survival time. Thus, it is critical to identify new molecular markers to select patients for therapy and additional therapeutic strategies to treat ReCa.
Global gene-expression profiles are valuable resources for discovering potential therapeutic drug targets in human cancers [27,28]. Cancer gene-expression profiles are accumulating and available for exploration and reuse [29] through databases such as the Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA) [30], and the Cancer Cell Line Encyclopedia (CCLE) [31,32]. The integration of these cancer profiles with publicly available open data containing the effect of drugs on gene expression [28,33,34] or growth-inhibitory activity in human cancer cell lines [27] provides a valuable framework to computationally identify novel therapeutic candidates [33,35]. Different studies have combined drug-induced gene expression profiles with a cancer-specific expression profile to select compounds with high potency to reverse the expression of cancer-associated genes [36,37,38,39,40,41,42]. These drug-disease relationships found among anticorrelated expression profiles can benefit from using robust cancer gene expression signatures, which can be obtained by systematically analyzing cancer transcriptome datasets from several sources [43]. A data-mining of tumor gene-expression datasets allows the comparison of similar studies for consistency and identification of gene expression patterns [44], leading to biological discoveries and more accurate predictions of drug candidates for cancer treatment.
In this study, we first followed well-established criteria to select and reanalyze publicly available gene expression datasets of ReCa. Then, six signatures were obtained and used in a drug-repositioning bioinformatics approach to identify novel US Food and Drug Administration (FDA)–approved or investigational candidate drugs to treat ReCa. Using this strategy, we found that topoisomerase and cyclin-dependent kinase (CDK) inhibitors potentially reverse ReCa signatures and inhibit the growth of colorectal cancer (CRC) cell lines closely representing ReCa signatures. Thus, we demonstrate the reversal of ReCa gene expression as a strategy to rapidly identify drugs that are FDA-approved or currently in clinical trials to treat the disease.
2. Methods
2.1. Acquisition and Processing of Publicly Available ReCa Transcriptomic Datasets
We manually curated the GEO repository (https://www.ncbi.nlm.nih.gov/geo/) on 7 March 2021 to select the ReCa transcriptome datasets. The search strategy used the Medical Subject Headings (MeSH; https://www.ncbi.nlm.nih.gov/mesh/) (accessed on 2 March 2021) unique ID D012004 for Rectal Neoplasms (tumor or cancer of the rectum) from PubMed (Tables S1 and S2). We applied the following filters to select the datasets: Homo sapiens, gene expression platforms (Affymetrix, Agilent, and Illumina), with at least 30 rectal tissue samples (excluding formalin-fixed, paraffin-embedded tissues) from pre-treatment patients, with the surrounding normal tissues, studies indexed in PubMed and published in peer-reviewed journals from 2010 to 2021, and with standardized and annotated metadata. We also compared TCGA (https://www.cancer.gov/tcga) (accessed on 20 March 2021) RNA sequencing data from primary rectal adenocarcinoma (READ) [45] with RNA-Seq data from normal samples available in The Genotype-Tissue Expression (GTEx) program [46].
2.2. Generation of ReCa Signatures
We performed differential gene expression analyses between tumor and normal tissues for each cDNA microarray dataset using the interactive web tool GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) (accessed on 10 March 2021), which is based on the limma package [47]. Next, we removed probes matching the same gene symbol by selecting the one with the lowest p-value. For the TCGA RNA-Seq dataset, the differential expression analysis between TCGA tumor and GTEx normal samples was performed using EdgeR [48] in the workspace Open Cancer TherApeutic Discovery (OCTAD) [49]. For this analysis, OCTAD uses a deep learning approach to select the top 50 highly correlated normal samples based on their gene expression profiles [50]. The final gene expression signature for each dataset was determined by applying the statistical cutoffs of Fold-change >1 and Adjusted p-value < 0.001.
2.3. Gene Set Enrichment Analysis
The final gene expression signature for each selected dataset and the list of differentially expressed genes (DEG) shared among all ReCa signatures were inputted into the EnrichR tool [51,52,53] for a comprehensive gene set enrichment analysis. Gene Ontology (Biological Process and Molecular Function), Kyoto Encyclopedia of Genes and Genomes (KEGG), NCI-Nature 2016 Pathway Interaction Database, ARCHS4 (Kinases Co-expression), and DisGeNET EnrichR libraries were used to identify genes compatible with drug targets potentially applicable to ReCa. The top five gene set enrichment terms for each library were included in consensus lists when satisfying the criteria of overrepresentation (Adjusted p-value < 0.05) in at least one signature. The enrichment results were represented by the p-value (Fisher’s exact test) and Z score (correction to the test) in a combined score computed by EnrichR [51,52]. To further explore commonly-enriched terms and their relationships, we used Metascape (https://metascape.org/) (accessed on 9 April 2021) [54] to perform a comparative analysis of all ReCa signatures. The enriched terms were represented by clusters with a similarity metric >0.3 determined by Kappa scores [55] and were displayed as nodes connected by edges in a network of overlapping enriched terms. Metascape selects the most statistically significant term within a cluster to represent the cluster. We also used Metascape [54] to identify protein-protein interaction (PPI) networks that were consistently altered in all ReCa signatures. Metascape uses PPI networks containing 3–500 proteins to detect densely connected network components using the Molecular Complex Detection (MCODE) algorithm [56].
2.4. Screening Drugs Targeting ReCa Using Gene Expression Signatures
The gene expression signature for each dataset was virtually screened for therapeutic targets using the OCTAD tool [49]. This platform matches cancer-specific expression signatures to compound-induced gene expression profiles (66,612 drug-induced gene expression profiles derived from 71 cell lines and 12,442 drugs) of the Library of Integrated Network-based Cellular Signatures (LINCS; L1000 dataset) [28,33,34]. The analysis is based on prioritizing small molecules with high potency to reverse the gene expression signature of the disease. OCTAD employs a summarization method to reduce bias and calculates the summarized Reversal Gene Expression Score (sRGES) [49]. The sRGES represents the reversal potency of a drug to a specific cancer signature. We selected the top 100 drug candidates based on the sRGES scores (lowest values < −0.25 in more than two drug-induced gene expression profiles) for the LINCS compounds (drugs in clinical trials or FDA-approved). These lists were compared with the results from the gene set enrichment analysis to generate a final list of drugs for further evaluation.
2.5. Genetic Dependencies of Target Drugs in CRC Cell Lines
To identify target genes for anticancer drugs and to discover the mechanistic basis of essentiality in specific cell types, we assessed the web tool shinyDepMap (https://labsyspharm.shinyapps.io/depmap) (accessed on 17 May 2021) [57]. This tool combines CRISPR and shRNA data available at the Cancer Dependency Map (DepMap; dataset version 19Q3) [58] to define, for each drug target gene, the growth reduction caused by knockdown/knockout (efficacy) and the selectivity of this effect across 423 cancer cell lines.
To systematically identify cancer vulnerabilities of selected drug targets, we further analyzed in vitro genetic dependencies in 53 CRC cell lines using DepMap CRISPR/Cas9 knockout screenings (DepMap 21Q2 Public + Score dataset) [59,60,61].
2.6. Drug Sensitivity in CRC Cell Lines
We also queried DepMap [58] to compare drug sensitivity between cancer cell lines to the selected drugs. We collected sensitivity data (PRISM Repurposing Primary and Secondary Screenings—19Q4) [27] for selected drugs showing high potency to reverse the ReCa-specific expression signatures in 35 CRC cell lines. PRISM is a 2-stage screening strategy that first screened 4518 drugs at a single dose (2.5 μM, in triplicate), and then 1448 drugs screened positive were re-screened in an 8-point dose-response (from 10 μM to 600 pM, in triplicate). Our drug sensitivity analysis based on these data prioritized drugs that effectively inhibited the growth of CRC cell lines with similar gene expression signatures to ReCa.
To measure this transcriptional similarity of the ReCa signatures and cancer cell lines to model anticancer drug sensitivity, we first used EnrichR to generate a consensus list based on the CCLE [31,32]. This CCLE consensus list includes the top 20 gene set enrichment terms (cell lines) from each signature that fulfilled the criteria of overrepresentation (Adjusted p-value < 0.05). We also compared the consensus list with tumor-cell line similarities estimated by Celligner (https://depmap.org/portal/celligner) (accessed on 6 May 2021) [62] and OCTAD [49]. Finally, the cancer cell lines were further classified into consensus molecular subtypes, according to Yu et al. 2019 [63].
2.7. Identification of Predictive Markers of Drug-Based Neoadjuvant Chemotherapy Efficacy
Copy number [log2 (relative to ploidy +1)] and RNA-Seq expression (TPM+1) data of CRC cell lines from the CCLE [31,32] were extracted from the DepMap [58] data portal (Copy Number 21Q2 Public and Expression 21Q2 Public datasets). Next, we calculated Pearson correlation coefficients for TOP2A and TOP2B copy number and gene expression with drug sensitivity data. This analysis was performed with topoisomerase inhibitors (doxorubicin, daunorubicin, idarubicin, epirubicin, teniposide, and mitoxantrone) in eight CRC cell lines, using the Correlation Matrix—online software: analysis and visualization (Statistical tools for high-throughput data analysis—STHDA; http://www.sthda.com/english/) (accessed on 19 May 2021).
Copy number alterations involving TOP2A were evaluated in 32 cases of locally advanced ReCa, as previously described [64]. Briefly, genomic profiling of the fresh-frozen pre-treatment biopsies was performed using the CytoScan HD array platform and analyzed with the Chromosome Analysis Suite (ChAS v.3.1, Affymetrix, Santa Clara, CA, USA). We considered significant alterations containing at least 25 probes altered for losses, 50 for gains, and a minimum of 5 Mb for cnLOH (copy-neutral loss of heterozygosity).
We analyzed the frequency of gene copy number and gene expression alterations of TOP2A in colorectal adenocarcinoma (TCGA, Firehose Legacy) patients using the cBioPortal (https://www.cbioportal.org/) (accessed on 20 May 2021) [65,66] database. Putative copy-number calls were determined using GISTIC 2.0 considered as −2: homozygous deletion, −1: hemizygous deletion, 0: neutral/no change, 1: gain, and 2: high-level amplification. Gene expression was represented by mRNA expression z-scores (RNA Seq V2 RSEM) compared to the expression distribution of each gene in tumors diploid for this gene. We included READ (n = 92), colon adenocarcinomas (COAD, n = 242), and mucinous adenocarcinomas of the colon and rectum (MUAD, n = 40).
2.8. Data Representation and Analysis
The intersection of DEG or drug lists among all signatures was analyzed using the Intervene Shiny App (https://intervene.shinyapps.io/intervene/) (accessed on 19 August 2021) [67]. This tool was also applied to determine the top DEG among the ReCa signatures based on the Manhattan distance. DEG shared by all signatures were displayed using a circos plot based on Metascape analysis [54]. The upstream regulatory network analysis of DEG shared among all signatures was generated with the eXpression2Kinases2 (X2K) Web (https://maayanlab.cloud/X2K/) (accessed on 14 April 2021) tool [68]. Heatmaps were created using the web tool Morpheus [69] (https://software.broadinstitute.org/morpheus). The ranking of the top 20 DEG and the volcano plots for each signature were generated in VolcanoseR (https://huygens.science.uva.nl/VolcaNoseR/) (accessed on 19 August 2021) [70].
3. Results
3.1. Acquisition and Processing of Publicly Available Transcriptomic Datasets for ReCa
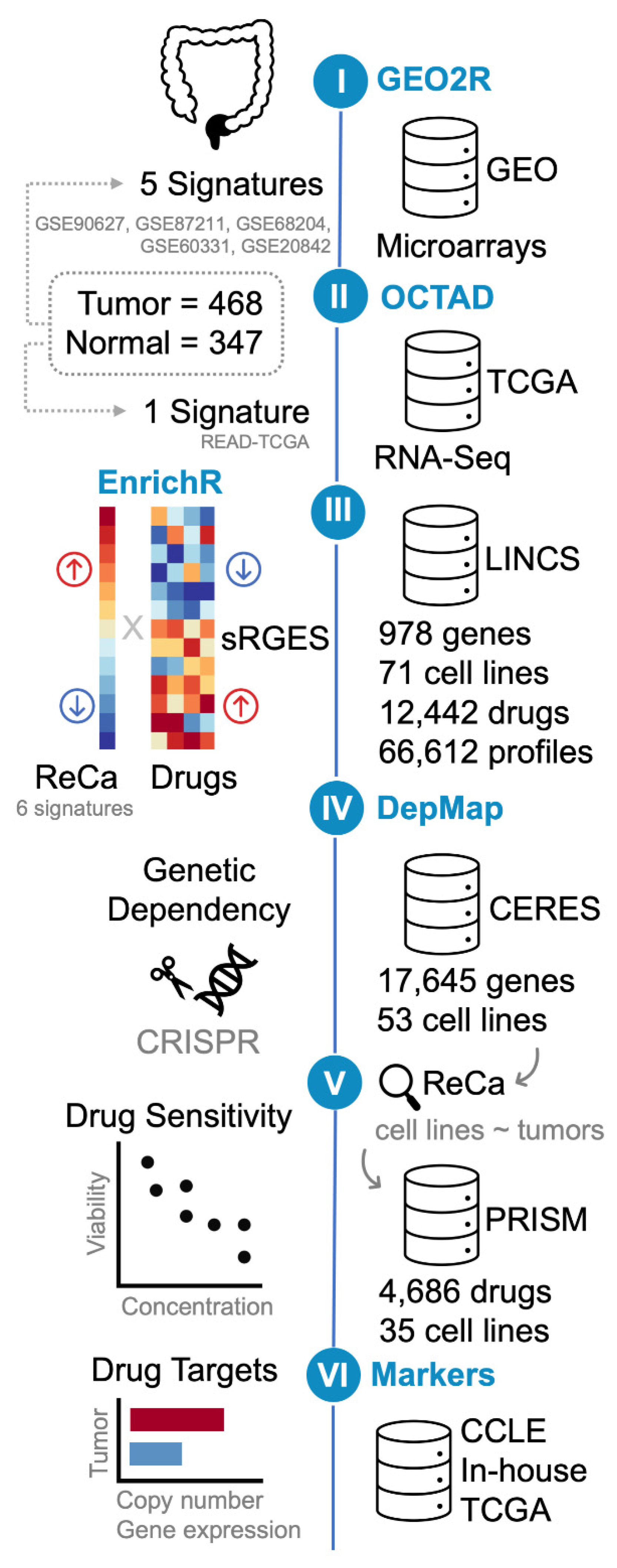
We explored and compared publicly available gene expression profiles of ReCa with those of multiple cancer cell lines treated with drugs in clinical trials or FDA-approved to identify drugs with potency to reverse gene expression signatures of the disease. The workflow of our analysis is summarized in Figure 1.
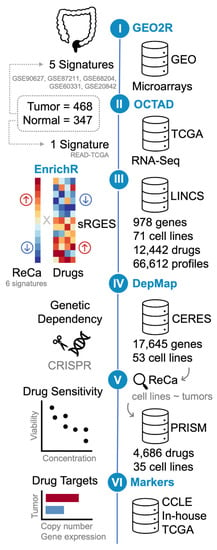
Figure 1.
Schematic representation of the bioinformatics drug-repositioning approach (steps I-IV) using open data to identify drugs with potency to reverse gene expression signatures of rectal cancer (ReCa). The signatures were defined as the list of differentially expressed genes (DEG; log2-fold change > 1 and Adjusted p-value < 0.001) between tumor and normal samples. Step I: The reanalysis of microarrays transcriptome data available at Gene Expression Omnibus (GEO; GSE90627 [71], GSE87211 [72], GSE68204 [73], GSE60331 [74], and GSE20842 [75]) using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) (accessed on 10 March 2021) generated five signatures. Step II: The reanalysis of The Cancer Genome Atlas (TCGA) RNA-Seq data from primary rectal adenocarcinoma (READ) [45] resulted in one additional signature, which was created using the workspace “Open Cancer TherApeutic Discovery” (OCTAD; http://octad.org) (accessed on 5 May 2021) [49]. Step III: gene-set enrichment analysis was performed using EnrichR (https://maayanlab.cloud/Enrichr/) (accessed on 29 March 2021) [51,52,53]. OCTAD was also used to compare the six ReCa signatures to all the Library of Integrated Network-based Cellular Signatures (LINCS) drug-induced gene expression profiles [28,33,34]. OCTAD calculates the drug reversal potency based on a summarization method that quantifies a summarized Reversal Gene Expression Score (sRGES) to match anticorrelated signatures and drug-induced expression profiles. OCTAD was also used to generate target and chemical structure enrichment. Steps IV and V: The Cancer Dependency Map (DepMap; https://depmap.org/) (accessed on 11 June 2021) [27,58] was queried for discoveries of cancer cell lines vulnerabilities. The in vitro genetic dependencies of selected drug targets in cancer cell lines were analyzed using DepMap CRISPR/Cas9 knockout screenings (DepMap 21Q2 Public + Score, CERES dataset) [60,61] (Step IV). Colorectal cancer cell lines that present the most correlated transcriptomic profiles with primary ReCa samples, as determined by EnrichR [51,52,53] Cancer Cell Line Encyclopedia (CCLE) [31,32] library, Celligner [62], OCTAD [49], and Yu et al. [63] were selected for further analysis. The DepMap PRISM drug repurposing resource was used to validate the predictions using drug sensitivity data (PRISM Repurposing Primary and Secondary 19Q4 Screenings) [27] for the selected drugs showing high potency to reverse the ReCa-specific expression signatures in selected cancer cell lines (Step V). Step VI: Copy number and gene expression alterations of selected drug targets were assessed in colorectal cancer (CRC) cell lines (CCLE [31,32]) and tissues (TCGA [45] and in-house [64]) datasets to identify potential predictive markers of drug efficacy.
A total of six ReCa data sets comparing tumor (n = 468) and normal tissue (n = 347) samples was selected. Five of these ReCa datasets were generated using microarrays (GSE90627 [71], GSE87211 [72], GSE68204 [73], GSE60331 [74], and GSE20842 [75]) and one using RNA sequencing (RNA-Seq; TCGA;READ [45]) (Supplementary Data S1). The ReCa sample set comprises pre-treatment biopsy specimens, and the majority is represented by locally advanced ReCa. The description of each ReCa dataset and the corresponding clinical-pathological information is detailed in Tables S3 and S4, respectively.
Each selected microarray dataset was analyzed by comparing the ReCa samples with the respective normal tissues using the web tool GEO2R. For the TCGA RNA-Seq dataset, ReCa samples were compared with RNA-Seq data from normal samples available at GTEx Project [46]. The GTEx normal samples were selected using a deep learning approach based on gene expression profiles, available at the workspace OCTAD (http://octad.org) (accessed on 5 May 2021) [49]. The top 50 GTEx normal samples highly correlated with TCGA tumor samples [50] (Figure S1) were selected to perform differential gene expression analysis with OCTAD. The list of DEG (fold-change > 1 and adjusted p-value < 0.001) between ReCa and normal samples was defined as a ReCa signature for each dataset (Supplementary Data S2, Figure S2).
Among all microarrays-based signatures, the number of DEG in ReCa, relative to normal samples, varied from 2019 to 3326 genes (Figure S3a). As expected, RNA-Seq technology identified the highest number (4604) of DEG for the disease (Figure S3a).
3.2. Integrative Transcriptomic Analysis Reveals Cell Cycle Genes as Potential Drug Targets for ReCa
The essential idea of our bioinformatics strategy was to identify drugs that potentially reverse the ReCa signatures by decreasing genes that are overexpressed while stimulating those under-expressed. The key to such success is identifying specific targeted therapies, selecting drugs that influence the action or activity of a particular signaling pathway [76].
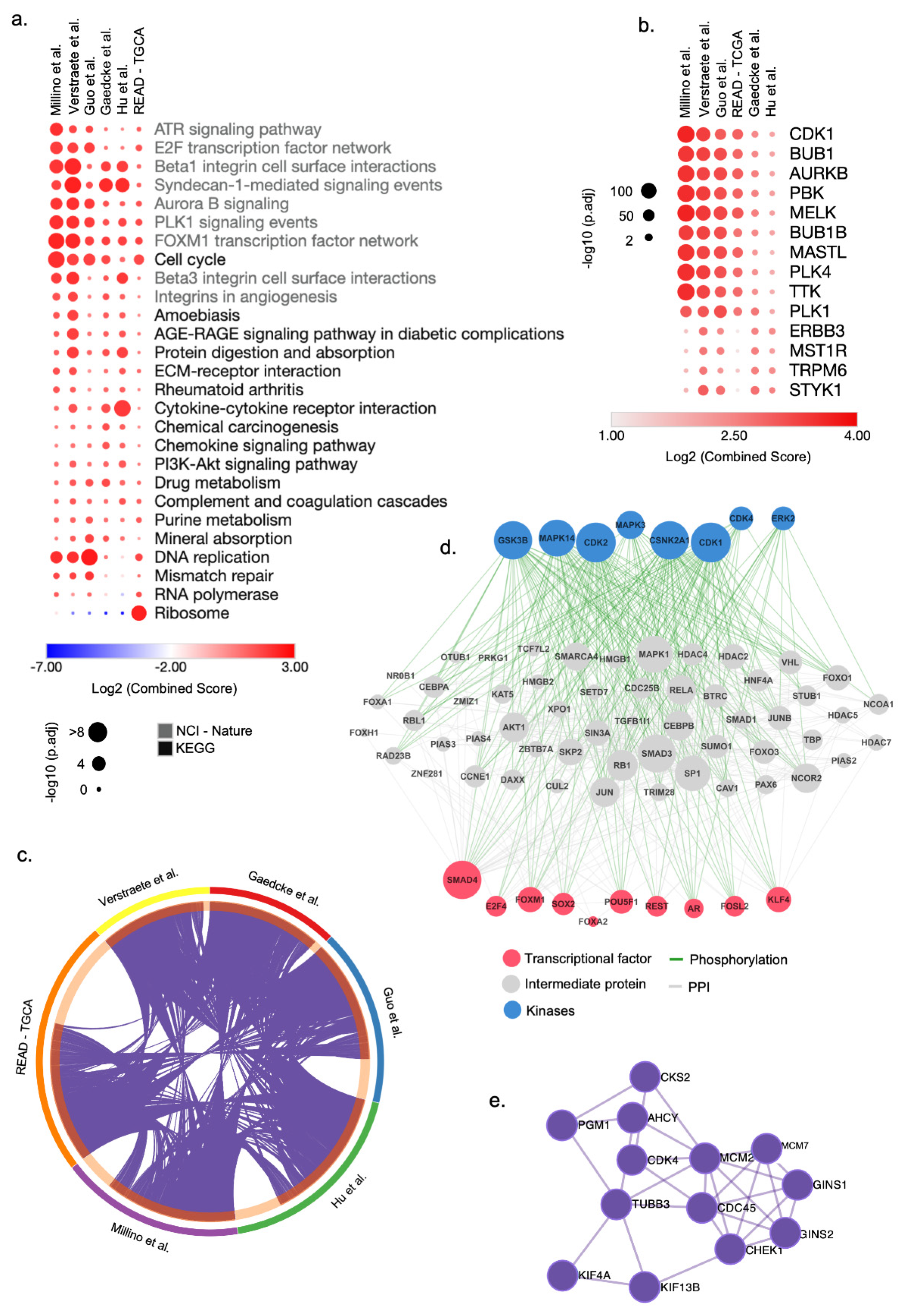
To this aim, we first selected genes significantly deregulated in each signature to perform enrichment analysis using EnrichR. To facilitate a multi-platform analysis, we generated a consensus list comprising the top five pathways or terms for each category when satisfying the criteria of over-representation (Adjusted p-value < 0.05) in at least one signature. We observed a comprehensive spectrum of pathways and functional modules associated with cell cycle genes over-represented in all ReCa signatures (Figure 2a, Figures S4 and S5). The list of pathways includes the ATR signaling pathway, E2F transcription factor network, Beta1 integrin cell surface interactions, Syndecan-1-mediated signaling events, Aurora B signaling events, and DNA replication (Figure 2a). The gene ontology of the ReCa signatures also revealed the enrichment of biological processes and molecular functions associated with the cell cycle (Figure S6). The ARCHS4 kinase analysis showed an enrichment of several cell cycle kinases genes such as CDK1, BUB1, AURKB, PBK, MELK, BUB1B, MASTL, PLK4, TKK, and PLK1 (Figure 2b), which constitute therapeutic targets for cancers [77]. BUB1 and MELK were confirmed consistently deregulated in all ReCa signatures.
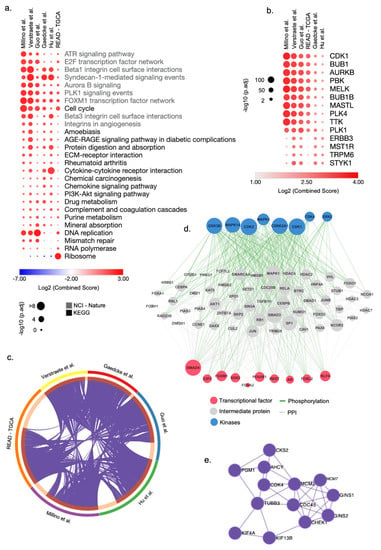
Figure 2.
Integrative transcriptomic analysis reveals cell cycle genes as potential drug targets for rectal cancer (ReCa). (a) Top-ranked combined scores for The Kyoto Encyclopedia of Genes and Genomes (KEGG) and NCI-Nature 2016 pathway categories determined by the gene set enrichment analysis tool EnrichR [51,52]. The top five pathways for each category were included in a consensus list when satisfying the criteria of over-representation (Adjusted p-value < 0.05) in at least one signature. Columns and rows were clustered based on the Euclidean distance of log2 combined score values. (b) Top-ranked combined scores for kinase genes co-expressed with each ReCa signature in the ARCHS4 [78] database determined by the gene set enrichment analysis tool EnrichR [51,52]. The top five gene set enrichment terms were included in a consensus list when satisfying the criteria of over-representation (Adjusted p-value < 0.05) in at least one signature. Columns and rows were clustered based on Euclidean distance of log2 combined score values. (c) Circos plot visualization of the differentially expressed genes (DEG) that overlap among the ReCa signatures: Gaedcke et al., GSE20842 [75]; Guo et al., GSE90627 [71]; Hu et al., GSE87211 [72]; Millino et al., GSE68204 [73]; Rectal Adenocarcinoma from The Cancer Genome Atlas (TCGA) [45]; and Verstraete et al., GSE60331 [74]. The purple lines in the Circos plot connect DEG among multiple signatures. Each color in the outer circle represents one signature. The dark orange in the inner circle represents the number of overlapping DEG among the signatures. (d) The eXpression2Kinases2 (X2K) upstream regulatory network analysis [68] for a signature of 372 genes differentially expressed among all six ReCa signatures. The network includes transcription factors, proteins, and kinases predicted to regulate the expression of these 372 genes. (e) Protein-protein interaction network of the top enriched terms associated with DNA replication identified in all ReCa signatures by the Molecular Complex Detection (MCODE) algorithm in Metascape (https://metascape.org/) (accessed on 9 April 2021) [54].
We found that most of the DEG are shared by at least two signatures (Figure 2c, Table S5), and 372 genes are deregulated in all six signatures (Figure S3a, Table S6). The upstream regulatory networks that regulate these 372 genes include transcription factors, proteins, and kinases associated with the cell cycle and DNA replication (Figure 2d,e). CLDN1, CDH3, FOXQ1, and KRT80, previously associated with CRC, were consistently placed in the top-ranked DEG among the signatures (Figures S2 and S3b). Interestingly, CDK4 and the topoisomerases TOP1MT and TOP2A were included in the list of genes deregulated shared by all six signatures (Table S6). Additionally, TOP1, the target of the topoisomerase inhibitor irinotecan, was deregulated in four (Gaedcke et al. [75], Guo et al. [71], Hu et al. [72], and Verstraete et al. [74]) of six signatures (Table S5).
3.3. Topoisomerase and CDK Inhibitors Are Candidate Targets for Drug Repositioning in ReCa
We used a systems-based approach, available at the OCTAD [49], to compare ReCa signatures and drug-induced gene expression profiles from cancer cell lines (LINCS; L1000 dataset) to predict new therapeutic candidates. This analysis revealed the top compounds (in clinical trials or FDA-approved) that present high potency in reversing the expression of ReCa signatures (Supplementary Data S3, Figure S7).
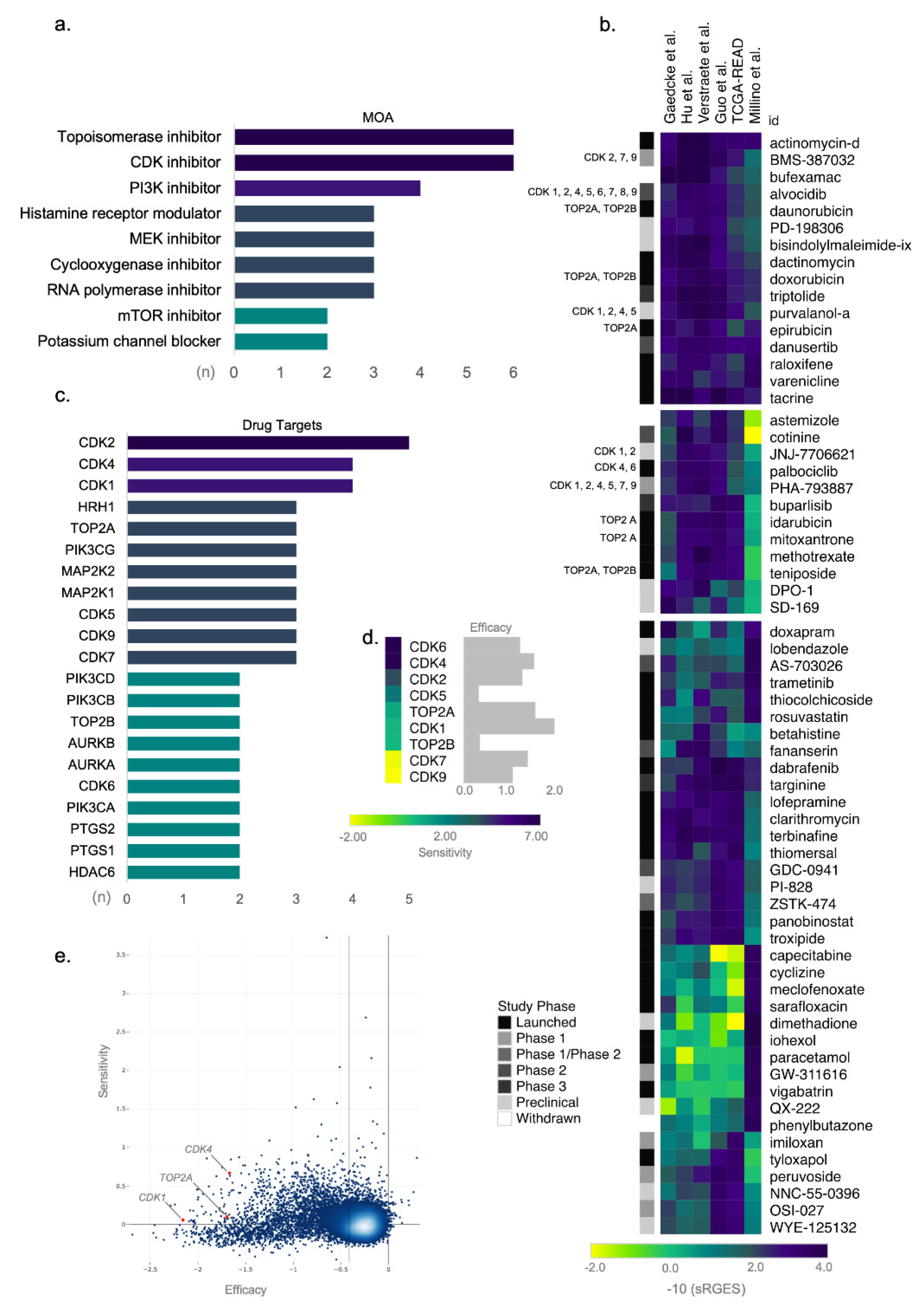
To prioritize drugs predicted to reverse the ReCa gene expression signatures, we selected the top 100 compounds with the highest reversal potency score determined by OCTAD among all signatures (Table S7). Drugs that appeared in more than one signature were included once, resulting in a final list of 64 drugs (Table S8). Drugs targeting cell cycle pathways such as topoisomerase and CDK inhibitors had the highest number of drugs with a similar mechanism of action (six each) in our final list of top compounds (Figure 3a).
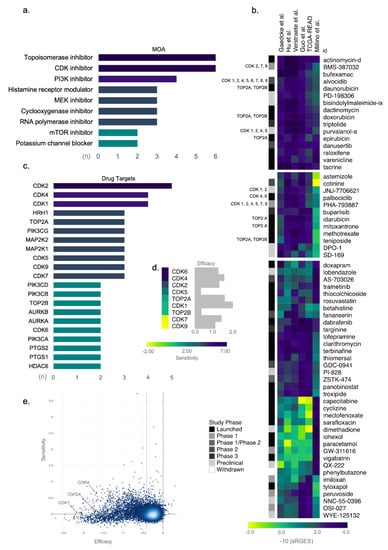
Figure 3.
Topoisomerase and cyclin-dependent kinase (CDK) inhibitors as candidate targets for drug repositioning in rectal cancer (ReCa). (a) Drugs sharing similar mechanism of action (MOA) included among the top 100 compounds with the highest summarized Reversal Gene Expression Score (sRGES) determined by Open Cancer TherApeutic Discovery (OCTAD; http://octad.org) (accessed on 5 May 2021). (b) Heatmap of representative scores of overall reversal potency of a compound (sRGES) to ReCa signatures. The sRGES were clustered by the k-means clustering algorithm, and only the three clusters with the highest sRGES were displayed in the heatmap. The clinical phase of the drug development process for each compound is indicated in grayscale on the left side of the heatmap. Compounds targeting CDK and topoisomerase inhibitors are in the same two clusters with the highest sRGES. CDK1, CDK2, CDK4, CDK5, CDK6, CDK7, CDK8, and CDK9 genes are indicated by 1, 2, 4, 5, 6, 7, 8, and 9, respectively. ReCa signatures are clustered by Euclidean distance between sRGES. (c) Drug targets for the top 100 compounds with the highest drug reversal potency score determined by OCTAD. We included only drug targets sharing the same specific target in the graph. (d) To predict the efficacy and selectivity of candidate drugs, the growth reduction caused by the loss of function of the CDK1, CDK2, CDK4, CDK5, CDK6, CDK7, CDK8, CDK9, TOP2A, and TOP2B genes (efficacy), and the selectivity of this effect across all cancer cell lines available at Cancer Dependency Map (DepMap) [58] were analyzed with the shinyDepMap tool (https://labsyspharm.shinyapps.io/depmap) (accessed on 17 May 2021) [57]. (e) Considering CDK and topoisomerase inhibitors, CDK1 (1), CDK4 (4), and TOP2A (A) showed high efficacy among 15.847 genes (blue dots). CDK4 also presents high selectivity and constitutes a promising drug target for synthetic lethality of cancer cells.
Topoisomerase and CKD inhibitors are also among the drugs with the highest drug reversal potency score (Figure 3b). The top-scored drugs predicted to reverse the ReCa gene expression are FDA-approved for cancer treatment, such as the topoisomerase inhibitors (daunorubicin, doxorubicin, epirubicin, idarubicin, mitoxantrone, and teniposide) and the CDK inhibitor (palbociclib) (Figure 3b). The list of these top-scored drugs also includes the CDK inhibitors currently in clinical trials (BMS-387032, alvocidib, purvalanol-a, JNJ-7706621, and PHA-793887) (Figure 3b). The six predicted topoisomerase inhibitors target TOP2A and TOP2B, whereas the CDK2, CDK4, and CDK1 are the main targets of the six CDK inhibitors (Figure 3c).
Next, we used the shinyDepMap tool to evaluate the most promising topoisomerases and CDK genes as therapeutic targets by predicting their efficacy and selectivity in cancer cell lines. Among the CDK and topoisomerase inhibitors, those targeting CDK1, CDK4, or TOP2A showed the highest efficacies (Figure 3d). CDK4 also presented high selectivity among 15.847 genes (Figure 3e), suggesting that this gene is a promising drug target for selective synthetic lethality of cancer cells. By explicitly analyzing CRISPR screening in 53 CRC cell lines, TOP2A, CDK1, CDK7, and CDK9 presented the highest efficacies demonstrated by the most negative or toxic CERES effects (Figure S8).
We investigated similarities between gene expression profiles of ReCa and cancer cell lines. Using this in silico analysis, we tested the effects of selected drugs in cancer cell lines with the highest correlations to ReCa signatures. We first selected genes significantly deregulated in each signature to perform enrichment analyses using the CCLE [31,32] library in EnrichR [51,52,53] (Figure S9a). Our selection of representative cancer cell lines as models of rectal tumor signatures further used Celligner [62] data, which confirmed the alignment of the enriched CCLE CRC cell lines with ReCa tumor samples (Figure S9a,b). These similarity analyses allowed us to select a list of CRC cell lines with drug sensitivity data in the PRISM Repurposing Primary 19Q4 (SW1463, HT115, and CW2) or Primary and Secondary 19Q4 Screenings (HT55, SNUC4, LS80, LS1034, HCC56, SW498, CL34, and SNU61), available at DepMap (https://depmap.org/) [27,58] (Supplementary Data S4). The gene expression profiles for the cell lines SW948, CL34, HT115, and SNUC4 from the L1000 dataset were highly correlated with TCGA-READ gene expression profiles (Figure S9c), according to OCTAD results. The consensus molecular subtypes (CMS) classification, as described by Yu et al. [63], revealed cell lines matching the subtypes CMS1 (CW2), CMS2 (HCC56, HT55, LS1034, SW1463, SW948), and CMS3 (LC34, LS180, HT115, and SNU61) (Figure S9d). Moreover, the cell lines HCC56, HT55, and CL34 integrate the cancer cell line panel TCGA-110-CL, proposed by Yu et al. [63] for pan-cancer studies.
We used primary and secondary PRISM data available at DepMap [27,58] to verify the drug sensitivity of 11 cancer cell lines that presented similar transcriptional profiles to ReCa signatures. Fifty-eight of 64 drugs with the highest drug reversal potency score had available data in the PRISM (Supplementary Data S4). PRISM repurposing primary screening (single dose) had data for these 11 cell lines, and the secondary screening (8-point dose-response) had data for eight of them (Figures S10a,b, Supplementary Data S4). Notably, drugs that induced the highest sensitivity in these selected cancer cell lines in both primary and secondary screenings are inhibitors of topoisomerase (doxorubicin, idarubicin, epirubicin, daunorubicin, mitoxantrone, and teniposide) and CDK (BMS-387032, JNJ-7706621, alvocidib, PHA-793887, and palbociclib) (Figure S10). Among these drugs, idarubicin, BMS-387032, doxorubicin, epirubicin, teniposide, and mitoxantrone induced high sensitivity even at low doses (Figure S10).
3.4. TOP2A Gene Expression and Copy Number Gene Are Potential Predictive Markers of Topoisomerase Inhibitors Efficacy in ReCa
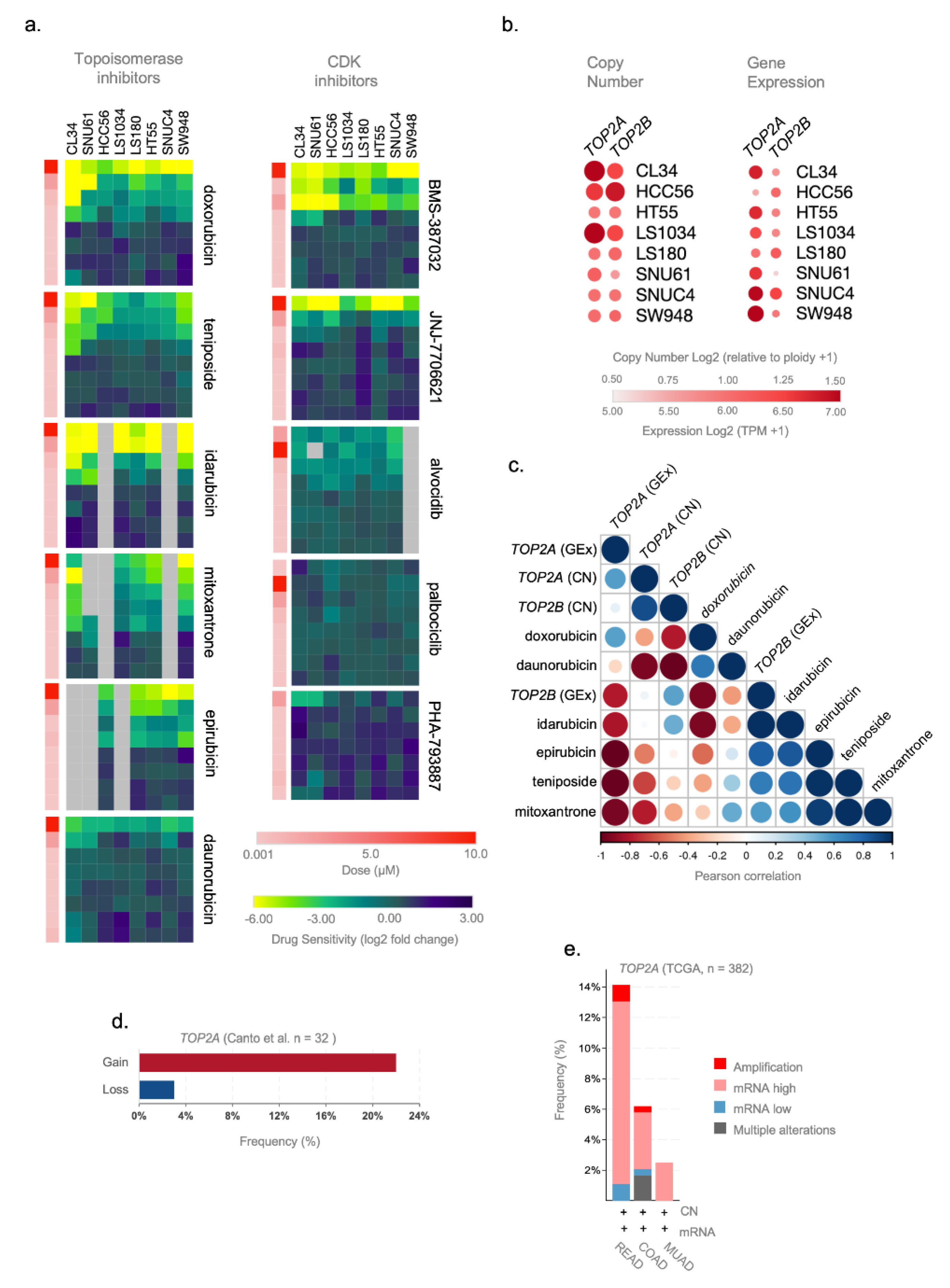
We used the secondary PRISM Repurposing dataset containing cell line drug-perturbation viability screens for 11 topoisomerases and CDK inhibitors tested in eight CRC cell lines (CL34, SNU61, HCC56, LS1034, LS180, HT55, SNUC4, and SW948) that presented similar transcriptional profiles to ReCa signatures. Figure 4a depicts the treatment of these cell lines with eight concentrations of topoisomerases and CDK inhibitors (4-fold dilution starting from 10μM). By comparing the cellular vulnerabilities to topoisomerases and CDK inhibitors, we found that these cells were more sensitive to low doses of FDA-approved topoisomerase inhibitors (Figure 4a, Figure S10). Three cell lines (CL34, SNU61, and HCC56) showed the highest sensitivity and similar response to topoisomerase inhibitors (Figure 4a). Low doses of the CDK inhibitors BMS-387032 and JNJ-7706621 also induced high sensitivity in the eight selected CRC cell lines, but the FDA has not yet approved these two drugs. Based on the specific cellular vulnerabilities to FDA-approved topoisomerase inhibitors, we next focused on predictors of anticancer sensitivity for drugs targeting TOP2A and TOP2B. The genetic predictions of drug response can speed up the development of “personalized” therapeutic regimens [31]. Therefore, we examine TOP2A and TOP2B genes’ copy number and expression levels in CRC cell lines and tissues. Using CCLE data, we found gains of TOP2A and TOP2B in the cell lines CL34, HCC56, and LS1034; and increased TOP2A gene expression level in the cell lines CL34, SNU61, HCC56, LS1034, HT55, SNUC4, and SW948 (Figure 4b). We found clear inverse correlations between TOP2A gene copy number or expression levels with the sensitivity for topoisomerase inhibitors but not for TOP2B. Significantly, gains of TOP2A inversely correlated with the sensitivity of doxorubicin, daunorubicin, idarubicin, epirubicin, teniposide, and mitoxantrone (Figure 4c). Moreover, the expression levels of TOP2A inversely correlated with the sensitivity of daunorubicin, idarubicin, epirubicin, teniposide, and mitoxantrone (Figure 4c). Considering that some ReCa patients presented TOP2A gains (Figure 4d,e) or increased expression in ReCa compared to other colorectal tumors (Figure 4e), these events might constitute specific predictive markers of topoisomerase inhibitors efficacy in ReCa patients.
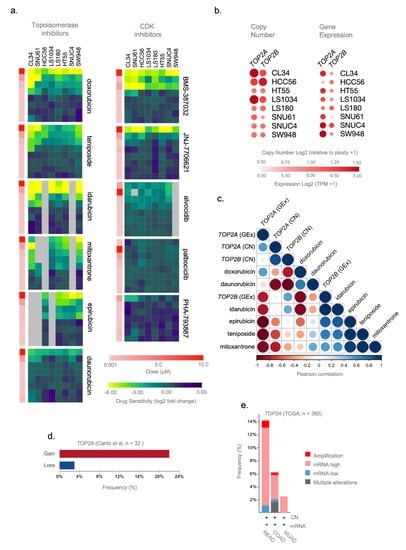
Figure 4.
TOP2A gene expression and copy number are potential predictive markers of the efficacy of topoisomerase inhibitors in rectal cancer (ReCa). (a) Heatmaps of sensitivity (log2 fold change) for Topoisomerase and cyclin-dependent kinase (CDK) inhibitors in eight colorectal cancer (CRC) cell lines to validate the predictions using the PRISM Repurposing Secondary 19Q4 Screen [27,58]. The data were obtained in CRC cell lines with transcriptomic profiles similar to rectal tumor samples (Figure S8) and were downloaded from the Cancer Dependency DepMap; https://depmap.org/) (accessed on 11 June 2021) [58]. Columns (cells) and rows (drugs) for each compound were clustered based on the Euclidean distance of the log2 fold change values. The drug dose is represented in color bars and increases from light (0.001 µM) to dark red (10.0 µM). (b) Relative expression (TPM+1) and gene copy number [log2 (relative to ploidy +1)] of TOP2A and TOP2B in eight CRC cell lines from CCLE [31,32] database. The data (Copy Number 21Q2 Public and Expression 21Q2 Public datasets) were obtained in CRC cell lines with transcriptomic profiles highly correlated with rectal tumor samples (Figure S8) and downloaded from DepMap [58]. (c) Pearson correlation plot showing a negative correlation between TOP2A and TOP2B copy number and gene expression (Copy Number 21Q2 Public and Expression 21Q2 Public datasets) and drug sensitivity of topoisomerase inhibitors (doxorubicin, daunorubicin, idarubicin, epirubicin, teniposide, and mitoxantrone) (PRISM Repurposing Secondary 19Q4 Screen) in CRC cell lines (CL34, HCC56, HT55, LS1034, LS180, SNU61, SNUC4, and SW948). These colorectal cell lines present transcriptomic profiles highly correlated to rectal tumor samples (Figure S8). The molecular [31,32] and drug sensitivity [27] data were downloaded from DepMap [58]. (d) Frequency of copy number alterations (gain/loss) of TOP2A gene in a cohort of locally advanced rectal adenocarcinoma patients (n = 33) [64]. (e) Frequency of copy number and gene expression alterations of TOP2A gene in colorectal adenocarcinoma (TCGA, Firehose Legacy) patients using the cBioPortal (https://www.cbioportal.org/) (accessed on 20 May 2021) database. The alterations include amplification (red), mRNA high (pink), mRNA low (blue), and multiple alterations (gray). Putative copy number calls were determined using GISTIC 2.0 and considered the following values: −2 = homozygous deletion; −1 = hemizygous deletion; 0 = neutral/no change; 1 = gain; 2 = high level amplification. Gene expression is represented by mRNA expression z-scores (RNA Seq V2 RSEM) compared to the expression distribution of each gene tumor that is diploid for this gene. The analysis was performed using rectal adenocarcinoma (READ, n = 92), colon adenocarcinoma (COAD, n = 242), and mucinous adenocarcinomas of the colon and rectum (MUAD, n = 40).
4. Discussion
The treatment option for most patients with locally advanced ReCa is neoadjuvant chemoradiation and surgery followed by adjuvant chemotherapy in selected cases [16,17]. However, only a few patients benefit from the treatment. Using a computational drug repositioning approach, we found that FDA-approved or investigational candidate drugs as new therapeutic options that may rapidly be translated to the clinics to treat ReCa. Among these drugs, topoisomerase (doxorubicin, idarubicin, epirubicin, daunorubicin, mitoxantrone, and teniposide) and CDK (BMS-387032, JNJ-7706621, alvocidib, PHA-793887, and palbociclib) inhibitors potentially reverse ReCa signatures and inhibit the growth of CRC cell lines with transcriptional profiles similar to ReCa. These cell lines presented higher sensitivity at low doses of topoisomerase inhibitors, which were correlated with the TOP2A gene expression and copy number. Therefore, we also propose TOP2A as a potential predictive marker of higher efficacy of topoisomerase inhibitors in ReCa patients. Together, our findings demonstrated that reversal gene expression analysis evinced drug targets and therapeutic alternatives for patients with ReCa. We consider that the computational workflow presented here can be applied with success to drug repositioning predictions in other cancer types.
Here, we present a global transcriptomic approach that used the OCTAD [49] tool to connect six ReCa signatures from GEO [71,72,73,74,75] and TCGA [45] datasets to 66,000 drug-induced gene expression profiles from the LINCS L1000 dataset [28,33,34]. Similar computational system biology approaches have been used to identify potential therapeutic targets for repositioning for hepatocellular carcinoma [38,42], Ewing sarcoma [39], basal cell carcinoma [40], renal cell cancer [41], small cell lung cancer, and neuroendocrine tumors [36]. Additionally, it has been demonstrated that the use of drug-induced gene expression profiles to determine the potency of a drug to reverse cancer gene expression signatures correlates with drug efficacy in preclinical models of breast, liver, and colon cancer [42]. The use of drug-induced gene expression profiles matched to inverse disease profiles has identified citalopram, an antidepressant, as a potential therapeutic option for patients with metastatic CRC [37]. Our approach generated a list of 64 FDA-approved or investigational candidate drugs that may potentially reverse the gene expression signature of ReCa. The topoisomerase and CKD inhibitors predominate among the FDA-approved drugs with the highest drug reversal potency score. Thus, the synergistic interaction of these drugs, when combined, should be better investigated.
To validate the effects of these 64 compounds in cancer cell lines as potential drugs to treat ReCa, we selected DepMap data from clinically relevant cancer cell lines, aiming to increase the likelihood of translating the preclinical findings. Recent studies have shown that cell lines differ in their ability to model primary tumors and that selecting cancer cell lines that recapitulate the pathobiology of tumors is critical [62,63]. Although some cancer cell lines are derived from ReCa, it is not possible to determine whether they were derived from patients undergoing multimodal therapy or from the actual anatomic rectum [79]. Therefore, we used cancer cell lines and tumor transcriptomics data [31,32,62,63] to select eight cell lines HT55, SNUC4, LS80, LS1034, HCC56, SW498, CL34, and SNU611 for dose-response screenings. These colorectal cell lines present gene expression profiles highly correlated with the gene expression signatures of ReCa samples.
Among our list of 64 compounds, inhibitors of topoisomerase (daunorubicin, doxorubicin, epirubicin, idarubicin, mitoxantrone, and teniposide) and CDK (palbociclib) are among the most potent drugs that induce cell sensitivity in selected cell lines. These results provide proof of the concept of our bioinformatics approach. Most notably, these drugs available for repurposing are FDA-approved or are in late-phase clinical trials for several malignancies. Although topoisomerases 2 and CDK4/6 inhibitors have emerged as a potent strategy for cancer treatment [80,81], few studies have tested these compounds in clinical trials for ReCa treatment. The CDK inhibitor palbociclib is under clinical trials phases 1 and 2 to treat CRC (ClinicalTrials.gov; NCT03981614, NCT01037790, NCT02465060, and NCT02897375). These studies warrant further attention considering that we found CDK4 consistently deregulated across all ReCa signatures. The topoisomerase 2 inhibitor doxorubicin has also been tested in a terminated phase III clinical trial that evaluated the effectiveness of chemoembolization in treating patients with CRC metastatic to the liver (ClinicalTrials.gov; NCT00023868). Unfortunately, the results of this study were not published.
Topoisomerase 1 inhibitors are relevant drugs for ReCa treatment [80]. The combination of topoisomerase 1 inhibitor irinotecan with 5-fluorouracil has been proven effective for treating CRC [82]. Moreover, previous studies have demonstrated improved therapy efficacy combining irinotecan to the capecitabine or 5-fluorouracil chemoradiotherapy of locally advanced ReCa [83,84,85,86]. We found changes in the expression of the TOP1 gene in four ReCa signatures, while TOP1MT and TOP2A were altered in all six signatures. Interestingly, two clinical trials (phases 1 and 2) that evaluated the use of topoisomerase 2 inhibitor epirubicin in CRCs highlighted that it induces better responses to rectal tumors [87,88].
The mechanisms involved in this specific response remain unclear and deserve further investigation. However, all cancer cell lines with transcriptional profiles comparable to ReCa signatures presented high sensitivity to topoisomerase inhibitors (idarubicin, doxorubicin, epirubicin, teniposide, and mitoxantrone), even at low doses. Based on the high sensitivity of the cell lines tested to topoisomerase inhibitors and that these drugs are all approved by the FDA, rectal cancer patients may also be benefited from this treatment. Moreover, the dysregulation of TOP2A found in all ReCa signatures suggests its role as a marker of anticancer sensitivity for these drugs. TOP2A amplification is a known predictive marker of anthracycline-based neoadjuvant chemotherapy efficacy in patients with breast cancer [89,90]. We found correlations between TOP2A gene imbalances and expression levels and the sensitivity of CRC cell lines for topoisomerase inhibitors. We also confirmed that TOP2A gene amplification and increased expression are frequently detected in ReCa samples [91,92,93,94]. Therefore, testing the TOP2A status may predict more accurately the patients that likely benefit from treatments based on topoisomerase inhibitors. Accordingly, the clinical benefit of epirubicin treatment was recently tested in patients with metastatic CRC resistant to oxaliplatin and TOP2A amplification [95]. However, only six patients were enrolled in the study, and the results were not conclusive. Based on our data, we propose that topoisomerase inhibitors identified here are promising candidates for repositioning to advance the ReCa treatment. These drugs may specifically benefit ReCa patients with amplification and increased expression of the TOP2A gene.
Besides predicting drug candidates, our comprehensive and integrated characterization of the six ReCa signatures revealed marked similarities in gene expression and pathways deregulated in the disease. Pathways associated with cell cycle and DNA replication genes were over-represented in all ReCa signatures. The list of oncogenic pathways that can be targeted to treat ReCa includes the ATR signaling pathway [96], E2F transcription factor network [97], Beta1 integrin cell surface interactions [98], Syndecan-1-mediated signaling events [99], Aurora B signaling events [100,101], and DNA replication [102,103]. We identified genes consistently altered in all ReCa signatures, such as the top-ranked DEG CLDN1, CDH3, FOXQ1, and KRT80, all previously associated with CRC [104,105,106,107]. The consistent deregulation of these pathways and genes such as CDK4 and TOP2A reinforces their relevance as potential therapeutic targets for ReCa.
In conclusion, we showed that mining and integrating publicly available open data of cancer gene expression profiles with the effect of drugs on gene expression or inhibitory activity in human cancer cell lines is a promising strategy for the computational repositioning of compounds to ReCa treatment. Based on the generated data, we propose that topoisomerase inhibitors (such as idarubicin, doxorubicin, epirubicin, teniposide, and mitoxantrone) should be tested in a specific cohort of ReCa patients with increased expression or amplification of the TOP2A gene. Alternatively, these drugs or their combinations with the top hits here identified can be screened in rectal cancer-derived organoids [108]. Pre-clinical studies using cancer-derived organoids to drug testing are valuable in predicting drug responses in a clinical setting. These in vitro models are known to more accurately recapitulate the tumoral biological heterogeneity associated with patient-specific cancers [109], allowing a diversity of drug-screening applications [110].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13215492/s1: Supplementary Data S1: Microarrays (GEO) and RNA-Seq (TCGA) data of rectal cancer datasets included in this study, Supplementary Data S2: List of differentially expressed genes (fold-change > 1 and adjusted p-value < 0.001) between rectal cancer and normal samples, Supplementary Data S3: OCTAD output files with the sRGES (summarized Reversal Gene Expression Score) for rectal cancer gene expression signatures, Supplementary Data S4: Sensitivity data (PRISM Repurposing Primary and Secondary Screenings—19Q4) for selected drugs showing high potency to reverse the rectal cancer-specific expression signatures in 35 colorectal cancer cell lines, Table S1: Medical Subject Headings (MeSH) unique ID D012004 for Rectal Neoplasms (tumors or cancer of the rectum) retrieved from PubMed (https://www.ncbi.nlm.nih.gov/mesh) (accessed on 2 March 2021), Table S2: Medical Subject Headings (MeSH) unique IDs and GEO datasets Study IDs for rectal cancer, Table S3: Description of the rectal cancer GEO datasets included in this study, Table S4: Clinical and pathological data of rectal cancer patients investigated in this study. Table S5: Differentially expressed genes (fold-change > 1 and adjusted p-value < 0.001) shared in rectal cancer signatures, Table S6: Differentially expressed genes (fold-change > 1 and adjusted p-value < 0.001) shared in six rectal cancer signatures, Table S7: Top 100 compounds with the highest reversal potency score (sRGES < −0.25; n > 2) determined by OCTAD among the six rectal cancer signatures, Table S8: Top 64 compounds (unique, from the top 100) with the highest reversal potency score determined by OCTAD among the six rectal cancer signatures, Figure S1: Selection of reference normal samples from The Genotype-Tissue Expression (GTEx) to generate rectal cancer (ReCa) gene expression signature for primary rectal adenocarcinoma (READ) samples from The Cancer Genome Atlas (TCGA) [45]. The GTEx normal samples that highly correlated with READ samples were selected in the workspace “Open Cancer TherApeutic Discovery” (OCTAD; http://octad.org) (accessed on 5 May 2021) [49]. This OCTAD analysis selected the top 50 normal samples (red dots) using the Spearman correlation test of top varying genes between each READ-TCGA sample and all GTEx normal samples (gray dots), Figure S2: Rectal cancer (ReCa)-specific gene expression signatures, Volcano plots showing fold changes (Log2 FC > 1) for differentially expressed genes (DEG; FDR-adjusted p-value < 0.01) between ReCa and normal samples. The volcano plots summarize results for Gaedcke et al. (GSE20842) [75], Millino et al. (GSE68204) [73], Verstraete et al. (GSE60331) [74], Hu et al. (GSE87211) [72], Guo et al. (GSE90627) [71], and TCGA-READ [45]. The top 20 ranking genes indicated in the volcano plots for each signature were calculated using the Manhattan distance. CLDN1, CDH3, FOXQ1, and KRT80, previously associated with CRCs, were consistently placed in the top-ranked DEG (green dots), Figure S3: Differentially expressed genes shared among all rectal cancer (ReCa) signatures identified as top-deregulated genes previously described in colorectal cancer (CRC). (a) Upset plot showing the number of differentially expressed genes (DEG; red bars) shared by all ReCa signatures. The blue bar graphs depict the number of DEG in each signature. The combinations of shared DEG among bar plots are indicated in the grid (bottom). (b) Upset plot showing the intersections of the top 20 DEG (red bars) among ReCa signatures determined based on the Manhattan distance. CLDN1, CDH3, FOXQ1, and KRT80 genes (bold), previously associated with CRCs, were consistently placed in the top-ranked DEG among the signatures, Figure S4: Network of overlapping enriched terms among rectal cancer (ReCa) signatures, performed by Metascape (https://metascape.org/) (accessed on 9 April 2021) [54]. (a) The enriched terms are colored according to the ID of each cluster. The clusters consist of enriched terms with a similarity metric > 0.3 determined by Kappa scores and are connected by edges. The nodes sharing the same cluster ID are generally next to each other. (b) The enriched terms are colored according to p-values. Terms containing more genes tend to have a more significant p-value, Figure S5: Protein-protein interaction network analysis based on overlapping significant differentially expressed genes (DEG) in all rectal cancer (ReCa) signatures, performed by Metascape (https://metascape.org/) (accessed on 9 April 2021) [54]. Networks containing between 3 and 500 proteins were used to detect densely connected network components using Molecular Complex Detection (MCODE) algorithm, Figure S6: Top-ranked combined scores for gene ontologies (GO) terms determined by the gene set enrichment analysis tool EnrichR [51,52,53]. The top five terms for each biological process (BP) and molecular function (MF) were included in a consensus list according to the adopted criteria of over-representation (Adjusted p-value < 0.05) in at least one signature. Columns and rows were clustered based on Euclidean distance based on log2 combined score values, Supplementary Figure S7: Identification of top compounds in clinical trials or FDA-approved that present high potency to reverse the expression of six rectal cancer (ReCa) signatures. The first column of each heatmap shows a ReCa signature: Gaedcke et al., GSE20842 [75]; Millino et al., GSE68204 [73]; Verstraete et al., GSE60331 [74]; Hu et al., GSE87211 [72]; Guo et al., GSE90627 [71]; and Rectal Adenocarcinoma (READ) from The Cancer Genome Atlas (TCGA) [45]. In this first column, genes that are upregulated and downregulated in ReCa, compared to normal samples, are shown in red and blue, respectively. The remaining columns of each heatmap show the top LINCS L1000 drug signatures [28,33,34] with a reverse expression profile of the corresponding ReCa signature (first column). In these drug signature columns, red and blue indicate genes with high and low expression induced by drug treatment, respectively, Figure S8: CERES [60] effects for CRISPR knockout screening of CDK1, CDK2, CDK4, CDK5, CDK6, CDK7, CDK8, CDK9, TOP2A, and TOP2B genes among 53 colorectal cancer (CRC) cell lines available at Cancer Dependency Map (DepMap; 21Q2 Public + Score dataset) [58]. Columns and rows were clustered using Euclidean distance based on the gene effect (CERES) values, Figure S9: Selection of representative cancer cell lines as models for rectal tumors. (a) Top-ranked combined scores for colorectal cancer (CRC) cell lines from Cancer Cell Line Encyclopedia (CCLE) [31,32] presenting the most correlated transcriptomic profiles with primary rectal cancer (ReCa) samples. This gene set enrichment analysis was performed using EnrichR (https://maayanlab.cloud/Enrichr/) (accessed on 13 April 2021) [51,52,53]. Cell lines with drug sensitivity data in the PRISM Repurposing Primary 19Q4 (gray arrowhead) or Primary and Secondary 19Q4 Screenings (black arrowhead) available at the Cancer Dependency Map (DepMap; https://depmap.org/) (accessed on 11 June 2021) [27,58] were selected for further analyses. The cell lines that closely resembled rectum adenocarcinomas (shown in b—connecting lines) were queried to discover cancer cell line vulnerabilities for selected drugs showing high potency to reverse the ReCa-specific expression signatures. (b) ReCa cell lines from CCLE [31,32] that closely resemble rectal adenocarcinomas, as determined by Celligner [62] (https://depmap.org/portal/celligner/) (accessed on 6 May 2021). This method applies an unsupervised approach that corrects differences when integrating large-scale cell line and tumor RNA-Seq datasets. The distance between a cell line and rectal adenocarcinomas, based on median Euclidean distances, decreases from blue to red according to the color bars. The cell lines with drug sensitivity data in the PRISM Repurposing Primary 19Q4 (gray arrowhead) or Primary and Secondary 19Q4 Screenings (black arrowhead) available at the DepMap [27,58] were selected for further analyses. (c) Spearman rank correlation between gene expression profiles from the Library of Integrated Network-based Cellular Signatures (LINCS) L1000 data set [28,33,34] and The Cancer Genome Atlas (TCGA) RNA-Seq data for primary rectal adenocarcinoma (READ) [45] samples was estimated by the workspace “Open Cancer TherApeutic Discovery” (OCTAD; http://octad.org) (accessed on 5 May 2021) [49]. This OCTAD analysis is based on the average of correlations between the LINCS cell lines and individual tumors. This analysis also revealed cell lines with drug sensitivity data in the PRISM Repurposing Primary 19Q4 (gray arrowhead) or Primary 19Q4 and Secondary 19Q4 Screenings (black arrowhead), available at the DepMap [27,58]. (d) Prediction of cancer cell line subtypes using gene expression-based consensus molecular subtypes (CMS) of colorectal adenocarcinoma tumors. The data obtained from Yu et al. [63] is based on correlations between transcriptomic profiles from TCGA [45] and the CCLE [31,32]. Selected cell lines (in bold) with drug sensitivity data in the PRISM Repurposing Primary 19Q4 or Primary 19Q4 and Secondary 19Q4 Screenings available at the DepMap [27,58] are indicated as gray or black arrowheads, respectively. The cell lines HCC56, HT55, and CL34 (indicated as asterisks) are included in the cancer cell line panel TCGA-110-C [63], Figure S10: Drug sensitivity for the 58 selected drugs showing high potency to reverse rectal cancer (ReCa)-specific expression signatures. The data were obtained from colorectal cancer (CRC) cell lines with transcriptomic profiles highly correlated with rectal tumor samples (Figure S9) (downloaded from the Cancer Dependency Map, DepMap; https://depmap.org/) (accessed on 11 June 2021) [27,58]. (a) Heatmap of drug sensitivity (log2 fold change) in 11 CRC cell lines to validate the predictions using the PRISM Repurposing Primary 19Q4 Screen [27,58]. (b) Heatmaps of drug sensitivity (log2 fold change) in eight CRC cell lines to validate the predictions using the PRISM Repurposing Secondary 19Q4 Screen [27,58]. Columns (cells) and rows (drugs) were clustered based on the Euclidean distance of the log2 fold change values. The complete heatmap on the right has 280 rows intervals; however, only the first top 74 (selection indicated by red dashed line), with the corresponding drug dose, are shown on the left for visualization purposes. According to the color bars, drug dose increases from light (0.001 µM) to dark red (10.0 µM).
Author Contributions
R.F.C., T.F.H., L.H.J. and S.R.R. developed the research concept. R.F.C. designed the analysis, workflow and collected the data. R.F.C., S.S.C. and L.M.d.C. analyzed the data and prepared the figures. R.F.C. wrote the manuscript with significant contributions of all authors. S.R.R. supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Region of Southern Denmark Research Fund (Efond. 519, J.nr. 20/14277 to SRR), Denmark, The Danish Colorectal Cancer Center (to SRR, TFN, and LHJ), Denmark, the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001 (CAPES-PrInt-UNESP project; to RFC) and Santander Universidades (Banco Santander, Brasil, S/A; to RFC).
Institutional Review Board Statement
This study was a retrospective analysis of existing publicly available datasets, and therefore, ethics approval is not required.
Informed Consent Statement
Patient consent was waived because this retrospective study was based on existing publicly available deindentified datasets.
Data Availability Statement
The datasets reanalyzed during the current study are available in the GEO (https://www.ncbi.nlm.nih.gov/geo/; GSE90627, GSE8721145, GSE6820446, GSE6033147, and GSE2084248), TCGA (https://portal.gdc.cancer.gov/; TCGA-READ), LINCS (https://lincsproject.org/), and DepMap (https://depmap.org/) repositories. The data generated during this study are included in this published article and its supplementary information files.
Acknowledgments
The results shown here are in part based upon data generated by the Genotype-Tissue Expression project (GTEx) (https://gtexportal.org/) and by the TCGA Research Network (http://cancergenome.nih.gov/). The GTEX data were obtained from OCTAD (http://octad.org/) on 23 March 2021. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. OCTAD also uses compound-induced gene expression profiles from cancer cell lines using data downloaded from the LINCS [28,33,34] Consortium, an NIH Common Fund program. The authors would also like to thank researchers at the Broad Institute who released or made data available to the public through The Cancer Dependency Map (DepMap; https://depmap.org/). DepMap is partially funded by Cancer Target Discovery and Development (CTD2), the Achilles consortium, and The Carlos Slim Foundation in Mexico through the Slim Initiative for Genomic Medicine.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.G.; Rudolph, J.; Bailey, D. Phenotypic Screening in Cancer Drug Discovery—Past, Present and Future. Nat. Rev. Drug Discov. 2014, 13, 588–602. [Google Scholar] [CrossRef] [PubMed]
- Eder, J.; Sedrani, R.; Wiesmann, C. The Discovery of First-in-Class Drugs: Origins and Evolution. Nat. Rev. Drug Discov. 2014, 13, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Munos, B. Lessons from 60 Years of Pharmaceutical Innovation. Nat. Rev. Drug Discov. 2009, 8, 959–968. [Google Scholar] [CrossRef]
- Paul, S.M.; Mytelka, D.S.; Dunwiddie, C.T.; Persinger, C.C.; Munos, B.H.; Lindborg, S.R.; Schacht, A.L. How to Improve R&D Productivity: The Pharmaceutical Industry’s Grand Challenge. Nat. Rev. Drug Discov. 2010, 9, 203–214. [Google Scholar] [CrossRef]
- Petsko, G.A. When Failure Should Be the Option. BMC Biol. 2010, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zheng, S.; Chen, B.; Butte, A.J.; Swamidass, S.J.; Lu, Z. A Survey of Current Trends in Computational Drug Repositioning. Brief. Bioinform. 2016, 17, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug Repositioning: Identifying and Developing New Uses for Existing Drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, L.; Xie, N.; Nice, E.C.; Zhang, T.; Cui, Y.; Huang, C. Overcoming Cancer Therapeutic Bottleneck by Drug Repurposing. Signal Transduct. Target. Ther. 2020, 5, 113. [Google Scholar] [CrossRef]
- Mottini, C.; Napolitano, F.; Li, Z.; Gao, X.; Cardone, L. Computer-Aided Drug Repurposing for Cancer Therapy: Approaches and Opportunities to Challenge Anticancer Targets. Semin. Cancer Biol. 2021, 68, 59–74. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dozois, E.J.; Boardman, L.A.; Suwanthanma, W.; Limburg, P.J.; Cima, R.R.; Bakken, J.L.; Vierkant, R.A.; Aakre, J.A.; Larson, D.W. Young-Onset Colorectal Cancer in Patients with No Known Genetic Predisposition: Can We Increase Early Recognition and Improve Outcome? Medicine 2008, 87, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Vuik, F.E.; Nieuwenburg, S.A.; Bardou, M.; Lansdorp-Vogelaar, I.; Dinis-Ribeiro, M.; Bento, M.J.; Zadnik, V.; Pellisé, M.; Esteban, L.; Kaminski, M.F.; et al. Increasing Incidence of Colorectal Cancer in Young Adults in Europe over the Last 25 Years. Gut 2019, 68, 1820–1826. [Google Scholar] [CrossRef] [PubMed]
- Keller, D.S.; Berho, M.; Perez, R.O.; Wexner, S.D.; Chand, M. The Multidisciplinary Management of Rectal Cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 414–429. [Google Scholar] [CrossRef] [PubMed]
- Glynne-Jones, R.; Wyrwicz, L.; Tiret, E.; Brown, G.; Rödel, C.; Cervantes, A.; Arnold, D.; ESMO Guidelines Committee. Rectal Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, iv22–iv40. [Google Scholar] [CrossRef]
- Deschner, B.W.; VanderWalde, N.A.; Grothey, A.; Shibata, D. Evolution and Current Status of the Multidisciplinary Management of Locally Advanced Rectal Cancer. JCO Oncol. Pract. 2021, 17, 383–402. [Google Scholar] [CrossRef]
- Marsh, P.J.; James, R.D.; Schofield, P.F. Adjuvant Preoperative Radiotherapy for Locally Advanced Rectal Carcinoma: Results of a Prospective, Randomized Trial. Dis. Colon Rectum 1994, 37, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.J. Simulating Watch and Wait for Rectal Cancer. Dis. Colon Rectum 2015, 58, 155–156. [Google Scholar] [CrossRef] [Green Version]
- Habr-Gama, A.; São Julião, G.P.; Perez, R.O. Nonoperative Management of Rectal Cancer. Hematol. Oncol. Clin. N. Am. 2015, 29, 135–151. [Google Scholar] [CrossRef]
- Bruheim, K.; Guren, M.G.; Skovlund, E.; Hjermstad, M.J.; Dahl, O.; Frykholm, G.; Carlsen, E.; Tveit, K.M. Late Side Effects and Quality of Life After Radiotherapy for Rectal Cancer. Int. J. Radiat. Oncol. 2010, 76, 1005–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmertsen, K.J.; Laurberg, S. Bowel Dysfunction after Treatment for Rectal Cancer. Acta Oncol. 2008, 47, 994–1003. [Google Scholar] [CrossRef]
- Zorcolo, L.; Rosman, A.S.; Restivo, A.; Pisano, M.; Nigri, G.R.; Fancellu, A.; Melis, M. Complete Pathologic Response after Combined Modality Treatment for Rectal Cancer and Long-Term Survival: A Meta-Analysis. Ann. Surg. Oncol. 2012, 19, 2822–2832. [Google Scholar] [CrossRef]
- Park, I.J.; You, Y.N.; Agarwal, A.; Skibber, J.M.; Rodriguez-Bigas, M.A.; Eng, C.; Feig, B.W.; Das, P.; Krishnan, S.; Crane, C.H.; et al. Neoadjuvant Treatment Response As an Early Response Indicator for Patients With Rectal Cancer. J. Clin. Oncol. 2012, 30, 1770–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chand, M.; Siddiqui, M.R.S.; Swift, I.; Brown, G. Systematic Review of Prognostic Importance of Extramural Venous Invasion in Rectal Cancer. World J. Gastroenterol. 2016, 22, 1721–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Geest, L.G.M.; Lam-Boer, J.; Koopman, M.; Verhoef, C.; Elferink, M.A.G.; De Wilt, J.H.W. Nationwide Trends in Incidence, Treatment and Survival of Colorectal Cancer Patients with Synchronous Metastases. Clin. Exp. Metastasis 2015, 32, 457–465. [Google Scholar] [CrossRef]
- Corsello, S.M.; Nagari, R.T.; Spangler, R.D.; Rossen, J.; Kocak, M.; Bryan, J.G.; Humeidi, R.; Peck, D.; Wu, X.; Tang, A.A.; et al. Discovering the Anticancer Potential of Non-Oncology Drugs by Systematic Viability Profiling. Nat. Cancer 2020, 1, 235–248. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Wang, Z.; Lachmann, A.; Ma’ayan, A. Mining Data and Metadata from the Gene Expression Omnibus. Biophys. Rev. 2019, 11, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M.; The Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modelling of Anticancer Drug Sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-Generation Characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Lamb, J. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [Green Version]
- Corsello, S.M.; Bittker, J.A.; Liu, Z.; Gould, J.; McCarren, P.; Hirschman, J.E.; Johnston, S.E.; Vrcic, A.; Wong, B.; Khan, M.; et al. The Drug Repurposing Hub: A next-Generation Drug Library and Information Resource. Nat. Med. 2017, 23, 405–408. [Google Scholar] [CrossRef] [Green Version]
- Sirota, M.; Dudley, J.T.; Kim, J.; Chiang, A.P.; Morgan, A.A.; Sweet-Cordero, A.; Sage, J.; Butte, A.J. Discovery and Preclinical Validation of Drug Indications Using Compendia of Public Gene Expression Data. Sci. Transl. Med. 2011, 3, 96ra77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahchan, N.S.; Dudley, J.T.; Mazur, P.K.; Flores, N.; Yang, D.; Palmerton, A.; Zmoos, A.-F.; Vaka, D.; Tran, K.Q.T.; Zhou, M.; et al. A Drug Repositioning Approach Identifies Tricyclic Antidepressants as Inhibitors of Small Cell Lung Cancer and Other Neuroendocrine Tumors. Cancer Discov. 2013, 3, 1364–1377. [Google Scholar] [CrossRef] [Green Version]
- Van Noort, V.; Schölch, S.; Iskar, M.; Zeller, G.; Ostertag, K.; Schweitzer, C.; Werner, K.; Weitz, J.; Koch, M.; Bork, P. Novel Drug Candidates for the Treatment of Metastatic Colorectal Cancer through Global Inverse Gene-Expression Profiling. Cancer Res. 2014, 74, 5690–5699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Wei, W.; Ma, L.; Yang, B.; Gill, R.M.; Chua, M.-S.; Butte, A.J.; So, S. Computational Discovery of Niclosamide Ethanolamine, a Repurposed Drug Candidate That Reduces Growth of Hepatocellular Carcinoma Cells In Vitro and in Mice by Inhibiting Cell Division Cycle 37 Signaling. Gastroenterology 2017, 152, 2022–2036. [Google Scholar] [CrossRef]
- Pessetto, Z.Y.; Chen, B.; Alturkmani, H.; Hyter, S.; Flynn, C.A.; Baltezor, M.; Ma, Y.; Rosenthal, H.G.; Neville, K.A.; Weir, S.J.; et al. In Silico and in Vitro Drug Screening Identifies New Therapeutic Approaches for Ewing Sarcoma. Oncotarget 2017, 8, 4079–4095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirza, A.N.; Fry, M.A.; Urman, N.M.; Atwood, S.X.; Roffey, J.; Ott, G.R.; Chen, B.; Lee, A.; Brown, A.S.; Aasi, S.Z.; et al. Combined Inhibition of Atypical PKC and Histone Deacetylase 1 Is Cooperative in Basal Cell Carcinoma Treatment. JCI Insight 2017, 2, e97071. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, L.F.; Bhasin, M.K.; De Vasconcellos, J.F.; Paccez, J.D.; Gu, X.; Kung, A.L.; Libermann, T.A. Computational Repositioning and Preclinical Validation of Pentamidine for Renal Cell Cancer. Mol. Cancer Ther. 2014, 13, 1929–1941. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Ma, L.; Paik, H.; Sirota, M.; Wei, W.; Chua, M.-S.; So, S.; Butte, A.J. Reversal of Cancer Gene Expression Correlates with Drug Efficacy and Reveals Therapeutic Targets. Nat. Commun. 2017, 8, 16022. [Google Scholar] [CrossRef]
- Hanash, S. Integrated Global Profiling of Cancer. Nat. Rev. Cancer 2004, 4, 638–644. [Google Scholar] [CrossRef]
- Gundersen, G.W.; Jagodnik, K.M.; Woodland, H.; Fernandez, N.F.; Sani, K.; Dohlman, A.B.; Ung, P.M.-U.; Monteiro, C.D.; Schlessinger, A.; Ma’ayan, A. GEN3VA: Aggregation and Analysis of Gene Expression Signatures from Related Studies. BMC Bioinform. 2016, 17, 461. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Statistics for Biology and Health; Gentleman, R., Carey, V.J., Huber, W., Irizarry, R.A., Dudoit, S., Eds.; Springer: New York, NY, USA, 2005; pp. 397–420. ISBN 978-0-387-25146-2. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinforma. Oxf. Engl. 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, B.; Glicksberg, B.S.; Newbury, P.; Chekalin, E.; Xing, J.; Liu, K.; Wen, A.; Chow, C.; Chen, B. OCTAD: An Open Workspace for Virtually Screening Therapeutics Targeting Precise Cancer Patient Groups Using Gene Expression Features. Nat. Protoc. 2021, 16, 728–753. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.Z.D.; Glicksberg, B.S.; Li, Y.; Chen, B. Selecting Precise Reference Normal Tissue Samples for Cancer Research Using a Deep Learning Approach. BMC Med. Genom. 2019, 12, 21. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An Automated Method for Finding Molecular Complexes in Large Protein Interaction Networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Shimada, K.; Bachman, J.A.; Muhlich, J.L.; Mitchison, T.J. ShinyDepMap, a Tool to Identify Targetable Cancer Genes and Their Functional Connections from Cancer Dependency Map Data. eLife 2021, 10, e57116. [Google Scholar] [CrossRef]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e16. [Google Scholar] [CrossRef] [Green Version]
- Depmap Release: Broad, D. Public_21q1. 2021. Available online: https://doi.org/10.6084/m9.figshare.13681534.v1 (accessed on 11 June 2021).
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational Correction of Copy Number Effect Improves Specificity of CRISPR–Cas9 Essentiality Screens in Cancer Cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [Green Version]
- Dempster, J.M.; Rossen, J.; Kazachkova, M.; Pan, J.; Kugener, G.; Root, D.E.; Tsherniak, A. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. BioRxiv 2019, 720243. [Google Scholar] [CrossRef]
- Warren, A.; Chen, Y.; Jones, A.; Shibue, T.; Hahn, W.C.; Boehm, J.S.; Vazquez, F.; Tsherniak, A.; McFarland, J.M. Global Computational Alignment of Tumor and Cell Line Transcriptional Profiles. Nat. Commun. 2021, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Chen, B.; Aran, D.; Charalel, J.; Yau, C.; Wolf, D.M.; Van ’t Veer, L.J.; Butte, A.J.; Goldstein, T.; Sirota, M. Comprehensive Transcriptomic Analysis of Cell Lines as Models of Primary Tumors across 22 Tumor Types. Nat. Commun. 2019, 10, 3574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- do Canto, L.M.; Larsen, S.J.; Catin Kupper, B.E.; Begnami, M.D.F.S.; Scapulatempo-Neto, C.; Petersen, A.H.; Aagaard, M.M.; Baumbach, J.; Aguiar, S.; Rogatto, S.R. Increased Levels of Genomic Instability and Mutations in Homologous Recombination Genes in Locally Advanced Rectal Carcinomas. Front. Oncol. 2019, 9, 395. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Mathelier, A. Intervene: A Tool for Intersection and Visualization of Multiple Gene or Genomic Region Sets. BMC Bioinform. 2017, 18, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, D.J.B.; Kuleshov, M.V.; Schilder, B.M.; Torre, D.; Duffy, M.E.; Keenan, A.B.; Lachmann, A.; Feldmann, A.S.; Gundersen, G.W.; Silverstein, M.C.; et al. EXpression2Kinases (X2K) Web: Linking Expression Signatures to Upstream Cell Signaling Networks. Nucleic Acids Res. 2018, 46, W171–W179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starruß, J.; de Back, W.; Brusch, L.; Deutsch, A. Morpheus: A User-Friendly Modeling Environment for Multiscale and Multicellular Systems Biology. Bioinformatics 2014, 30, 1331–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR Is a Web App for Creating, Exploring, Labeling and Sharing Volcano Plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zeng, W.; Feng, L.; Yu, X.; Li, P.; Zhang, K.; Zhou, Z.; Cheng, S. Integrated Transcriptomic Analysis of Distance-Related Field Cancerization in Rectal Cancer Patients. Oncotarget 2017, 8, 61107–61117. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Gaedcke, J.; Emons, G.; Beissbarth, T.; Grade, M.; Jo, P.; Yeager, M.; Chanock, S.J.; Wolff, H.; Camps, J.; et al. Colorectal Cancer Susceptibility Loci as Predictive Markers of Rectal Cancer Prognosis after Surgery. Genes Chromosomes Cancer 2018, 57, 140–149. [Google Scholar] [CrossRef]
- Millino, C.; Maretto, I.; Pacchioni, B.; Digito, M.; De Paoli, A.; Canzonieri, V.; D’Angelo, E.; Agostini, M.; Rizzolio, F.; Giordano, A.; et al. Gene and MicroRNA Expression Are Predictive of Tumor Response in Rectal Adenocarcinoma Patients Treated With Preoperative Chemoradiotherapy: Combined Mirnas and Gene Expression. J. Cell. Physiol. 2017, 232, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Verstraete, M.; Debucquoy, A.; Dekervel, J.; Van Pelt, J.; Verslype, C.; Devos, E.; Chiritescu, G.; Dumon, K.; D’Hoore, A.; Gevaert, O.; et al. Combining Bevacizumab and Chemoradiation in Rectal Cancer. Translational Results of the AXEBeam Trial. Br. J. Cancer 2015, 112, 1314–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaedcke, J.; Grade, M.; Jung, K.; Camps, J.; Jo, P.; Emons, G.; Gehoff, A.; Sax, U.; Schirmer, M.; Becker, H.; et al. Mutated KRAS Results in Overexpression of DUSP4, a MAP-Kinase Phosphatase, and SMYD3, a Histone Methyltransferase, in Rectal Carcinomas. Genes Chromosomes Cancer 2010, 49, 1024–1034. [Google Scholar] [CrossRef] [Green Version]
- Benson, J.D.; Chen, Y.-N.P.; Cornell-Kennon, S.A.; Dorsch, M.; Kim, S.; Leszczyniecka, M.; Sellers, W.R.; Lengauer, C. Validating Cancer Drug Targets. Nature 2006, 441, 451–456. [Google Scholar] [CrossRef]
- Lapenna, S.; Giordano, A. Cell Cycle Kinases as Therapeutic Targets for Cancer. Nat. Rev. Drug Discov. 2009, 8, 547–566. [Google Scholar] [CrossRef]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive Mining of Publicly Available RNA-Seq Data from Human and Mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganesh, K.; Wu, C.; O’Rourke, K.P.; Szeglin, B.C.; Zheng, Y.; Sauvé, C.-E.G.; Adileh, M.; Wasserman, I.; Marco, M.R.; Kim, A.S.; et al. A Rectal Cancer Organoid Platform to Study Individual Responses to Chemoradiation. Nat. Med. 2019, 25, 1607–1614. [Google Scholar] [CrossRef]
- Hevener, E.; Verstak, T.A.; Lutat, K.E.; Riggsbee, D.L.; Mooney, J.W. Recent Developments in Topoisomerase-Targeted Cancer Chemotherapy. Acta Pharm. Sin. B 2018, 8, 844–861. [Google Scholar] [CrossRef]
- Schettini, F.; De Santo, I.; Rea, C.G.; De Placido, P.; Formisano, L.; Giuliano, M.; Arpino, G.; De Laurentiis, M.; Puglisi, F.; De Placido, S.; et al. CDK 4/6 Inhibitors as Single Agent in Advanced Solid Tumors. Front. Oncol. 2018, 8, 608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, D.C.; Chalmers, A.J.; El-Khamisy, S.F. Topoisomerase I Inhibition in Colorectal Cancer: Biomarkers and Therapeutic Targets. Br. J. Cancer 2012, 106, 18–24. [Google Scholar] [CrossRef] [Green Version]
- Willeke, F.; Horisberger, K.; Kraus-Tiefenbacher, U.; Wenz, F.; Leitner, A.; Hochhaus, A.; Grobholz, R.; Willer, A.; Kähler, G.; Post, S.; et al. A Phase II Study of Capecitabine and Irinotecan in Combination with Concurrent Pelvic Radiotherapy (CapIri-RT) as Neoadjuvant Treatment of Locally Advanced Rectal Cancer. Br. J. Cancer 2007, 96, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Glynne-Jones, R.; Falk, S.; Maughan, T.S.; Meadows, H.M.; Sebag-Montefiore, D. A Phase I/II Study of Irinotecan When Added to 5-Fluorouracil and Leucovorin and Pelvic Radiation in Locally Advanced Rectal Cancer: A Colorectal Clinical Oncology Group Study. Br. J. Cancer 2007, 96, 551–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gollins, S.; Sun Myint, A.; Haylock, B.; Wise, M.; Saunders, M.; Neupane, R.; Essapen, S.; Samuel, L.; Dougal, M.; Lloyd, A.; et al. Preoperative Chemoradiotherapy Using Concurrent Capecitabine and Irinotecan in Magnetic Resonance Imaging–Defined Locally Advanced Rectal Cancer: Impact on Long-Term Clinical Outcomes. J. Clin. Oncol. 2011, 29, 1042–1049. [Google Scholar] [CrossRef]
- Wang, J.; Fan, J.; Li, C.; Yang, L.; Wan, J.; Zhang, H.; Zhang, Z.; Zhu, J. The Impact of Chemotherapy Completion on the Efficacy of Irinotecan in the Preoperative Chemoradiotherapy of Locally Advanced Rectal Cancer: An Expanded Analysis of the CinClare Phase III Trial. Clin. Colorectal Cancer 2020, 19, e58–e69. [Google Scholar] [CrossRef]
- Kolarić, K.; Potrebica, V.; Stanovnik, M. Controlled Phase III Clinical Study of 4-Epi-Doxorubicin + 5-Fluorouracil versus 5-Fluorouracil Alone in Metastatic Gastric and Rectosigmoid Cancer. Oncology 1986, 43, 73–77. [Google Scholar] [CrossRef]
- Ferrazzi, E.; Pappagallo, G.L.; Nicoletto, O.; Fornasiero, A.; Refatti, F.; Cartei, G.; Vinante, O.; Fiorentino, M.V. Phase II Evaluation of 4’epi-Doxorubicin in Patients with Metastatic Colorectal Carcinoma. Tumori 1984, 70, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Leo, A.D.; Desmedt, C.; Bartlett, J.M.S.; Piette, F.; Ejlertsen, B.; Pritchard, K.I.; Larsimont, D.; Poole, C.; Isola, J.; Earl, H.; et al. HER2 and TOP2A as Predictive Markers for Anthracycline-Containing Chemotherapy Regimens as Adjuvant Treatment of Breast Cancer: A Meta-Analysis of Individual Patient Data. Lancet Oncol. 2011, 12, 1134–1142. [Google Scholar] [CrossRef]
- Du, Y.; Zhou, Q.; Yin, W.; Zhou, L.; Di, G.; Shen, Z.; Shao, Z.; Lu, J. The Role of Topoisomerase IIα in Predicting Sensitivity to Anthracyclines in Breast Cancer Patients: A Meta-Analysis of Published Literatures. Breast Cancer Res. Treat. 2011, 129, 839–848. [Google Scholar] [CrossRef]
- Al-Kuraya, K.; Novotny, H.; Bavi, P.; Siraj, A.K.; Uddin, S.; Ezzat, A.; Sanea, N.A.; Al-Dayel, F.; Al-Mana, H.; Sheikh, S.S.; et al. HER2, TOP2A, CCND1, EGFR and C-MYC Oncogene Amplification in Colorectal Cancer. J. Clin. Pathol. 2006, 60, 768–772. [Google Scholar] [CrossRef]
- Sønderstrup, I.M.H.; Nygård, S.B.; Poulsen, T.S.; Linnemann, D.; Stenvang, J.; Nielsen, H.J.; Bartek, J.; Brünner, N.; Nørgaard, P.; Riis, L. Topoisomerase-1 and -2A Gene Copy Numbers Are Elevated in Mismatch Repair-Proficient Colorectal Cancers. Mol. Oncol. 2015, 9, 1207–1217. [Google Scholar] [CrossRef]
- Nygård, S.B.; Christensen, I.J.; Smith, D.H.; Nielsen, S.L.; Jensen, N.F.; Nielsen, H.J.; Vainer, B.; Brünner, N. Underpinning the Repurposing of Anthracyclines towards Colorectal Cancer: Assessment of Topoisomerase II Alpha Gene Copy Number Alterations in Colorectal Cancer. Scand. J. Gastroenterol. 2013, 48, 1436–1443. [Google Scholar] [CrossRef]
- Coss, A.; Tosetto, M.; Fox, E.J.; Sapetto-Rebow, B.; Gorman, S.; Kennedy, B.N.; Lloyd, A.T.; Hyland, J.M.; O’Donoghue, D.P.; Sheahan, K.; et al. Increased Topoisomerase IIα Expression in Colorectal Cancer Is Associated with Advanced Disease and Chemotherapeutic Resistance via Inhibition of Apoptosis. Cancer Lett. 2009, 276, 228–238. [Google Scholar] [CrossRef]
- Tarpgaard, L.S.; Qvortrup, C.; Nielsen, S.L.; Stenvang, J.; Detlefsen, S.; Brünner, N.; Pfeiffer, P. New Use for Old Drugs: Epirubicin in Colorectal Cancer. Acta Oncol. 2021, 60, 954–956. [Google Scholar] [CrossRef] [PubMed]
- Karnitz, L.M.; Zou, L. Molecular Pathways: Targeting ATR in Cancer Therapy. Clin. Cancer Res. 2015, 21, 4780–4785. [Google Scholar] [CrossRef] [Green Version]
- Kent, L.N.; Leone, G. The Broken Cycle: E2F Dysfunction in Cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, A.; Aghi, M.K.; Carbonell, W.S. Β1 Integrin: Critical Path to Antiangiogenic Therapy Resistance and Beyond. Cancer Res. 2014, 74, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Stewart, M.D.; Sanderson, R.D. Heparan Sulfate in the Nucleus and Its Control of Cellular Functions. Matrix Biol. 2014, 35, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Girdler, F.; Gascoigne, K.E.; Eyers, P.A.; Hartmuth, S.; Crafter, C.; Foote, K.M.; Keen, N.J.; Taylor, S.S. Validating Aurora B as an Anti-Cancer Drug Target. J. Cell Sci. 2006, 119, 3664–3675. [Google Scholar] [CrossRef] [Green Version]
- Pohl, A.; Azuma, M.; Zhang, W.; Yang, D.; Ning, Y.; Winder, T.; Danenberg, K.; Lenz, H.-J. Pharmacogenetic Profiling of Aurora Kinase B Is Associated with Overall Survival in Metastatic Colorectal Cancer. Pharm. J. 2011, 11, 93–99. [Google Scholar] [CrossRef] [Green Version]
- Pillaire, M.-J.; Selves, J.; Gordien, K.; Gouraud, P.-A.; Gentil, C.; Danjoux, M.; Do, C.; Negre, V.; Bieth, A.; Guimbaud, R.; et al. A ‘DNA Replication’ Signature of Progression and Negative Outcome in Colorectal Cancer. Oncogene 2010, 29, 876–887. [Google Scholar] [CrossRef]
- Solier, S.; Zhang, Y.-W.; Ballestrero, A.; Pommier, Y.; Zoppoli, G. DNA Damage Response Pathways and Cell Cycle Checkpoints in Colorectal Cancer: Current Concepts and Future Perspectives for Targeted Treatment. Curr. Cancer Drug Targets 2012, 12, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Miyoshi, N.; Ishii, H.; Mimori, K.; Tanaka, F.; Sekimoto, M.; Doki, Y.; Mori, M. Expression of CLDN1 in Colorectal Cancer: A Novel Marker for Prognosis. Int. J. Oncol. 2011, 39, 791–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, K.; Hirata, S.; Irie, A.; Senju, S.; Ikuta, Y.; Yokomine, K.; Harao, M.; Inoue, M.; Tsunoda, T.; Nakatsuru, S.; et al. Identification of a Novel Tumor-Associated Antigen, Cadherin 3/P-Cadherin, as a Possible Target for Immunotherapy of Pancreatic, Gastric, and Colorectal Cancers. Clin. Cancer Res. 2008, 14, 6487–6495. [Google Scholar] [CrossRef] [Green Version]
- Kaneda, H.; Arao, T.; Tanaka, K.; Tamura, D.; Aomatsu, K.; Kudo, K.; Sakai, K.; De Velasco, M.A.; Matsumoto, K.; Fujita, Y.; et al. FOXQ1 Is Overexpressed in Colorectal Cancer and Enhances Tumorigenicity and Tumor Growth. Cancer Res. 2010, 70, 2053–2063. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Fan, X.; Chen, J.; Xie, X.; Yu, H. Small Interfering RNA-mediated Knockdown of KRT80 Suppresses Colorectal Cancer Proliferation. Exp. Ther. Med. 2020, 20, 176. [Google Scholar] [CrossRef] [PubMed]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-Clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- LeSavage, B.L.; Suhar, R.A.; Broguiere, N.; Lutolf, M.P.; Heilshorn, S.C. Next-Generation Cancer Organoids. Nat. Mater. 2021. [Google Scholar] [CrossRef]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of Patient-Derived Cancer Organoids for Drug-Screening Applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).