Detection of Human Cholangiocarcinoma Markers in Serum Using Infrared Spectroscopy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Sera
2.2. ATR-FTIR Spectroscopy for Serum Analysis
2.3. ATR-FTIR Spectral Preprocessing and Analysis
2.4. Method Evaluation and Calculation
3. Results
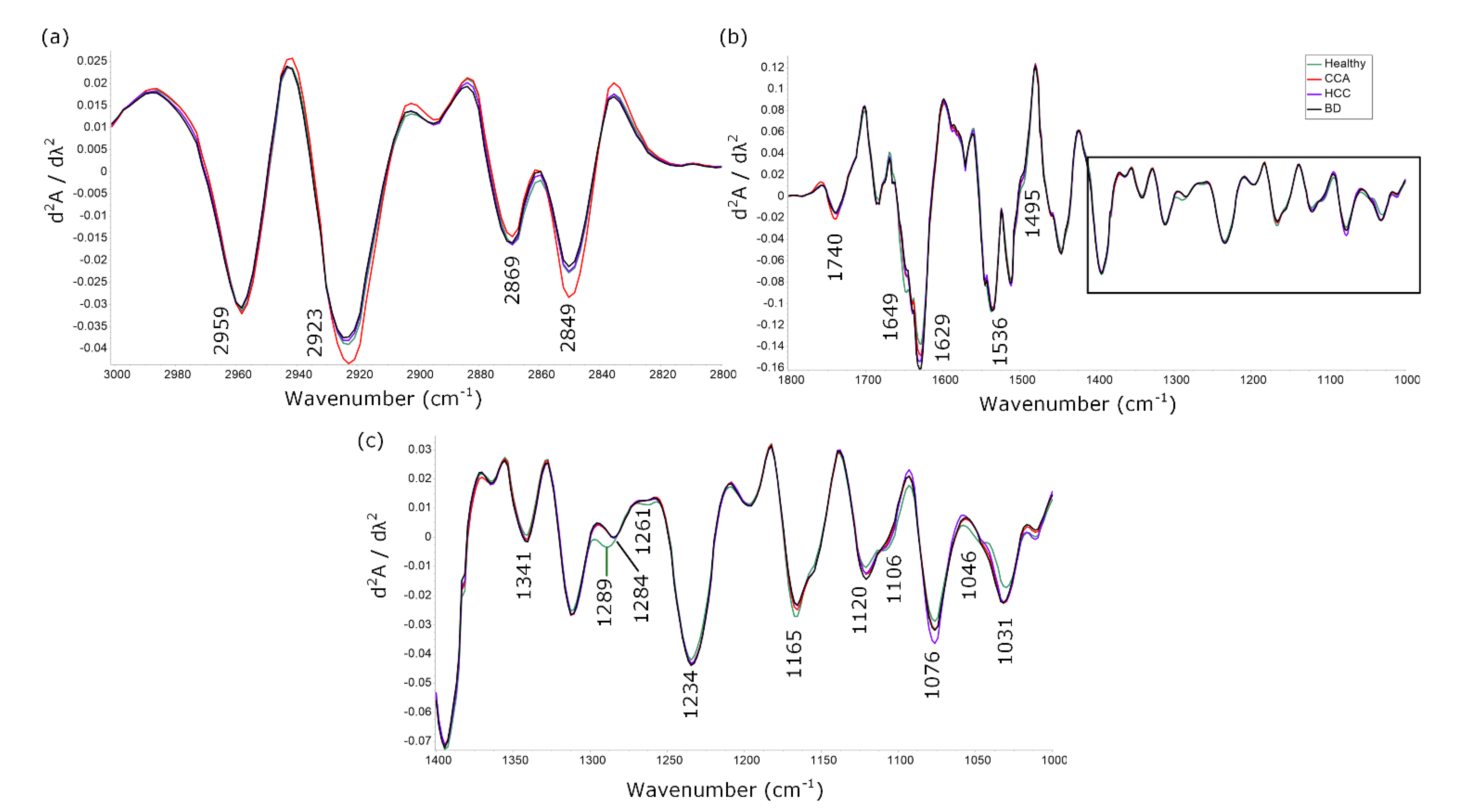
3.1. Characteristic Peaks of Healthy, CCA, HCC and BD Spectra
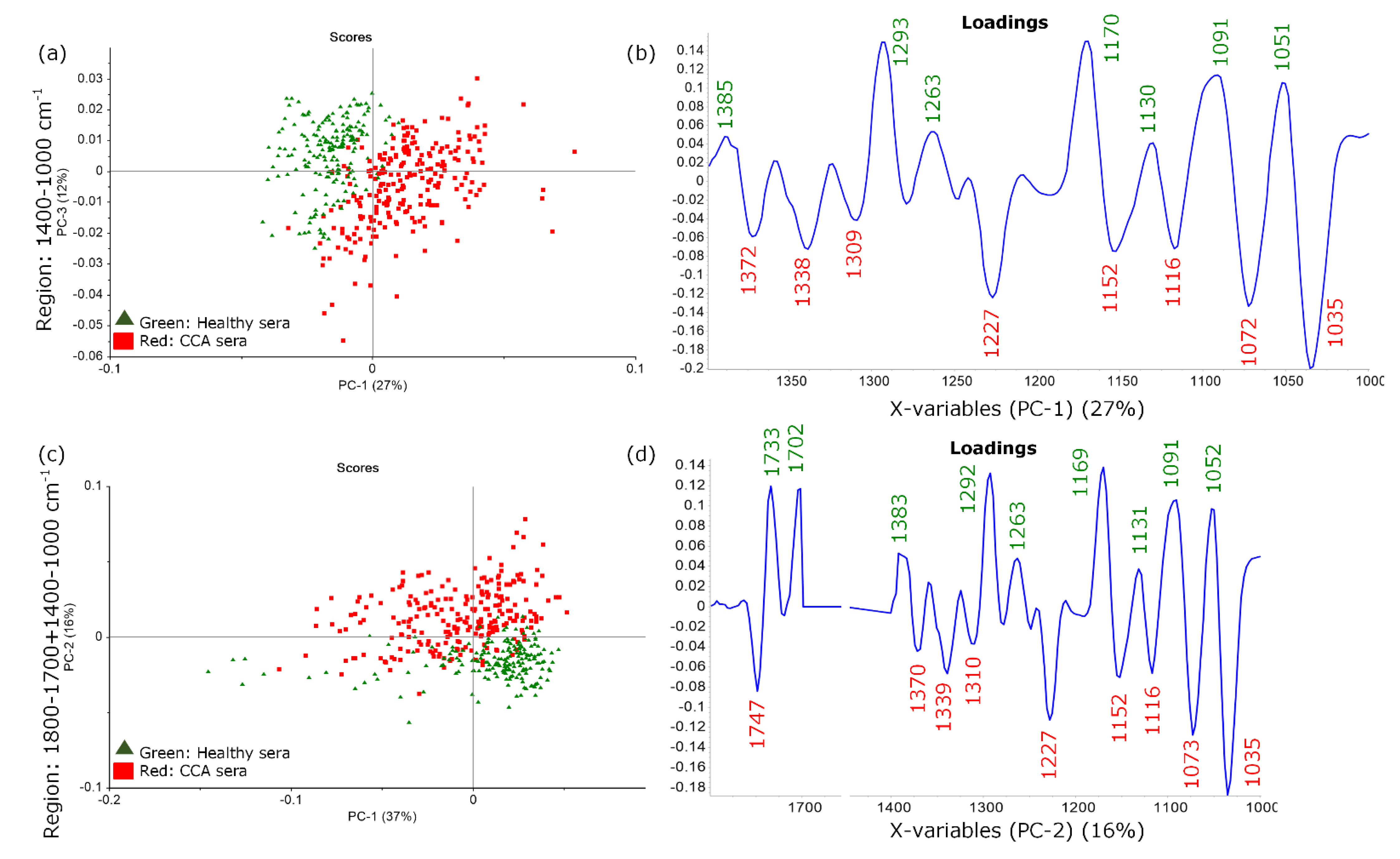
3.2. CCA Spectral Discrimination Using Unsupervised Analysis: Principal Component Analysis (PCA)
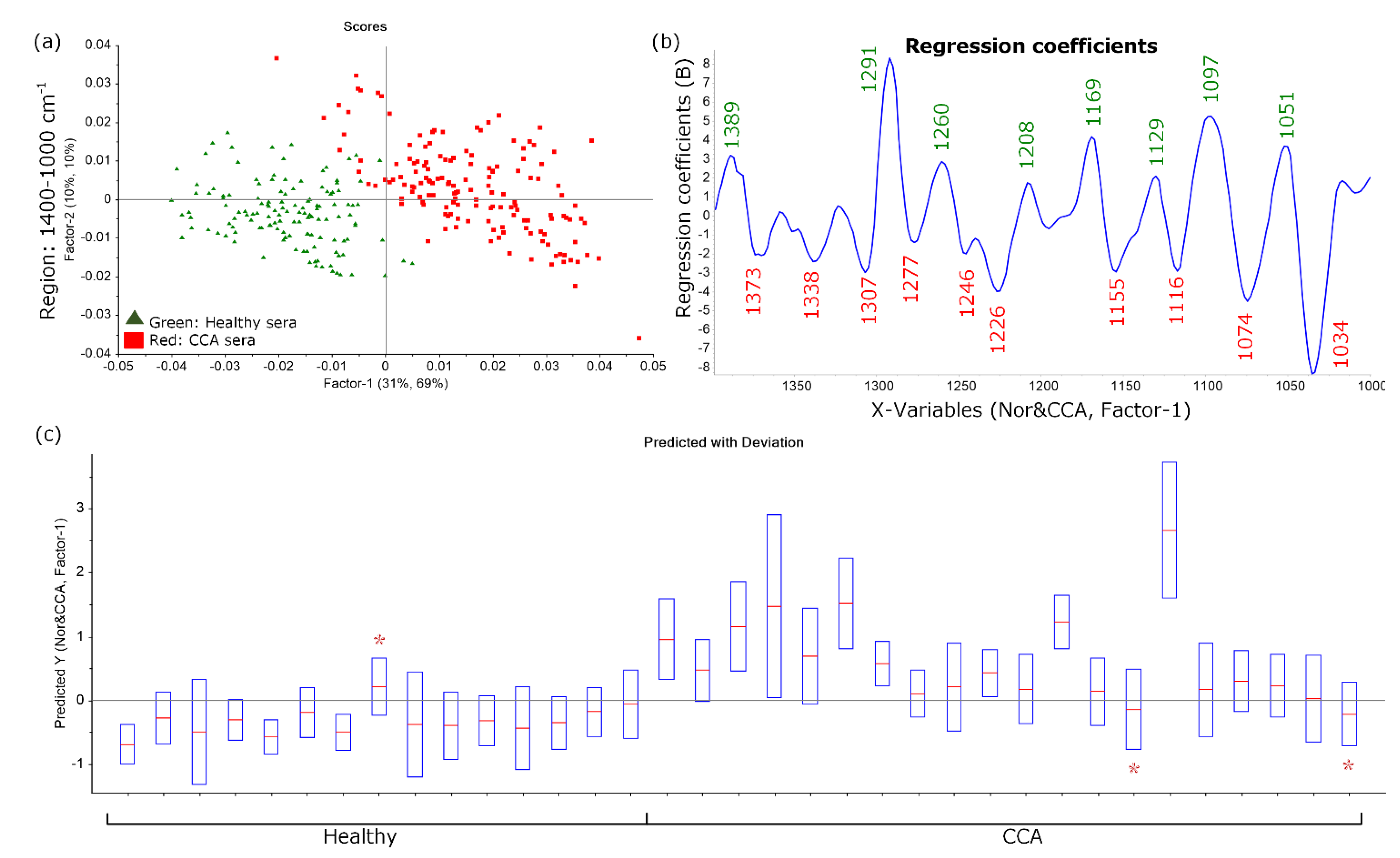
3.3. Establishment and Evaluation of CCA Predictive Model Using Partial Least Squares Discriminant Analysis (PLS-DA)
3.4. Advanced Machine Modelling of CCA Serum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banales, J.M.; Cardinale, V.; Carpino, G.; Marzioni, M.; Andersen, J.B.; Invernizzi, P.; Lind, G.E.; Folseraas, T.; Forbes, S.J.; Fouassier, L.; et al. Expert consensus document: Cholangiocarcinoma: Current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 261–280. [Google Scholar] [CrossRef]
- Alsaleh, M.; Leftley, Z.; Barbera, T.A.; Sithithaworn, P.; Khuntikeo, N.; Loilome, W.; Yongvanit, P.; Cox, I.J.; Chamodol, N.; Syms, R.R.A.; et al. Cholangiocarcinoma: A guide for the nonspecialist. Int. J. Gen. Med. 2019, 12, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Kirstein, M.M.; Vogel, A. Epidemiology and Risk Factors of Cholangiocarcinoma. Visc. Med. 2016, 32, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Tavolari, S.; Brandi, G. Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 2019, 39, 19–31. [Google Scholar] [CrossRef]
- Sripa, B.; Kaewkes, S.; Sithithaworn, P.; Mairiang, E.; Laha, T.; Smout, M.; Pairojkul, C.; Bhudhisawasdi, V.; Tesana, S.; Thinkamrop, B.; et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007, 4, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Hennedige, T.P.; Neo, W.T.; Venkatesh, S.K. Imaging of malignancies of the biliary tract- an update. Cancer Imaging 2014, 14, 1–21. [Google Scholar] [CrossRef]
- Zabron, A.; Edwards, R.J.; Khan, S.A. The challenge of cholangiocarcinoma: Dissecting the molecular mechanisms of an insidious cancer. DMM Dis. Model. Mech. 2013, 6, 281–292. [Google Scholar] [CrossRef]
- Zeng, X.; Tao, H. Diagnostic and prognostic serum marker of cholangiocarcinoma (Review). Oncol. Lett. 2015, 9, 3–8. [Google Scholar] [CrossRef]
- Kimawaha, P.; Jusakul, A.; Junsawang, P.; Thanan, R.; Titapun, A.; Khuntikeo, N.; Techasen, A. Establishment of a potential serum biomarker panel for the diagnosis and prognosis of cholangiocarcinoma using decision tree algorithms. Diagnostics 2021, 11, 589. [Google Scholar] [CrossRef]
- Roy, S.; Perez-Guaita, D.; Bowden, S.; Heraud, P.; Wood, B.R. Spectroscopy goes viral: Diagnosis of hepatitis B and C virus infection from human sera using ATR-FTIR spectroscopy. Clin. Spectrosc. 2019, 1, 100001. [Google Scholar] [CrossRef]
- Sala, A.; Anderson, D.J.; Brennan, P.M.; Butler, H.J.; Cameron, J.M.; Jenkinson, M.D.; Rinaldi, C.; Theakstone, A.G.; Baker, M.J. Biofluid diagnostics by FTIR spectroscopy: A platform technology for cancer detection. Cancer Lett. 2020, 477, 122–130. [Google Scholar] [CrossRef]
- Su, K.Y.; Lee, W.L. Fourier transform infrared spectroscopy as a cancer screening and diagnostic tool: A review and prospects. Cancers 2020, 12, 115. [Google Scholar] [CrossRef]
- Berisha, S.; Lotfollahi, M.; Jahanipour, J.; Gurcan, I.; Walsh, M.; Bhargava, R.; van Nguyen, H.; Mayerich, D. Deep learning for FTIR histology: Leveraging spatial and spectral features with convolutional neural networks. Analyst 2019, 144, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Lasch, P.; Stämmler, M.; Zhang, M.; Baranska, M.; Bosch, A.; Majzner, K. FT-IR Hyperspectral Imaging and Artificial Neural Network Analysis for Identification of Pathogenic Bacteria. Anal. Chem. 2018, 90, 8896–8904. [Google Scholar] [CrossRef] [PubMed]
- Backhaus, J.; Mueller, R.; Formanski, N.; Szlama, N.; Meerpohl, H.G.; Eidt, M.; Bugert, P. Diagnosis of breast cancer with infrared spectroscopy from serum samples. Vib. Spectrosc. 2010, 52, 173–177. [Google Scholar] [CrossRef]
- Toraman, S.; Girgin, M.; Üstündağ, B.; Türkoğlu, İ. Classification of the likelihood of colon cancer with machine learning techniques using FTIR signals obtained from plasma. Turkish J. Electr. Eng. Comput. Sci. 2019, 27, 1765–1779. [Google Scholar] [CrossRef]
- Butler, H.J.; Brennan, P.M.; Cameron, J.M.; Finlayson, D.; Hegarty, M.G.; Jenkinson, M.D.; Palmer, D.S.; Smith, B.R.; Baker, M.J. Development of high-throughput ATR-FTIR technology for rapid triage of brain cancer. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chatchawal, P.; Wongwattanakul, M.; Tippayawat, P.; Jearanaikoon, N.; Jumniansong, A.; Boonmars, T.; Jearanaikoon, P.; Wood, B.R. Monitoring the Progression of Liver Fluke-Induced Cholangiocarcinoma in a Hamster Model Using Synchrotron FTIR Microspectroscopy and Focal Plane Array Infrared Imaging. Anal. Chem. 2020, 92, 15361–15369. [Google Scholar] [CrossRef]
- Hackshaw, K.V.; Miller, J.S.; Aykas, D.P.; Rodriguez-Saona, L. Vibrational spectroscopy for identification of metabolites in biologic samples. Molecules 2020, 25, 4725. [Google Scholar] [CrossRef] [PubMed]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Elmi, F.; Movaghar, A.F.; Elmi, M.M.; Alinezhad, H.; Nikbakhsh, N. Application of FT-IR spectroscopy on breast cancer serum analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 187, 87–91. [Google Scholar] [CrossRef]
- Krafft, C.; Shapoval, L.; Sobottka, S.B.; Schackert, G.; Salzer, R. Identification of primary tumors of brain metastases by infrared spectroscopic imaging and linear discriminant analysis. Technol. Cancer Res. Treat. 2006, 5, 291–298. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Morais, C.L.M.; Raglan, O.; Lima, K.M.G.; Paraskevaidis, E.; Martin-Hirsch, P.L.; Kyrgiou, M.; Martin, F.L. Aluminium foil as an alternative substrate for the spectroscopic interrogation of endometrial cancer. J. Biophotonics 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Zawadzka, A.M.; Schilling, B.; Cusack, M.P.; Sahu, A.K.; Drake, P.; Fisher, S.J.; Benz, C.C.; Gibson, B.W. Phosphoprotein Secretome of Tumor Cells as a Source of Candidates for Breast Cancer Biomarkers in Plasma. Mol. Cell. Proteom. 2014, 13, 1034. [Google Scholar] [CrossRef]
- Bergquist, J.R.; Ivanics, T.; Storlie, C.B.; Groeschl, R.T.; Tee, M.C.; Habermann, E.B.; Smoot, R.L.; Kendrick, M.L.; Farnell, M.B.; Roberts, L.R.; et al. Implications of CA19-9 elevation for survival, staging, and treatment sequencing in intrahepatic cholangiocarcinoma: A national cohort analysis. J. Surg. Oncol. 2016, 114, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Shen, X.; Lei, J.; Xu, Q.; Yu, Y.; Li, R.; Wu, E.; Ma, Q. Hyperglycemia, a neglected factor during cancer progression. Biomed. Res. Int. 2014. [Google Scholar] [CrossRef]
- Zois, C.E.; Harris, A.L. Glycogen metabolism has a key role in the cancer microenvironment and provides new targets for cancer therapy. J. Mol. Med. 2016, 94, 137–154. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xu, H.; Dai, J.; Peng, J.; Wang, W.; Xia, L.; Zhou, F. Prognostic Significance of Serum Lactic Acid, Lactate Dehydrogenase, and Albumin Levels in Patients with Metastatic Colorectal Cancer. Biomed. Res. Int. 2018, 2018. [Google Scholar] [CrossRef]
- Willumsen, N.; Ali, S.M.; Leitzel, K.; Drabick, J.J.; Yee, N.; Polimera, H.V.; Nagabhairu, V.; Krecko, L.; Ali, A.; Maddukuri, A.; et al. Collagen fragments quantified in serum as measures of desmoplasia associate with survival outcome in patients with advanced pancreatic cancer. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Brassart-Pasco, S.; Brézillon, S.; Brassart, B.; Ramont, L.; Oudart, J.B.; Monboisse, J.C. Tumor Microenvironment: Extracellular Matrix Alterations Influence Tumor Progression. Front. Oncol. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Prakobwong, S.; Charoensuk, L.; Hiraku, Y.; Pinlaor, P.; Pairojkul, C. Plasma hydroxyproline, MMP-7 and collagen I as novel predictive risk markers of hepatobiliary disease-associated cholangiocarcinoma. Int. J. Cancer 2012, 131, 416–424. [Google Scholar] [CrossRef]
- De Bruyne, S.; Speeckaert, M.M.; Delanghe, J.R. Applications of mid-infrared spectroscopy in the clinical laboratory setting. Crit. Rev. Clin. Lab. Sci. 2018, 55, 1–20. [Google Scholar] [CrossRef]
- Holman, H.Y.N.; Martin, M.C.; Blakely, E.A.; Bjornstad, K.; Mckinney, W.R. IR spectroscopic characteristics of cell cycle and cell death probed by synchrotron radiation based Fourier transform IR spectromicroscopy. Biopolym. Biospectroscopy Sect. 2000, 57, 329–335. [Google Scholar] [CrossRef]
- Notarstefano, V.; Sabbatini, S.; Conti, C.; Pisani, M.; Astolfi, P.; Pro, C.; Rubini, C.; Vaccari, L.; Giorgini, E. Investigation of human pancreatic cancer tissues by Fourier Transform Infrared Hyperspectral Imaging. J. Biophotonics 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Stelling, A.L.; Toher, D.; Uckermann, O.; Tavkin, J.; Leipnitz, E.; Schweizer, J.; Cramm, H.; Steiner, G.; Geiger, K.D.; Kirsch, M. Infrared Spectroscopic Studies of Cells and Tissues: Triple Helix Proteins as a Potential Biomarker for Tumors. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef]
- Liu, K.Z.; Dixon, I.M.C.; Mantsch, H.H. Distribution of collagen deposition in cardiomyopathic hamster hearts determined by infrared microscopy. Cardiovasc. Pathol. 1999, 8, 41–47. [Google Scholar] [CrossRef]
- Liu, K.; Man, A.; Shaw, R.A. Molecular determination of liver fibrosis by synchrotron infrared microspectroscopy. Biochim. Biophys. Acta Biomembr. 2006, 1758, 960–967. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barnas, E.; Skret-Magierlo, J.; Skret, A.; Kaznowska, E.; Depciuch, J.; Szmuc, K.; Łach, K.; Krawczyk-Marć, I.; Cebulski, J. Simultaneous FTIR and Raman Spectroscopy in Endometrial Atypical Hyperplasia and Cancer. Int. J. Mol. Sci. 2020, 21, 4828. [Google Scholar] [CrossRef] [PubMed]
- Belbachir, K.; Noreen, R.; Gouspillou, G.; Petibois, C. Collagen types analysis and differentiation by FTIR spectroscopy. Anal. Bioanal. Chem. 2009, 395, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Callery, E.L.; Morais, C.L.M.; Paraskevaidi, M.; Brusic, V.; Vijayadurai, P.; Anantharachagan, A.; Martin, F.L.; Rowbottom, A.W. New approach to investigate Common Variable Immunodeficiency patients using spectrochemical analysis of blood. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Al-Jorani, K.; Rüther, A.; Martin, M.; Haputhanthri, R.; Deacon, G.B.; Li, H.L.; Wood, B.R. The application of ATR-FTIR spectroscopy and the reversible DNA conformation as a sensor to test the effectiveness of platinum(II) anticancer drugs. Sensors 2018, 18, 4297. [Google Scholar] [CrossRef] [PubMed]
- Thumanu, K.; Sangrajrang, S.; Khuhaprema, T.; Kalalak, A.; Tanthanuch, W.; Pongpiachan, S.; Heraud, P. Diagnosis of liver cancer from blood sera using FTIR microspectroscopy: A preliminary study. J. Biophotonics 2014, 7, 222–231. [Google Scholar] [CrossRef]
- Sitnikova, V.E.; Kotkova, M.A.; Nosenko, T.N.; Kotkova, T.N.; Martynova, D.M.; Uspenskaya, M.V. Breast cancer detection by ATR-FTIR spectroscopy of blood serum and multivariate data-analysis. Talanta 2020, 214, 120857. [Google Scholar] [CrossRef] [PubMed]
- Cameron, J.M.; Butler, H.J.; Smith, B.R.; Hegarty, M.G.; Jenkinson, M.D.; Syed, K.; Brennan, P.M.; Ashton, K.; Dawson, T.; Palmer, D.S.; et al. Developing infrared spectroscopic detection for stratifying brain tumour patients: Glioblastoma multiforme: Vs. lymphoma. Analyst 2019, 144, 6736–6750. [Google Scholar] [CrossRef]
- Giannini, E.G.; Testa, R.; Savarino, V. Liver enzyme alteration: A guide for clinicians. Cmaj 2005, 172, 367–379. [Google Scholar] [CrossRef]
- Zhang, X.; Thiéfin, G.; Gobinet, C.; Untereiner, V.; Taleb, I.; Bernard-Chabert, B.; Heurgué, A.; Truntzer, C.; Ducoroy, P.; Hillon, P.; et al. Profiling serologic biomarkers in cirrhotic patients via high-throughput Fourier transform infrared spectroscopy: Toward a new diagnostic tool of hepatocellular carcinoma. Transl. Res. 2013, 162, 279–286. [Google Scholar] [CrossRef]
- Khan, S.A.; Davidson, B.R.; Goldin, R.; Pereira, S.P.; Rosenberg, W.M.C.; Taylor-Robinson, S.D.; Thillainayagam, A.V.; Thomas, H.C.; Thursz, M.R.; Wasan, H.; et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: Consensus document. Gut 2002, 51, 1–9. [Google Scholar] [CrossRef]
- Slattery, J.M.; Sahani, D.V. What Is the Current State-of-the-Art Imaging for Detection and Staging of Cholangiocarcinoma? Oncologist 2006, 11, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Blechacz, B.R.A.; Gores, G.J. Cholangiocarcinoma: Advances in Pathogenesis, Diagnosis, and Treatment. Hepatology 2008, 48, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Breitenstein, S.; Apestegui, C.; Clavien, P.A. Positron emission tomography (PET) for cholangiocarcinoma. Hpb 2008, 10, 120–121. [Google Scholar] [CrossRef] [PubMed]
- Tshering, G.; Dorji, P.W.; Chaijaroenkul, W.; Na-Bangchang, K. Biomarkers for the diagnosis of cholangiocarcinoma: A systematic review. Am. J. Trop. Med. Hyg. 2018, 98, 1788–1797. [Google Scholar] [CrossRef]
- Beers, B.E.V.A.N. Diagnosis of cholangiocarcinoma. HPB 2008, 10, 87–93. [Google Scholar] [CrossRef]
- Tolek, A.; Wongkham, C.; Proungvitaya, S.; Silsirivanit, A.; Roytrakul, S.; Khuntikeo, N.; Wongkham, S. Serum α1β-glycoprotein and afamin ratio as potential diagnostic and prognostic markers in cholangiocarcinoma. Exp. Biol. Med. 2012, 237, 1142–1149. [Google Scholar] [CrossRef] [PubMed]
| Index Test | Clinical Diagnoses | |
|---|---|---|
| (Predictive Model) | CCA | Other Condition |
| CCA | a | b |
| Other condition | c | d |
| Models | Spectral Range | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3000–2800 cm−1 | 1800–1000 cm−1 | 1400–1000 cm−1 | 1800–1700 + 1400–1000 cm−1 | 3000–2800 + 1800–1000 cm−1 | ||||||||||||
| %Acc | %Sen | %Spec | %Acc | %Sen | %Spec | %Acc | %Sen | %Spec | %Acc | %Sen | %Spec | %Acc | %Sen | %Spec | ||
| PLS-DA | Healthy/CCA | 62 | 70 | 53 | 80 | 90 | 67 | 91 | 90 | 93 | 83 | 90 | 73 | 80 | 90 | 67 |
| SVM | Healthy/CCA | 86 | 85 | 87 | 94 | 95 | 93 | 94 | 95 | 93 | 94 | 95 | 93 | 94 | 95 | 93 |
| CCA/HCC | 73 | 95 | 0 | 81 | 100 | 17 | 85 | 100 | 33 | 81 | 100 | 17 | 81 | 100 | 17 | |
| CCA/BD | 73 | 95 | 0 | 77 | 90 | 33 | 73 | 85 | 33 | 77 | 90 | 33 | 77 | 90 | 33 | |
| RF | Healthy/CCA | 71 | 85 | 53 | 97 | 100 | 93 | 94 | 100 | 87 | 94 | 100 | 87 | 97 | 100 | 93 |
| CCA/HCC | 73 | 95 | 0 | 81 | 100 | 17 | 81 | 95 | 33 | 77 | 90 | 33 | 85 | 100 | 33 | |
| CCA/BD | 81 | 95 | 33 | 73 | 85 | 33 | 77 | 90 | 33 | 77 | 90 | 33 | 77 | 90 | 33 | |
| NN | Healthy/CCA | 82 | 90 | 73 | 97 | 95 | 100 | 97 | 95 | 100 | 97 | 95 | 100 | 100 | 100 | 100 |
| CCA/HCC | 84 | 95 | 50 | 92 | 95 | 83 | 92 | 100 | 67 | 88 | 100 | 50 | 88 | 100 | 50 | |
| CCA/BD | 80 | 85 | 66 | 81 | 80 | 33 | 73 | 70 | 83 | 81 | 85 | 67 | 81 | 80 | 83 | |
| Biomolecule | Molecular Vibration | Wavenumber (cm−1) | References | |||
|---|---|---|---|---|---|---|
| PCA | PLS-DA | |||||
| Human Serum | Hamster Serum | Human Serum | Hamster Serum | |||
| Lipid | C=O | 1747 | 1745 | 1743 | 1736 | [34,35] |
| Collagen | Amide III and CH2 wagging | 1380–1200 | 1380–1200 | 1380–1200 | 1380–1200 | [36,37,38,39,40] |
| CH2 vibration | 1339 | ~1337 | 1337 | ~1337 | ||
| 1035 | 1030 | 1034 | 1030 | [41] | ||
| Nucleic acid or Protein | 1073 | 1077 | 1074 | 1076 | [23,25,34,42] | |
| Polysaccharide | C-O stretching | 1152 | 1156 | 1154 | 1153 | [22,23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatchawal, P.; Wongwattanakul, M.; Tippayawat, P.; Kochan, K.; Jearanaikoon, N.; Wood, B.R.; Jearanaikoon, P. Detection of Human Cholangiocarcinoma Markers in Serum Using Infrared Spectroscopy. Cancers 2021, 13, 5109. https://doi.org/10.3390/cancers13205109
Chatchawal P, Wongwattanakul M, Tippayawat P, Kochan K, Jearanaikoon N, Wood BR, Jearanaikoon P. Detection of Human Cholangiocarcinoma Markers in Serum Using Infrared Spectroscopy. Cancers. 2021; 13(20):5109. https://doi.org/10.3390/cancers13205109
Chicago/Turabian StyleChatchawal, Patutong, Molin Wongwattanakul, Patcharaporn Tippayawat, Kamilla Kochan, Nichada Jearanaikoon, Bayden R. Wood, and Patcharee Jearanaikoon. 2021. "Detection of Human Cholangiocarcinoma Markers in Serum Using Infrared Spectroscopy" Cancers 13, no. 20: 5109. https://doi.org/10.3390/cancers13205109
APA StyleChatchawal, P., Wongwattanakul, M., Tippayawat, P., Kochan, K., Jearanaikoon, N., Wood, B. R., & Jearanaikoon, P. (2021). Detection of Human Cholangiocarcinoma Markers in Serum Using Infrared Spectroscopy. Cancers, 13(20), 5109. https://doi.org/10.3390/cancers13205109