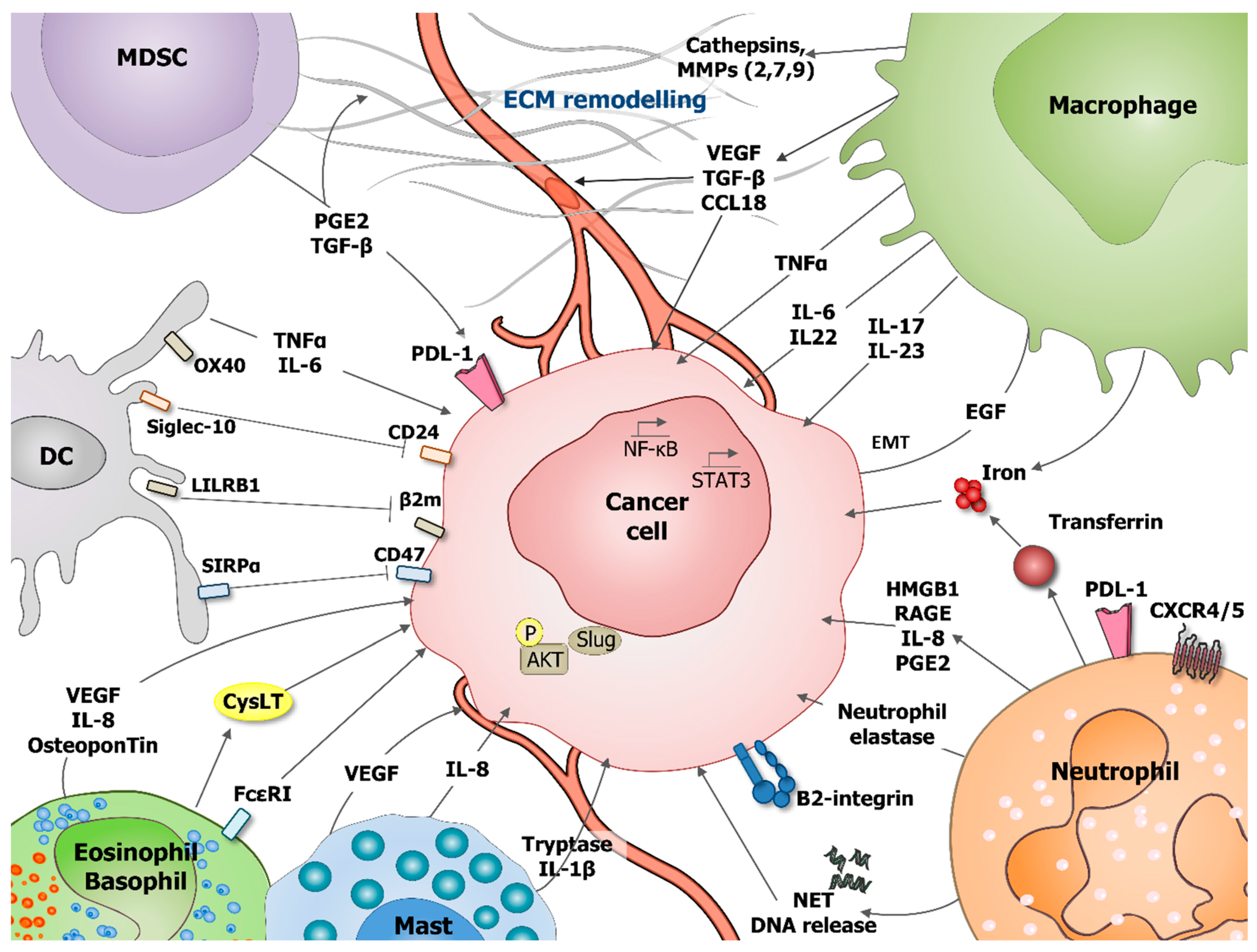
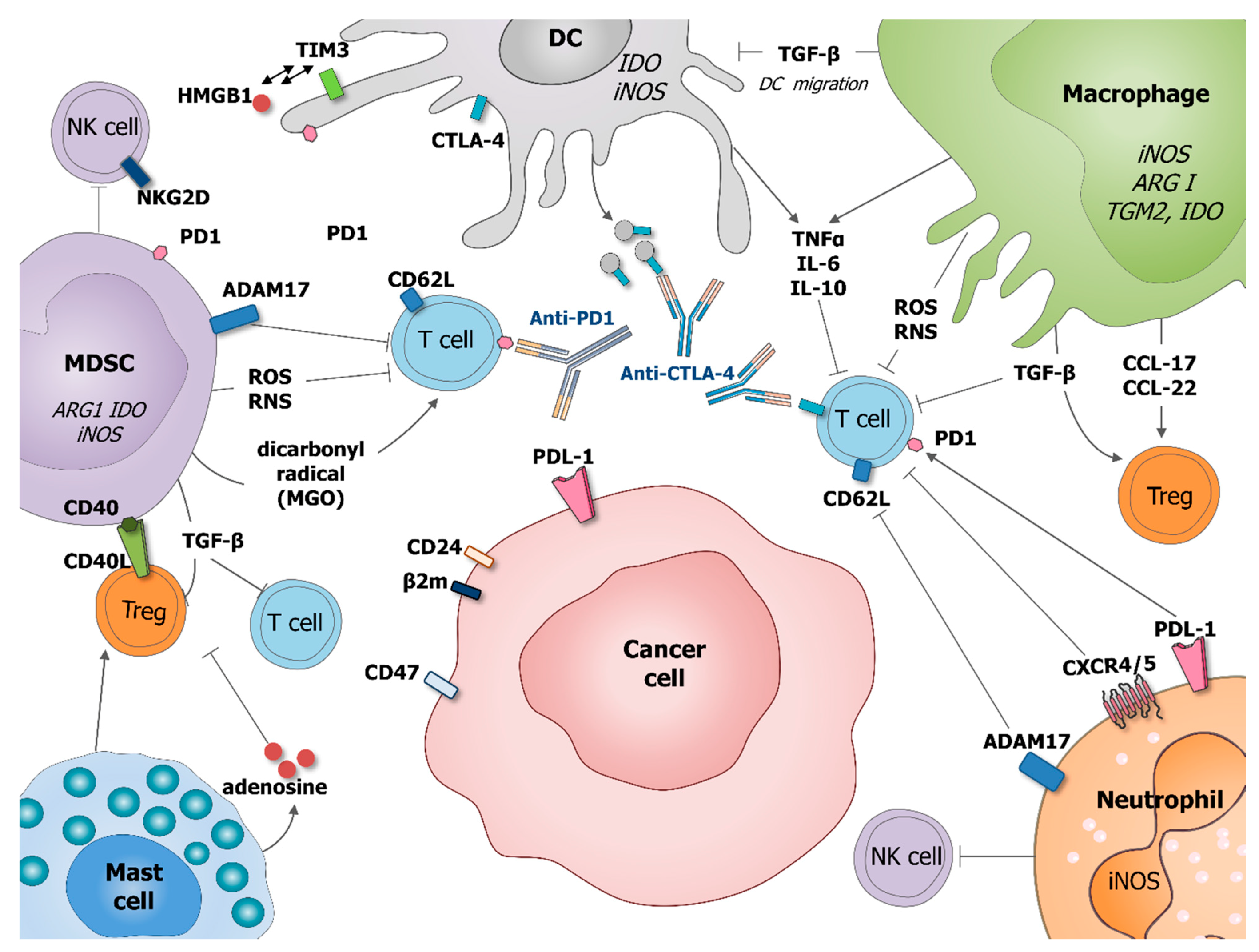
Cancer Cells Resistance Shaping by Tumor Infiltrating Myeloid Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Macrophages
2.1. Origin and Physiological Roles of Macrophages
2.2. Pathological Functions of Macrophages
2.3. Macrophages in Cancer
2.3.1. Origin and Functions of TAM
2.3.2. How Can TAM Shape Cancer Cell Resistance?
3. Dendritic Cells
3.1. Origin and Roles of Dendritic Cells
3.2. Dendritic Cells in Cancer
4. Neutrophils
4.1. Origin and Physiological Roles of Neutrophils
4.2. Neutrophils in Cancer
5. MDSC
5.1. Origins and Roles of MDSCs
5.2. MDSCs in Cancer
6. Mast Cells
6.1. Origin and Physiological Roles of Mast Cells
6.2. Mast Cells in Cancer
7. Eosinophils and Basophils
7.1. Origin and Functions
7.2. Basophils and Eosinophils in Cancer
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of Tumor Microenvironment in Tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage Biology in Development, Homeostasis and Disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Schultze, J.L.; Murray, P.J.; Ochando, J.; Biswas, S.K. New Insights into the Multidimensional Concept of Macrophage Ontogeny, Activation and Function. Nat. Immunol. 2016, 17, 34–40. [Google Scholar] [CrossRef]
- Ajami, B.; Bennett, J.L.; Krieger, C.; Tetzlaff, W.; Rossi, F.M.V. Local Self-Renewal Can Sustain CNS Microglia Maintenance and Function throughout Adult Life. Nat. Neurosci. 2007, 10, 1538–1543. [Google Scholar] [CrossRef]
- Hettinger, J.; Richards, D.M.; Hansson, J.; Barra, M.M.; Joschko, A.-C.; Krijgsveld, J.; Feuerer, M. Origin of Monocytes and Macrophages in a Committed Progenitor. Nat. Immunol. 2013, 14, 821–830. [Google Scholar] [CrossRef]
- Geissmann, F.; Jung, S.; Littman, D.R. Blood Monocytes Consist of Two Principal Subsets with Distinct Migratory Properties. Immunity 2003, 19, 71–82. [Google Scholar] [CrossRef]
- Passlick, B.; Flieger, D.; Ziegler-Heitbrock, H.W. Identification and Characterization of a Novel Monocyte Subpopulation in Human Peripheral Blood. Blood 1989, 74, 2527–2534. [Google Scholar] [CrossRef]
- Patel, A.A.; Zhang, Y.; Fullerton, J.N.; Boelen, L.; Rongvaux, A.; Maini, A.A.; Bigley, V.; Flavell, R.A.; Gilroy, D.W.; Asquith, B.; et al. The Fate and Lifespan of Human Monocyte Subsets in Steady State and Systemic Inflammation. J. Exp. Med. 2017, 214, 1913–1923. [Google Scholar] [CrossRef]
- Theret, M.; Mounier, R.; Rossi, F. The Origins and Non-Canonical Functions of Macrophages in Development and Regeneration. Development 2019, 146, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gautier, E.L.; Yvan-Charvet, L. Understanding Macrophage Diversity at the Ontogenic and Transcriptomic Levels. Immunol. Rev. 2014, 262, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Amit, I.; Winter, D.R.; Jung, S. The Role of the Local Environment and Epigenetics in Shaping Macrophage Identity and Their Effect on Tissue Homeostasis. Nat. Immunol. 2016, 17, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Clauss, S.; Xiao, L.; Aguirre, A.D.; King, K.R.; Hanley, A.; Hucker, W.J.; Wülfers, E.M.; Seemann, G.; Courties, G.; et al. Macrophages Facilitate Electrical Conduction in the Heart. Cell 2017, 169, 510–522.e20. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Red Eagle, A.; Vats, D.; Morel, C.R.; Goforth, M.H.; Subramanian, V.; Mukundan, L.; Ferrante, A.W.; Chawla, A. Alternative M2 Activation of Kupffer Cells by PPARdelta Ameliorates Obesity-Induced Insulin Resistance. Cell Metab. 2008, 7, 496–507. [Google Scholar] [CrossRef]
- Kohyama, M.; Ise, W.; Edelson, B.T.; Wilker, P.R.; Hildner, K.; Mejia, C.; Frazier, W.A.; Murphy, T.L.; Murphy, K.M. Role for Spi-C in the Development of Red Pulp Macrophages and Splenic Iron Homeostasis. Nature 2009, 457, 318–321. [Google Scholar] [CrossRef] [PubMed]
- de Back, D.Z.; Kostova, E.B.; van Kraaij, M.; van den Berg, T.K.; van Bruggen, R. Of Macrophages and Red Blood Cells: A Complex Love Story. Front. Physiol. 2014, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, T.; Galligan, C.; Takahashi, M.; Chen, K.; Liu, M.; Tessarollo, L.; Wang, J.M. Non-Myeloid Cells Are Major Contributors to Innate Immune Responses via Production of Monocyte Chemoattractant Protein-1/CCL2. Front. Immunol. 2014, 4, 482. [Google Scholar] [CrossRef] [PubMed]
- Boring, L.; Gosling, J.; Chensue, S.W.; Kunkel, S.L.; Farese, R.V.; Broxmeyer, H.E.; Charo, I.F. Impaired Monocyte Migration and Reduced Type 1 (Th1) Cytokine Responses in C-C Chemokine Receptor 2 Knockout Mice. J. Clin. Investig. 1997, 100, 2552–2561. [Google Scholar] [CrossRef]
- Anbazhagan, K.; Duroux-Richard, I.; Jorgensen, C.; Apparailly, F. Transcriptomic Network Support Distinct Roles of Classical and Non-Classical Monocytes in Human. Int. Rev. Immunol. 2014, 33, 470–489. [Google Scholar] [CrossRef]
- Laskin, D.L.; Sunil, V.R.; Gardner, C.R.; Laskin, J.D. Macrophages and Tissue Injury: Agents of Defense or Destruction? Annu. Rev. Pharmacol. Toxicol. 2011, 51, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D.; Kincaid, K.; Alt, J.M.; Heilman, M.J.; Hill, A.M. M-1/M-2 Macrophages and the Th1/Th2 Paradigm. J. Immunol. 2000, 164, 6166–6173. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Sica, A.; Mantovani, A.; Locati, M. Macrophage Activation and Polarization. Front. Biosci. 2008, 13, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Schmidt, S.V.; Sander, J.; Draffehn, A.; Krebs, W.; Quester, I.; De Nardo, D.; Gohel, T.D.; Emde, M.; Schmidleithner, L.; et al. Transcriptome-Based Network Analysis Reveals a Spectrum Model of Human Macrophage Activation. Immunity 2014, 40, 274–288. [Google Scholar] [CrossRef] [PubMed]
- Lavin, Y.; Winter, D.; Blecher-Gonen, R.; David, E.; Keren-Shaul, H.; Merad, M.; Jung, S.; Amit, I. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell 2014, 159, 1312–1326. [Google Scholar] [CrossRef]
- Mould, K.J.; Jackson, N.D.; Henson, P.M.; Seibold, M.; Janssen, W.J. Single Cell RNA Sequencing Identifies Unique Inflammatory Airspace Macrophage Subsets. JCI Insight 2019, 4, 1–17. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The Role of Pattern-Recognition Receptors in Innate Immunity: Update on Toll-like Receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Vannella, K.M.; Wynn, T.A. Mechanisms of Organ Injury and Repair by Macrophages. Annu. Rev. Physiol. 2017, 79, 593–617. [Google Scholar] [CrossRef]
- Olingy, C.E.; San Emeterio, C.L.; Ogle, M.E.; Krieger, J.R.; Bruce, A.C.; Pfau, D.D.; Jordan, B.T.; Peirce, S.M.; Botchwey, E.A. Non-Classical Monocytes Are Biased Progenitors of Wound Healing Macrophages during Soft Tissue Injury. Sci. Rep. 2017, 7, 447. [Google Scholar] [CrossRef]
- Martinez, F.O.; Helming, L.; Milde, R.; Varin, A.; Melgert, B.N.; Draijer, C.; Thomas, B.; Fabbri, M.; Crawshaw, A.; Ho, L.P.; et al. Genetic Programs Expressed in Resting and IL-4 Alternatively Activated Mouse and Human Macrophages: Similarities and Differences. Blood 2013, 121, e57–e69. [Google Scholar] [CrossRef] [PubMed]
- Vogel, D.Y.S.; Vereyken, E.J.F.; Glim, J.E.; Heijnen, P.D.A.M.; Moeton, M.; van der Valk, P.; Amor, S.; Teunissen, C.E.; van Horssen, J.; Dijkstra, C.D. Macrophages in Inflammatory Multiple Sclerosis Lesions Have an Intermediate Activation Status. J. Neuroinflamm. 2013, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, J.S.; Fuentes-Duculan, J.; Suárez-Fariñas, M.; Pierson, K.C.; Pitts-Kiefer, A.; Fan, L.; Belkin, D.A.; Wang, C.Q.F.; Bhuvanendran, S.; Johnson-Huang, L.M.; et al. Tumor-Associated Macrophages in the Cutaneous SCC Microenvironment Are Heterogeneously Activated. J. Investig. Dermatol. 2011, 131, 1322–1330. [Google Scholar] [CrossRef] [PubMed]
- Franklin, R.A.; Liao, W.; Sarkar, A.; Kim, M.V.; Bivona, M.R.; Liu, K.; Pamer, E.G.; Li, M.O. The Cellular and Molecular Origin of Tumor-Associated Macrophages. Science 2014, 344, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Linde, N.; Lederle, W.; Depner, S.; van Rooijen, N.; Gutschalk, C.M.; Mueller, M.M. Vascular Endothelial Growth Factor-Induced Skin Carcinogenesis Depends on Recruitment and Alternative Activation of Macrophages. J. Pathol. 2012, 227, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Nandi, B.; Shapiro, M.; Samur, M.K.; Pai, C.; Frank, N.Y.; Yoon, C.; Prabhala, R.H.; Munshi, N.C.; Gold, J.S. Stromal CCR6 Drives Tumor Growth in a Murine Transplantable Colon Cancer through Recruitment of Tumor-Promoting Macrophages. Oncoimmunology 2016, 5, e1189052. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, J.; Kong, X.; Li, E.; Liu, Y.; Du, X.; Kang, Z.; Tang, Y.; Kuang, Y.; Yang, Z.; et al. CD47 Promotes Tumor Invasion and Metastasis in Non-Small Cell Lung Cancer. Sci. Rep. 2016, 6, 29719. [Google Scholar] [CrossRef]
- Griffith, T.S.; Wiley, S.R.; Kubin, M.Z.; Sedger, L.M.; Maliszewski, C.R.; Fanger, N.A. Monocyte-Mediated Tumoricidal Activity via the Tumor Necrosis Factor-Related Cytokine, TRAIL. J. Exp. Med. 1999, 189, 1343–1354. [Google Scholar] [CrossRef]
- De, I.; Steffen, M.D.; Clark, P.A.; Patros, C.J.; Sokn, E.; Bishop, S.M.; Litscher, S.; Maklakova, V.I.; Kuo, J.S.; Rodriguez, F.J.; et al. CSF1 Overexpression Promotes High-Grade Glioma Formation without Impacting the Polarization Status of Glioma-Associated Microglia and Macrophages. Cancer Res. 2016, 76, 2552–2560. [Google Scholar] [CrossRef]
- Li, D.; Ji, H.; Niu, X.; Yin, L.; Wang, Y.; Gu, Y.; Wang, J.; Zhou, X.; Zhang, H.; Zhang, Q. Tumor-Associated Macrophages Secrete CC-Chemokine Ligand 2 and Induce Tamoxifen Resistance by Activating PI3K/Akt/MTOR in Breast Cancer. Cancer Sci. 2020, 111, 47–58. [Google Scholar] [CrossRef]
- Zhao, P.; Gao, D.; Wang, Q.; Song, B.; Shao, Q.; Sun, J.; Ji, C.; Li, X.; Li, P.; Qu, X. Response Gene to Complement 32 (RGC-32) Expression on M2-Polarized and Tumor-Associated Macrophages Is M-CSF-Dependent and Enhanced by Tumor-Derived IL-4. Cell Mol. Immunol 2015, 12, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.-R.; Lee, J.H.; Hensel, J.A.; Sawant, A.B.; Davis, B.H.; Lee, C.M.; Deshane, J.S.; Ponnazhagan, S. Prostate Cancer-Derived Cathelicidin-Related Antimicrobial Peptide Facilitates Macrophage Differentiation and Polarization of Immature Myeloid Progenitors to Protumorigenic Macrophages. Prostate 2016, 76, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, C.; Tewari, B.N.; Kanchan, R.K.; Baghel, K.S.; Nautiyal, N.; Shrivastava, R.; Kaur, H.; Bhatt, M.L.B.; Bhadauria, S. Macrophages Are Recruited to Hypoxic Tumor Areas and Acquire a Pro-Angiogenic M2-Polarized Phenotype via Hypoxic Cancer Cell Derived Cytokines Oncostatin M and Eotaxin. Oncotarget 2014, 5, 5350–5368. [Google Scholar] [CrossRef] [PubMed]
- Gocheva, V.; Wang, H.-W.; Gadea, B.B.; Shree, T.; Hunter, K.E.; Garfall, A.L.; Berman, T.; Joyce, J.A. IL-4 Induces Cathepsin Protease Activity in Tumor-Associated Macrophages to Promote Cancer Growth and Invasion. Genes Dev. 2010, 24, 241–255. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Barreto, J.B.; Andreu, P.; Vasquez, L.; Tawfik, D.; Kolhatkar, N.; Coussens, L.M. CD4(+) T Cells Regulate Pulmonary Metastasis of Mammary Carcinomas by Enhancing Protumor Properties of Macrophages. Cancer Cell 2009, 16, 91–102. [Google Scholar] [CrossRef]
- Pedroza-Gonzalez, A.; Xu, K.; Wu, T.-C.; Aspord, C.; Tindle, S.; Marches, F.; Gallegos, M.; Burton, E.C.; Savino, D.; Hori, T.; et al. Thymic Stromal Lymphopoietin Fosters Human Breast Tumor Growth by Promoting Type 2 Inflammation. J. Exp. Med. 2011, 208, 479–490. [Google Scholar] [CrossRef]
- Solinas, G.; Schiarea, S.; Liguori, M.; Fabbri, M.; Pesce, S.; Zammataro, L.; Pasqualini, F.; Nebuloni, M.; Chiabrando, C.; Mantovani, A.; et al. Tumor-Conditioned Macrophages Secrete Migration-Stimulating Factor: A New Marker for M2-Polarization, Influencing Tumor Cell Motility. J. Immunol. 2010, 185, 642–652. [Google Scholar] [CrossRef]
- Sica, A.; Saccani, A.; Bottazzi, B.; Polentarutti, N.; Vecchi, A.; van Damme, J.; Mantovani, A. Autocrine Production of IL-10 Mediates Defective IL-12 Production and NF-Kappa B Activation in Tumor-Associated Macrophages. J. Immunol. 2000, 164, 762–767. [Google Scholar] [CrossRef]
- Sánchez-Martín, L.; Estecha, A.; Samaniego, R.; Sánchez-Ramón, S.; Vega, M.Á.; Sánchez-Mateos, P. The Chemokine CXCL12 Regulates Monocyte-Macrophage Differentiation and RUNX3 Expression. Blood 2011, 117, 88–97. [Google Scholar] [CrossRef]
- Doedens, A.L.; Stockmann, C.; Rubinstein, M.P.; Liao, D.; Zhang, N.; DeNardo, D.G.; Coussens, L.M.; Karin, M.; Goldrath, A.W.; Johnson, R.S. Macrophage Expression of Hypoxia-Inducible Factor-1 Alpha Suppresses T-Cell Function and Promotes Tumor Progression. Cancer Res. 2010, 70, 7465–7475. [Google Scholar] [CrossRef]
- Kim, S.; Takahashi, H.; Lin, W.-W.; Descargues, P.; Grivennikov, S.; Kim, Y.; Luo, J.-L.; Karin, M. Carcinoma-Produced Factors Activate Myeloid Cells through TLR2 to Stimulate Metastasis. Nature 2009, 457, 102–106. [Google Scholar] [CrossRef]
- Deligne, C.; Murdamoothoo, D.; Gammage, A.N.; Gschwandtner, M.; Erne, W.; Loustau, T.; Marzeda, A.M.; Carapito, R.; Paul, N.; Velazquez-Quesada, I.; et al. Matrix-Targeting Immunotherapy Controls Tumor Growth and Spread by Switching Macrophage Phenotype. Cancer Immunol. Res. 2020, 8, 368–382. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Wang, K.; Mucida, D.; Stewart, C.A.; Schnabl, B.; Jauch, D.; Taniguchi, K.; Yu, G.-Y.; Osterreicher, C.H.; Hung, K.E.; et al. Adenoma-Linked Barrier Defects and Microbial Products Drive IL-23/IL-17-Mediated Tumour Growth. Nature 2012, 491, 254–258. [Google Scholar] [CrossRef]
- Kong, L.; Zhou, Y.; Bu, H.; Lv, T.; Shi, Y.; Yang, J. Deletion of Interleukin-6 in Monocytes/Macrophages Suppresses the Initiation of Hepatocellular Carcinoma in Mice. J. Exp. Clin. Cancer Res. 2016, 35, 131. [Google Scholar] [CrossRef]
- Jung, M.; Mertens, C.; Tomat, E.; Brüne, B. Iron as a Central Player and Promising Target in Cancer Progression. Int. J. Mol. Sci. 2019, 20, 273. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Wang, S.; Li, W.; Wu, D.; Chen, W. Tumor-Associated Macrophages Promote the Metastasis of Ovarian Carcinoma Cells by Enhancing CXCL16/CXCR6 Expression. Pathol. Res. Pract. 2018, 214, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, J.; Chen, S.; Lu, M.; Luo, X.; Yao, S.; Liu, S.; Qin, Y.; Chen, H. Tumor-Associated Macrophages Provide a Suitable Microenvironment for Non-Small Lung Cancer Invasion and Progression. Lung Cancer 2011, 74, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Liu, H.; Li, M.; Li, B.; Gao, W.; Shao, Q.; Fan, B.; Zhao, F.; Wang, Q.; Xie, Q.; et al. Increased Metabolites of 5-Lipoxygenase from Hypoxic Ovarian Cancer Cells Promote Tumor-Associated Macrophage Infiltration. Oncogene 2015, 34, 1241–1252. [Google Scholar] [CrossRef]
- Chen, J.; Yao, Y.; Gong, C.; Yu, F.; Su, S.; Chen, J.; Liu, B.; Deng, H.; Wang, F.; Lin, L.; et al. CCL18 from Tumor-Associated Macrophages Promotes Breast Cancer Metastasis via PITPNM3. Cancer Cell 2011, 19, 541–555. [Google Scholar] [CrossRef]
- Liu, L.; Wang, X.; Li, X.; Wu, X.; Tang, M.; Wang, X. Upregulation of IGF1 by Tumor-Associated Macrophages Promotes the Proliferation and Migration of Epithelial Ovarian Cancer Cells. Oncol. Rep. 2018, 39, 818–826. [Google Scholar] [CrossRef]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y.; Huang, J.-D.; Song, E. Microvesicles Secreted by Macrophages Shuttle Invasion-Potentiating MicroRNAs into Breast Cancer Cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Pollard, J.W. Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell 2010, 141, 39. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Chen, Y.-S.; Yao, Y.-D.; Chen, J.-Q.; Chen, J.-N.; Huang, S.-Y.; Zeng, Y.-J.; Yao, H.-R.; Zeng, S.-H.; Fu, Y.-S.; et al. CCL18 from Tumor-Associated Macrophages Promotes Angiogenesis in Breast Cancer. Oncotarget 2015, 6, 34758–34773. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhang, W.; Zhong, W.-Q.; Liu, Z.-J.; Li, H.-M.; Yu, Z.-L.; Zhao, Y.-F. Tumor Associated Macrophages Induce Epithelial to Mesenchymal Transition via the EGFR/ERK1/2 Pathway in Head and Neck Squamous Cell Carcinoma. Oncol. Rep. 2018, 40, 2558. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.C.; Gabriely, G.; Rothhammer, V.; Mascanfroni, I.D.; Wheeler, M.A.; Chao, C.-C.; Gutiérrez-Vázquez, C.; Kenison, J.; Tjon, E.C.; Barroso, A.; et al. Control of Tumor-Associated Macrophages and T Cells in Glioblastoma via AHR and CD39. Nat. Neurosci. 2019, 22, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Tabata, K.; Kimura, T.; Yachie-Kinoshita, A.; Ozawa, Y.; Yamada, K.; Ito, J.; Tachino, S.; Hori, Y.; Matsuki, M.; et al. Lenvatinib plus Anti-PD-1 Antibody Combination Treatment Activates CD8+ T Cells through Reduction of Tumor-Associated Macrophage and Activation of the Interferon Pathway. PLoS ONE 2019, 14, e0212513. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Minamiya, Y.; Kawai, H.; Saito, S.; Saito, H.; Nakagawa, T.; Imai, K.; Hirokawa, M.; Ogawa, J. Tumor-Derived TGFbeta-1 Induces Dendritic Cell Apoptosis in the Sentinel Lymph Node. J. Immunol. 2006, 176, 5637–5643. [Google Scholar] [CrossRef]
- Solinas, G.; Germano, G.; Mantovani, A.; Allavena, P. Tumor-Associated Macrophages (TAM) as Major Players of the Cancer-Related Inflammation. J. Leukoc. Biol. 2009, 86, 1065–1073. [Google Scholar] [CrossRef]
- Lievense, L.A.; Cornelissen, R.; Bezemer, K.; Kaijen-Lambers, M.E.H.; Hegmans, J.P.J.J.; Aerts, J.G.J.V. Pleural Effusion of Patients with Malignant Mesothelioma Induces Macrophage-Mediated T Cell Suppression. J. Thorac. Oncol. 2016, 11, 1755–1764. [Google Scholar] [CrossRef]
- Campesato, L.F.; Budhu, S.; Tchaicha, J.; Weng, C.-H.; Gigoux, M.; Cohen, I.J.; Redmond, D.; Mangarin, L.; Pourpe, S.; Liu, C.; et al. Blockade of the AHR Restricts a Treg-Macrophage Suppressive Axis Induced by L-Kynurenine. Nat. Commun. 2020, 11, 4011. [Google Scholar] [CrossRef]
- Chen, C.; Qu, Q.-X.; Shen, Y.; Mu, C.-Y.; Zhu, Y.-B.; Zhang, X.-G.; Huang, J.-A. Induced Expression of B7-H4 on the Surface of Lung Cancer Cell by the Tumor-Associated Macrophages: A Potential Mechanism of Immune Escape. Cancer Lett. 2012, 317, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Kuang, D.-M.; Zhao, Q.; Peng, C.; Xu, J.; Zhang, J.-P.; Wu, C.; Zheng, L. Activated Monocytes in Peritumoral Stroma of Hepatocellular Carcinoma Foster Immune Privilege and Disease Progression through PD-L1. J. Exp. Med. 2009, 206, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Prima, V.; Kaliberova, L.N.; Kaliberov, S.; Curiel, D.T.; Kusmartsev, S. COX2/MPGES1/PGE2 Pathway Regulates PD-L1 Expression in Tumor-Associated Macrophages and Myeloid-Derived Suppressor Cells. Proc. Natl. Acad. Sci. USA 2017, 114, 1117–1122. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Ramakrishnan, R.; Altiok, S.; Youn, J.-I.; Cheng, P.; Celis, E.; Pisarev, V.; Sherman, S.; Sporn, M.B.; Gabrilovich, D. Tumor-Infiltrating Myeloid Cells Induce Tumor Cell Resistance to Cytotoxic T Cells in Mice. J. Clin. Investig. 2011, 121, 4015–4029. [Google Scholar] [CrossRef]
- Zheng, P.; Chen, L.; Yuan, X.; Luo, Q.; Liu, Y.; Xie, G.; Ma, Y.; Shen, L. Exosomal Transfer of Tumor-Associated Macrophage-Derived MiR-21 Confers Cisplatin Resistance in Gastric Cancer Cells. J. Exp. Clin. Cancer Res. 2017, 36, 53. [Google Scholar] [CrossRef]
- Fu, X.-T.; Song, K.; Zhou, J.; Shi, Y.-H.; Liu, W.-R.; Shi, G.-M.; Gao, Q.; Wang, X.-Y.; Ding, Z.-B.; Fan, J. Tumor-Associated Macrophages Modulate Resistance to Oxaliplatin via Inducing Autophagy in Hepatocellular Carcinoma. Cancer Cell Int. 2019, 19, 71. [Google Scholar] [CrossRef]
- Burger, J.A.; Tsukada, N.; Burger, M.; Zvaifler, N.J.; Dell’Aquila, M.; Kipps, T.J. Blood-Derived Nurse-like Cells Protect Chronic Lymphocytic Leukemia B Cells from Spontaneous Apoptosis through Stromal Cell-Derived Factor-1. Blood 2000, 96, 2655–2663. [Google Scholar] [CrossRef]
- Boissard, F.; Fournié, J.-J.; Quillet-Mary, A.; Ysebaert, L.; Poupot, M. Nurse-like Cells Mediate Ibrutinib Resistance in Chronic Lymphocytic Leukemia Patients. Blood Cancer J. 2015, 5, e355. [Google Scholar] [CrossRef]
- Boissard, F.; Laurent, C.; Ramsay, A.G.; Quillet-Mary, A.; Fournié, J.-J.; Poupot, M.; Ysebaert, L. Nurse-like Cells Impact on Disease Progression in Chronic Lymphocytic Leukemia. Blood Cancer J. 2016, 6, e381. [Google Scholar] [CrossRef]
- Boissard, F.; Tosolini, M.; Ligat, L.; Quillet-Mary, A.; Lopez, F.; Fournié, J.-J.; Ysebaert, L.; Poupot, M. Nurse-like Cells Promote CLL Survival through LFA-3/CD2 Interactions. Oncotarget 2017, 8, 52225–52236. [Google Scholar] [CrossRef]
- Caligaris-Cappio, F. Role of the Microenvironment in Chronic Lymphocytic Leukaemia. Br. J. Haematol. 2003, 123, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Nishio, M.; Endo, T.; Tsukada, N.; Ohata, J.; Kitada, S.; Reed, J.C.; Zvaifler, N.J.; Kipps, T.J. Nurselike Cells Express BAFF and APRIL, Which Can Promote Survival of Chronic Lymphocytic Leukemia Cells via a Paracrine Pathway Distinct from That of SDF-1alpha. Blood 2005, 106, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Dammeijer, F.; Lievense, L.A.; Kaijen-Lambers, M.E.; van Nimwegen, M.; Bezemer, K.; Hegmans, J.P.; van Hall, T.; Hendriks, R.W.; Aerts, J.G. Depletion of Tumor-Associated Macrophages with a CSF-1R Kinase Inhibitor Enhances Antitumor Immunity and Survival Induced by DC Immunotherapy. Cancer Immunol. Res. 2017, 5, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Peranzoni, E.; Lemoine, J.; Vimeux, L.; Feuillet, V.; Barrin, S.; Kantari-Mimoun, C.; Bercovici, N.; Guérin, M.; Biton, J.; Ouakrim, H.; et al. Macrophages Impede CD8 T Cells from Reaching Tumor Cells and Limit the Efficacy of Anti-PD-1 Treatment. Proc. Natl. Acad. Sci. USA 2018, 115, E4041–E4050. [Google Scholar] [CrossRef] [PubMed]
- Panni, R.Z.; Herndon, J.M.; Zuo, C.; Hegde, S.; Hogg, G.D.; Knolhoff, B.L.; Breden, M.A.; Li, X.; Krisnawan, V.E.; Khan, S.Q.; et al. Agonism of CD11b Reprograms Innate Immunity to Sensitize Pancreatic Cancer to Immunotherapies. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Lebdai, S.; Gigoux, M.; Alvim, R.; Somma, A.; Nagar, K.; Azzouzi, A.R.; Cussenot, O.; Merghoub, T.; Wolchok, J.D.; Scherz, A.; et al. Potentiating Vascular-Targeted Photodynamic Therapy through CSF-1R Modulation of Myeloid Cells in a Preclinical Model of Prostate Cancer. Oncoimmunology 2019, 8, e1581528. [Google Scholar] [CrossRef]
- Guilliams, M.; Ginhoux, F.; Jakubzick, C.; Naik, S.H.; Onai, N.; Schraml, B.U.; Segura, E.; Tussiwand, R.; Yona, S. Dendritic Cells, Monocytes and Macrophages: A Unified Nomenclature Based on Ontogeny. Nat. Rev. Immunol. 2014, 14, 571–578. [Google Scholar] [CrossRef]
- Askenase, M.H.; Han, S.-J.; Byrd, A.L.; da Fonseca, D.M.; Bouladoux, N.; Wilhelm, C.; Konkel, J.E.; Hand, T.W.; Lacerda-Queiroz, N.; Su, X.; et al. Bone-Marrow-Resident NK Cells Prime Monocytes for Regulatory Function during Infection. Immunity 2015, 42, 1130–1142. [Google Scholar] [CrossRef]
- Vu Manh, T.-P.; Bertho, N.; Hosmalin, A.; Schwartz-Cornil, I.; Dalod, M. Investigating Evolutionary Conservation of Dendritic Cell Subset Identity and Functions. Front. Immunol. 2015, 6, 260. [Google Scholar] [CrossRef]
- Brewitz, A.; Eickhoff, S.; Dähling, S.; Quast, T.; Bedoui, S.; Kroczek, R.A.; Kurts, C.; Garbi, N.; Barchet, W.; Iannacone, M.; et al. CD8+ T Cells Orchestrate PDC-XCR1+ Dendritic Cell Spatial and Functional Cooperativity to Optimize Priming. Immunity 2017, 46, 205–219. [Google Scholar] [CrossRef]
- Anderson, D.A.; Dutertre, C.-A.; Ginhoux, F.; Murphy, K.M. Genetic Models of Human and Mouse Dendritic Cell Development and Function. Nat. Rev. Immunol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Victora, G.D.; Schwickert, T.A.; Guermonprez, P.; Meredith, M.M.; Yao, K.; Chu, F.-F.; Randolph, G.J.; Rudensky, A.Y.; Nussenzweig, M. In Vivo Analysis of Dendritic Cell Development and Homeostasis. Science 2009, 324, 392–397. [Google Scholar] [CrossRef] [PubMed]
- McKenna, H.J.; Stocking, K.L.; Miller, R.E.; Brasel, K.; De Smedt, T.; Maraskovsky, E.; Maliszewski, C.R.; Lynch, D.H.; Smith, J.; Pulendran, B.; et al. Mice Lacking Flt3 Ligand Have Deficient Hematopoiesis Affecting Hematopoietic Progenitor Cells, Dendritic Cells, and Natural Killer Cells. Blood 2000, 95, 3489–3497. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, H.S.; Cisse, B.; Bunin, A.; Lewis, K.L.; Reizis, B. Continuous Expression of the Transcription Factor E2-2 Maintains the Cell Fate of Mature Plasmacytoid Dendritic Cells. Immunity 2010, 33, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Dai, D.; Horton, B.; Gajewski, T.F. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell 2017, 31, 711–723.e4. [Google Scholar] [CrossRef]
- Garris, C.S.; Arlauckas, S.P.; Kohler, R.H.; Trefny, M.P.; Garren, S.; Piot, C.; Engblom, C.; Pfirschke, C.; Siwicki, M.; Gungabeesoon, J.; et al. Successful Anti-PD-1 Cancer Immunotherapy Requires T Cell-Dendritic Cell Crosstalk Involving the Cytokines IFN-γ and IL-12. Immunity 2018, 49, 1148–1161.e7. [Google Scholar] [CrossRef]
- Laoui, D.; Keirsse, J.; Morias, Y.; Van Overmeire, E.; Geeraerts, X.; Elkrim, Y.; Kiss, M.; Bolli, E.; Lahmar, Q.; Sichien, D.; et al. The Tumour Microenvironment Harbours Ontogenically Distinct Dendritic Cell Populations with Opposing Effects on Tumour Immunity. Nat. Commun. 2016, 7, 13720. [Google Scholar] [CrossRef]
- Veglia, F.; Gabrilovich, D.I. Dendritic Cells in Cancer: The Role Revisited. Curr. Opin. Immunol. 2017, 45, 43–51. [Google Scholar] [CrossRef]
- Santini, S.M.; Di Pucchio, T.; Lapenta, C.; Parlato, S.; Logozzi, M.; Belardelli, F. The Natural Alliance between Type I Interferon and Dendritic Cells and Its Role in Linking Innate and Adaptive Immunity. J. Interferon Cytokine Res. 2002, 22, 1071–1080. [Google Scholar] [CrossRef]
- Vanderheyde, N.; Aksoy, E.; Amraoui, Z.; Vandenabeele, P.; Goldman, M.; Willems, F. Tumoricidal Activity of Monocyte-Derived Dendritic Cells: Evidence for a Caspase-8-Dependent, Fas-Associated Death Domain-Independent Mechanism. J. Immunol. 2001, 167, 3565–3569. [Google Scholar] [CrossRef]
- Sisirak, V.; Faget, J.; Vey, N.; Blay, J.-Y.; Ménétrier-Caux, C.; Caux, C.; Bendriss-Vermare, N. Plasmacytoid Dendritic Cells Deficient in IFNα Production Promote the Amplification of FOXP3+ Regulatory T Cells and Are Associated with Poor Prognosis in Breast Cancer Patients. Oncoimmunology 2013, 2, e22338. [Google Scholar] [CrossRef]
- Pinto, A.; Rega, A.; Crother, T.R.; Sorrentino, R. Plasmacytoid Dendritic Cells and Their Therapeutic Activity in Cancer. Oncoimmunology 2012, 1, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Aspord, C.; Leccia, M.-T.; Charles, J.; Plumas, J. Melanoma Hijacks Plasmacytoid Dendritic Cells to Promote Its Own Progression. Oncoimmunology 2014, 3, e27402. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Das, D.S.; Song, Y.; Macri, V.; Richardson, P.; Brooks, C.L.; Chauhan, D.; Anderson, K.C. A Novel Agent SL-401 Induces Anti-Myeloma Activity by Targeting Plasmacytoid Dendritic Cells, Osteoclastogenesis and Cancer Stem-like Cells. Leukemia 2017, 31, 2652–2660. [Google Scholar] [CrossRef] [PubMed]
- Blazar, B.R.; Lindberg, F.P.; Ingulli, E.; Panoskaltsis-Mortari, A.; Oldenborg, P.A.; Iizuka, K.; Yokoyama, W.M.; Taylor, P.A. CD47 (Integrin-Associated Protein) Engagement of Dendritic Cell and Macrophage Counterreceptors Is Required to Prevent the Clearance of Donor Lymphohematopoietic Cells. J. Exp. Med. 2001, 194, 541–549. [Google Scholar] [CrossRef]
- Barkal, A.A.; Weiskopf, K.; Kao, K.S.; Gordon, S.R.; Rosental, B.; Yiu, Y.Y.; George, B.M.; Markovic, M.; Ring, N.G.; Tsai, J.M.; et al. Engagement of MHC Class I by the Inhibitory Receptor LILRB1 Suppresses Macrophages and Is a Target of Cancer Immunotherapy. Nat. Immunol. 2018, 19, 76–84. [Google Scholar] [CrossRef]
- Chen, G.-Y.; Tang, J.; Zheng, P.; Liu, Y. CD24 and Siglec-10 Selectively Repress Tissue Damage-Induced Immune Responses. Science 2009, 323, 1722–1725. [Google Scholar] [CrossRef]
- Halpert, M.M.; Konduri, V.; Liang, D.; Chen, Y.; Wing, J.B.; Paust, S.; Levitt, J.M.; Decker, W.K. Dendritic Cell-Secreted Cytotoxic T-Lymphocyte-Associated Protein-4 Regulates the T-Cell Response by Downmodulating Bystander Surface B7. Stem. Cells Dev. 2016, 25, 774–787. [Google Scholar] [CrossRef]
- Yao, S.; Wang, S.; Zhu, Y.; Luo, L.; Zhu, G.; Flies, S.; Xu, H.; Ruff, W.; Broadwater, M.; Choi, I.-H.; et al. PD-1 on Dendritic Cells Impedes Innate Immunity against Bacterial Infection. Blood 2009, 113, 5811–5818. [Google Scholar] [CrossRef]
- Karyampudi, L.; Lamichhane, P.; Krempski, J.; Kalli, K.R.; Behrens, M.D.; Vargas, D.M.; Hartmann, L.C.; Janco, J.M.T.; Dong, H.; Hedin, K.E.; et al. PD-1 Blunts the Function of Ovarian Tumor-Infiltrating Dendritic Cells by Inactivating NF-ΚB. Cancer Res. 2016, 76, 239–250. [Google Scholar] [CrossRef]
- Schwartz, J.A.; Clayton, K.L.; Mujib, S.; Zhang, H.; Rahman, A.K.M.N.-U.; Liu, J.; Yue, F.Y.; Benko, E.; Kovacs, C.; Ostrowski, M.A. Tim-3 Is a Marker of Plasmacytoid Dendritic Cell Dysfunction during HIV Infection and Is Associated with the Recruitment of IRF7 and P85 into Lysosomes and with the Submembrane Displacement of TLR9. J. Immunol. 2017, 198, 3181–3194. [Google Scholar] [CrossRef]
- de Mingo Pulido, Á.; Gardner, A.; Hiebler, S.; Soliman, H.; Rugo, H.S.; Krummel, M.F.; Coussens, L.M.; Ruffell, B. TIM-3 Regulates CD103+ Dendritic Cell Function and Response to Chemotherapy in Breast Cancer. Cancer Cell 2018, 33, 60–74.e6. [Google Scholar] [CrossRef]
- Chiba, S.; Baghdadi, M.; Akiba, H.; Yoshiyama, H.; Kinoshita, I.; Dosaka-Akita, H.; Fujioka, Y.; Ohba, Y.; Gorman, J.V.; Colgan, J.D.; et al. Tumor-Infiltrating DCs Suppress Nucleic Acid-Mediated Innate Immune Responses through Interactions between the Receptor TIM-3 and the Alarmin HMGB1. Nat. Immunol. 2012, 13, 832–842. [Google Scholar] [CrossRef]
- Menetrier-Caux, C.; Montmain, G.; Dieu, M.C.; Bain, C.; Favrot, M.C.; Caux, C.; Blay, J.Y. Inhibition of the Differentiation of Dendritic Cells from CD34(+) Progenitors by Tumor Cells: Role of Interleukin-6 and Macrophage Colony-Stimulating Factor. Blood 1998, 92, 4778–4791. [Google Scholar] [CrossRef]
- Oyama, T.; Ran, S.; Ishida, T.; Nadaf, S.; Kerr, L.; Carbone, D.P.; Gabrilovich, D.I. Vascular Endothelial Growth Factor Affects Dendritic Cell Maturation through the Inhibition of Nuclear Factor-Kappa B Activation in Hemopoietic Progenitor Cells. J. Immunol. 1998, 160, 1224–1232. [Google Scholar]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Cañellas, A.; Hernando-Momblona, X.; et al. TGFβ Drives Immune Evasion in Genetically Reconstituted Colon Cancer Metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef]
- Paul, F.; Arkin, Y.; Giladi, A.; Jaitin, D.A.; Kenigsberg, E.; Keren-Shaul, H.; Winter, D.; Lara-Astiaso, D.; Gury, M.; Weiner, A.; et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 2015, 163, 1663–1677. [Google Scholar] [CrossRef]
- Bartels, M.; Govers, A.M.; Fleskens, V.; Lourenço, A.R.; Pals, C.E.; Vervoort, S.J.; van Gent, R.; Brenkman, A.B.; Bierings, M.B.; Ackerman, S.J.; et al. Acetylation of C/EBPε Is a Prerequisite for Terminal Neutrophil Differentiation. Blood 2015, 125, 1782–1792. [Google Scholar] [CrossRef]
- Cowland, J.B.; Borregaard, N. Granulopoiesis and Granules of Human Neutrophils. Immunol. Rev. 2016, 273, 11–28. [Google Scholar] [CrossRef]
- Ma, Y.; Yabluchanskiy, A.; Iyer, R.P.; Cannon, P.L.; Flynn, E.R.; Jung, M.; Henry, J.; Cates, C.A.; Deleon-Pennell, K.Y.; Lindsey, M.L. Temporal Neutrophil Polarization Following Myocardial Infarction. Cardiovasc. Res. 2016, 110, 51–61. [Google Scholar] [CrossRef]
- Jiao, J.; Dragomir, A.-C.; Kocabayoglu, P.; Rahman, A.H.; Chow, A.; Hashimoto, D.; Leboeuf, M.; Kraus, T.; Moran, T.; Carrasco-Avino, G.; et al. Central Role of Conventional Dendritic Cells in Regulation of Bone Marrow Release and Survival of Neutrophils. J. Immunol. 2014, 192, 3374–3382. [Google Scholar] [CrossRef]
- Phillipson, M.; Kubes, P. The Neutrophil in Vascular Inflammation. Nat. Med. 2011, 17, 1381–1390. [Google Scholar] [CrossRef]
- Petri, B.; Phillipson, M.; Kubes, P. The Physiology of Leukocyte Recruitment: An in Vivo Perspective. J. Immunol. 2008, 180, 6439–6446. [Google Scholar] [CrossRef]
- Sheshachalam, A.; Srivastava, N.; Mitchell, T.; Lacy, P.; Eitzen, G. Granule Protein Processing and Regulated Secretion in Neutrophils. Front. Immunol. 2014, 5, 448. [Google Scholar] [CrossRef]
- Kolaczkowska, E.; Jenne, C.N.; Surewaard, B.G.J.; Thanabalasuriar, A.; Lee, W.-Y.; Sanz, M.-J.; Mowen, K.; Opdenakker, G.; Kubes, P. Molecular Mechanisms of NET Formation and Degradation Revealed by Intravital Imaging in the Liver Vasculature. Nat. Com. 2015, 6, 6673. [Google Scholar] [CrossRef]
- Jorch, S.K.; Kubes, P. An Emerging Role for Neutrophil Extracellular Traps in Noninfectious Disease. Nat. Med. 2017, 23, 279–287. [Google Scholar] [CrossRef]
- Dalli, J.; Montero-Melendez, T.; Norling, L.V.; Yin, X.; Hinds, C.; Haskard, D.; Mayr, M.; Perretti, M. Heterogeneity in Neutrophil Microparticles Reveals Distinct Proteome and Functional Properties. Mol. Cell Prot. 2013, 12, 2205–2219. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. Regulation of the Inflammatory Response in Cardiac Repair. Circul. Res. 2012, 110, 159–173. [Google Scholar] [CrossRef]
- Wang, J. Neutrophils in Tissue Injury and Repair. Cell Tissue Res. 2018, 371, 531–539. [Google Scholar] [CrossRef]
- Sagiv, J.Y.; Michaeli, J.; Assi, S.; Mishalian, I.; Kisos, H.; Levy, L.; Damti, P.; Lumbroso, D.; Polyansky, L.; Sionov, R.V.; et al. Phenotypic Diversity and Plasticity in Circulating Neutrophil Subpopulations in Cancer. Cell Rep. 2015, 10, 562–573. [Google Scholar] [CrossRef]
- Fridlender, Z.G.; Albelda, S.M. Tumor-Associated Neutrophils: Friend or Foe? Carcinogenesis 2012, 33, 949–955. [Google Scholar] [CrossRef]
- Andzinski, L.; Wu, C.-F.; Lienenklaus, S.; Kröger, A.; Weiss, S.; Jablonska, J. Delayed Apoptosis of Tumor Associated Neutrophils in the Absence of Endogenous IFN-β. Int. J. Cancer 2015, 136, 572–583. [Google Scholar] [CrossRef]
- Andzinski, L.; Kasnitz, N.; Stahnke, S.; Wu, C.-F.; Gereke, M.; von Köckritz-Blickwede, M.; Schilling, B.; Brandau, S.; Weiss, S.; Jablonska, J. Type I IFNs Induce Anti-Tumor Polarization of Tumor Associated Neutrophils in Mice and Human. Int. J. Cancer 2016, 138, 1982–1993. [Google Scholar] [CrossRef]
- Ohms, M.; Möller, S.; Laskay, T. An Attempt to Polarize Human Neutrophils Toward N1 and N2 Phenotypes In Vitro. Front. Immunol. 2020, 11, 532. [Google Scholar] [CrossRef]
- Triner, D.; Devenport, S.N.; Ramakrishnan, S.K.; Ma, X.; Frieler, R.A.; Greenson, J.K.; Inohara, N.; Nunez, G.; Colacino, J.A.; Mortensen, R.M.; et al. Neutrophils Restrict Tumor-Associated Microbiota to Reduce Growth and Invasion of Colon Tumors in Mice. Gastroenterology 2019, 156, 1467–1482. [Google Scholar] [CrossRef]
- Mahiddine, K.; Blaisdell, A.; Ma, S.; Créquer-Grandhomme, A.; Lowell, C.A.; Erlebacher, A. Relief of Tumor Hypoxia Unleashes the Tumoricidal Potential of Neutrophils. J. Clin. Investig. 2020, 130, 389–403. [Google Scholar] [CrossRef]
- Mensurado, S.; Rei, M.; Lança, T.; Ioannou, M.; Gonçalves-Sousa, N.; Kubo, H.; Malissen, M.; Papayannopoulos, V.; Serre, K.; Silva-Santos, B. Tumor-Associated Neutrophils Suppress pro-Tumoral IL-17+ Γδ T Cells through Induction of Oxidative Stress. PLoS Biol. 2018, 16, e2004990. [Google Scholar] [CrossRef]
- Costanzo-Garvey, D.L.; Keeley, T.; Case, A.J.; Watson, G.F.; Alsamraae, M.; Yu, Y.; Su, K.; Heim, C.E.; Kielian, T.; Morrissey, C.; et al. Neutrophils Are Mediators of Metastatic Prostate Cancer Progression in Bone. Cancer Immunol. Immunother. 2020, 69, 1113–1130. [Google Scholar] [CrossRef]
- Martin, A.; Seignez, C.; Racoeur, C.; Isambert, N.; Mabrouk, N.; Scagliarini, A.; Reveneau, S.; Arnould, L.; Bettaieb, A.; Jeannin, J.-F.; et al. Tumor-Derived Granzyme B-Expressing Neutrophils Acquire Antitumor Potential after Lipid A Treatment. Oncotarget 2018, 9, 28364–28378. [Google Scholar] [CrossRef]
- Sionov, R.V.; Fainsod-Levi, T.; Zelter, T.; Polyansky, L.; Pham, C.T.; Granot, Z. Neutrophil Cathepsin G and Tumor Cell RAGE Facilitate Neutrophil Anti-Tumor Cytotoxicity. Oncoimmunology 2019, 8, e1624129. [Google Scholar] [CrossRef]
- Eruslanov, E.B.; Bhojnagarwala, P.S.; Quatromoni, J.G.; Stephen, T.L.; Ranganathan, A.; Deshpande, C.; Akimova, T.; Vachani, A.; Litzky, L.; Hancock, W.W.; et al. Tumor-Associated Neutrophils Stimulate T Cell Responses in Early-Stage Human Lung Cancer. J. Clin. Investig. 2014, 124, 5466–5480. [Google Scholar] [CrossRef]
- Hagerling, C.; Gonzalez, H.; Salari, K.; Wang, C.-Y.; Lin, C.; Robles, I.; van Gogh, M.; Dejmek, A.; Jirström, K.; Werb, Z. Immune Effector Monocyte-Neutrophil Cooperation Induced by the Primary Tumor Prevents Metastatic Progression of Breast Cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 21704–21714. [Google Scholar] [CrossRef]
- Governa, V.; Trella, E.; Mele, V.; Tornillo, L.; Amicarella, F.; Cremonesi, E.; Muraro, M.G.; Xu, H.; Droeser, R.; Däster, S.R.; et al. The Interplay Between Neutrophils and CD8+ T Cells Improves Survival in Human Colorectal Cancer. Clin. Cancer Res. 2017, 23, 3847–3858. [Google Scholar] [CrossRef]
- Yu, L.; Zhong, L.; Xiong, L.; Dan, W.; Li, J.; Ye, J.; Wan, P.; Luo, X.; Chu, X.; Liu, C.; et al. Neutrophil Elastase-Mediated Proteolysis of the Tumor Suppressor P200 CUX1 Promotes Cell Proliferation and Inhibits Cell Differentiation in APL. Life Sci. 2020, 242, 117229. [Google Scholar] [CrossRef]
- Hattar, K.; Franz, K.; Ludwig, M.; Sibelius, U.; Wilhelm, J.; Lohmeyer, J.; Savai, R.; Subtil, F.S.B.; Dahlem, G.; Eul, B.; et al. Interactions between Neutrophils and Non-Small Cell Lung Cancer Cells: Enhancement of Tumor Proliferation and Inflammatory Mediator Synthesis. Cancer Immunol. Immunother. 2014, 63, 1297–1306. [Google Scholar] [CrossRef]
- Zha, C.; Meng, X.; Li, L.; Mi, S.; Qian, D.; Li, Z.; Wu, P.; Hu, S.; Zhao, S.; Cai, J.; et al. Neutrophil Extracellular Traps Mediate the Crosstalk between Glioma Progression and the Tumor Microenvironment via the HMGB1/RAGE/IL-8 Axis. Cancer Biol. Med. 2020, 17, 154–168. [Google Scholar] [CrossRef]
- Miller-Ocuin, J.L.; Liang, X.; Boone, B.A.; Doerfler, W.R.; Singhi, A.D.; Tang, D.; Kang, R.; Lotze, M.T.; Zeh, H.J. DNA Released from Neutrophil Extracellular Traps (NETs) Activates Pancreatic Stellate Cells and Enhances Pancreatic Tumor Growth. Oncoimmunology 2019, 8, e1605822. [Google Scholar] [CrossRef]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils Escort Circulating Tumour Cells to Enable Cell Cycle Progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef]
- Liang, W.; Li, Q.; Ferrara, N. Metastatic Growth Instructed by Neutrophil-Derived Transferrin. Proc. Natl. Acad. Sci. USA 2018, 115, 11060–11065. [Google Scholar] [CrossRef]
- Spicer, J.D.; McDonald, B.; Cools-Lartigue, J.J.; Chow, S.C.; Giannias, B.; Kubes, P.; Ferri, L.E. Neutrophils Promote Liver Metastasis via Mac-1-Mediated Interactions with Circulating Tumor Cells. Cancer Res. 2012, 72, 3919–3927. [Google Scholar] [CrossRef]
- Huh, S.J.; Liang, S.; Sharma, A.; Dong, C.; Robertson, G.P. Transiently Entrapped Circulating Tumor Cells Interact with Neutrophils to Facilitate Lung Metastasis Development. Cancer Res. 2010, 70, 6071–6082. [Google Scholar] [CrossRef] [PubMed]
- Singel, K.L.; Emmons, T.R.; Khan, A.N.H.; Mayor, P.C.; Shen, S.; Wong, J.T.; Morrell, K.; Eng, K.H.; Mark, J.; Bankert, R.B.; et al. Mature Neutrophils Suppress T Cell Immunity in Ovarian Cancer Microenvironment. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.; Botta, C.; Zabaleta, A.; Puig, N.; Cedena, M.-T.; Goicoechea, I.; Alameda, D.; San José-Eneriz, E.; Merino, J.; Rodríguez-Otero, P.; et al. Immunogenomic Identification and Characterization of Granulocytic Myeloid-Derived Suppressor Cells in Multiple Myeloma. Blood 2020, 136, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Forsthuber, A.; Lipp, K.; Andersen, L.; Ebersberger, S.; Graña-Castro, O.; Ellmeier, W.; Petzelbauer, P.; Lichtenberger, B.M.; Loewe, R. CXCL5 as Regulator of Neutrophil Function in Cutaneous Melanoma. J. Investig. Dermatol. 2019, 139, 186–194. [Google Scholar] [CrossRef]
- Khou, S.; Popa, A.; Luci, C.; Bihl, F.; Meghraoui-Kheddar, A.; Bourdely, P.; Salavagione, E.; Cosson, E.; Rubod, A.; Cazareth, J.; et al. Tumor-Associated Neutrophils Dampen Adaptive Immunity and Promote Cutaneous Squamous Cell Carcinoma Development. Cancers 2020, 12, 1860. [Google Scholar] [CrossRef]
- Shang, A.; Wang, W.; Gu, C.; Chen, C.; Zeng, B.; Yang, Y.; Ji, P.; Sun, J.; Wu, J.; Lu, W.; et al. Long Non-Coding RNA HOTTIP Enhances IL-6 Expression to Potentiate Immune Escape of Ovarian Cancer Cells by Upregulating the Expression of PD-L1 in Neutrophils. J. Exp. Clin. Cancer Res. 2019, 38, 411. [Google Scholar] [CrossRef]
- Sceneay, J.; Chow, M.T.; Chen, A.; Halse, H.M.; Wong, C.S.F.; Andrews, D.M.; Sloan, E.K.; Parker, B.S.; Bowtell, D.D.; Smyth, M.J.; et al. Primary Tumor Hypoxia Recruits CD11b+/Ly6Cmed/Ly6G+ Immune Suppressor Cells and Compromises NK Cell Cytotoxicity in the Premetastatic Niche. Cancer Res. 2012, 72, 3906–3911. [Google Scholar] [CrossRef]
- Weber, R.; Fleming, V.; Hu, X.; Nagibin, V.; Groth, C.; Altevogt, P.; Utikal, J.; Umansky, V. Myeloid-Derived Suppressor Cells Hinder the Anti-Cancer Activity of Immune Checkpoint Inhibitors. Front. Immunol. 2018, 9, 1310. [Google Scholar] [CrossRef]
- Khaled, Y.S.; Ammori, B.J.; Elkord, E. Myeloid-Derived Suppressor Cells in Cancer: Recent Progress and Prospects. Immunol. Cell Biol. 2013, 91, 493–502. [Google Scholar] [CrossRef]
- Agrati, C.; Sacchi, A.; Bordoni, V.; Cimini, E.; Notari, S.; Grassi, G.; Casetti, R.; Tartaglia, E.; Lalle, E.; D’Abramo, A.; et al. Expansion of Myeloid-Derived Suppressor Cells in Patients with Severe Coronavirus Disease (COVID-19). Cell Death Differ. 2020, 27, 3196–3207. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated Regulation of Myeloid Cells by Tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Dumitru, C.A.; Moses, K.; Trellakis, S.; Lang, S.; Brandau, S. Neutrophils and Granulocytic Myeloid-Derived Suppressor Cells: Immunophenotyping, Cell Biology and Clinical Relevance in Human Oncology. Cancer Immunol. Immunother. 2012, 61, 1155–1167. [Google Scholar] [CrossRef]
- Talmadge, J.E.; Gabrilovich, D.I. History of Myeloid-Derived Suppressor Cells. Nat. Rev. Cancer 2013, 13, 739–752. [Google Scholar] [CrossRef]
- Bronte, V.; Brandau, S.; Chen, S.-H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for Myeloid-Derived Suppressor Cell Nomenclature and Characterization Standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef]
- Poschke, I.; Kiessling, R. On the Armament and Appearances of Human Myeloid-Derived Suppressor Cells. Clin. Immunol. 2012, 144, 250–268. [Google Scholar] [CrossRef]
- Youn, J.-I.; Gabrilovich, D.I. The Biology of Myeloid-Derived Suppressor Cells: The Blessing and the Curse of Morphological and Functional Heterogeneity. Eur. J. Immunol. 2010, 40, 2969–2975. [Google Scholar] [CrossRef]
- Meirow, Y.; Kanterman, J.; Baniyash, M. Paving the Road to Tumor Development and Spreading: Myeloid-Derived Suppressor Cells Are Ruling the Fate. Front. Immunol. 2015, 6, 523. [Google Scholar] [CrossRef]
- Lahat, N.; Rahat, M.A.; Ballan, M.; Weiss-Cerem, L.; Engelmayer, M.; Bitterman, H. Hypoxia Reduces CD80 Expression on Monocytes but Enhances Their LPS-Stimulated TNF-Alpha Secretion. J. Leukoc. Biol. 2003, 74, 197–205. [Google Scholar] [CrossRef]
- Huang, M.; Wu, R.; Chen, L.; Peng, Q.; Li, S.; Zhang, Y.; Zhou, L.; Duan, L. S100A9 Regulates MDSCs-Mediated Immune Suppression via the RAGE and TLR4 Signaling Pathways in Colorectal Carcinoma. Front. Immunol. 2019, 10, 2243. [Google Scholar] [CrossRef]
- Zhao, F.; Hoechst, B.; Duffy, A.; Gamrekelashvili, J.; Fioravanti, S.; Manns, M.P.; Greten, T.F.; Korangy, F. S100A9 a New Marker for Monocytic Human Myeloid-Derived Suppressor Cells. Immunology 2012, 136, 176–183. [Google Scholar] [CrossRef]
- Condamine, T.; Dominguez, G.A.; Youn, J.-I.; Kossenkov, A.V.; Mony, S.; Alicea-Torres, K.; Tcyganov, E.; Hashimoto, A.; Nefedova, Y.; Lin, C.; et al. Lectin-Type Oxidized LDL Receptor-1 Distinguishes Population of Human Polymorphonuclear Myeloid-Derived Suppressor Cells in Cancer Patients. Sci. Immunol. 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Bruderek, K.; Skrzeczynska-Moncznik, J.; Osiecka, O.; Hu, X.; Rundgren, I.M.; Lin, A.; Santegoets, K.; Horzum, U.; Godinho-Santos, A.; et al. Differential Expansion of Circulating Human MDSC Subsets in Patients with Cancer, Infection and Inflammation. J. Immunother. Cancer 2020, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Alshetaiwi, H.; Pervolarakis, N.; McIntyre, L.L.; Ma, D.; Nguyen, Q.; Rath, J.A.; Nee, K.; Hernandez, G.; Evans, K.; Torosian, L.; et al. Defining the Emergence of Myeloid-Derived Suppressor Cells in Breast Cancer Using Single-Cell Transcriptomics. Sci. Immunol. 2020, 5, 2–28. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, S.; Gupta, K.; Pisarev, V.; Kinarsky, L.; Sherman, S.; Kang, L.; Herber, D.L.; Schneck, J.; Gabrilovich, D.I. Altered Recognition of Antigen Is a Mechanism of CD8+ T Cell Tolerance in Cancer. Nat. Med. 2007, 13, 828–835. [Google Scholar] [CrossRef]
- Raber, P.L.; Thevenot, P.; Sierra, R.; Wyczechowska, D.; Halle, D.; Ramirez, M.E.; Ochoa, A.C.; Fletcher, M.; Velasco, C.; Wilk, A.; et al. Subpopulations of Myeloid-Derived Suppressor Cells Impair T Cell Responses through Independent Nitric Oxide-Related Pathways. Int. J. Cancer 2014, 134, 2853–2864. [Google Scholar] [CrossRef]
- Zhou, L.; Miao, K.; Yin, B.; Li, H.; Fan, J.; Zhu, Y.; Ba, H.; Zhang, Z.; Chen, F.; Wang, J.; et al. Cardioprotective Role of Myeloid-Derived Suppressor Cells in Heart Failure. Circulation 2018, 138, 181–197. [Google Scholar] [CrossRef]
- Crook, K.R.; Jin, M.; Weeks, M.F.; Rampersad, R.R.; Baldi, R.M.; Glekas, A.S.; Shen, Y.; Esserman, D.A.; Little, P.; Schwartz, T.A.; et al. Myeloid-Derived Suppressor Cells Regulate T Cell and B Cell Responses during Autoimmune Disease. J. Leukoc. Biol. 2015, 97, 573–582. [Google Scholar] [CrossRef]
- Kurkó, J.; Vida, A.; Ocskó, T.; Tryniszewska, B.; Rauch, T.A.; Glant, T.T.; Szekanecz, Z.; Mikecz, K. Suppression of Proteoglycan-Induced Autoimmune Arthritis by Myeloid-Derived Suppressor Cells Generated in Vitro from Murine Bone Marrow. PLoS ONE 2014, 9, e111815. [Google Scholar] [CrossRef]
- Ioannou, M.; Alissafi, T.; Lazaridis, I.; Deraos, G.; Matsoukas, J.; Gravanis, A.; Mastorodemos, V.; Plaitakis, A.; Sharpe, A.; Boumpas, D.; et al. Crucial Role of Granulocytic Myeloid-Derived Suppressor Cells in the Regulation of Central Nervous System Autoimmune Disease. J. Immunol. 2012, 188, 1136–1146. [Google Scholar] [CrossRef]
- Bergenfelz, C.; Leandersson, K. The Generation and Identity of Human Myeloid-Derived Suppressor Cells. Front. Oncol. 2020, 10, 109. [Google Scholar] [CrossRef]
- Bernard, V.; Semaan, A.; Huang, J.; San Lucas, F.A.; Mulu, F.C.; Stephens, B.M.; Guerrero, P.A.; Huang, Y.; Zhao, J.; Kamyabi, N.; et al. Single-Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. Clin. Cancer Res. 2019, 25, 2194–2205. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Donthireddy, L.; Marvel, D.; Condamine, T.; Wang, F.; Lavilla-Alonso, S.; Hashimoto, A.; Vonteddu, P.; Behera, R.; Goins, M.A.; et al. Cancer-Associated Fibroblasts Neutralize the Anti-Tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors. Cancer Cell 2017, 32, 654–668.e5. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Sun, W.; Shang, W.; Li, Y.; Zhang, D.; Wang, T.; Zhang, X.; Zhang, S.; Zhang, Y.; Yang, R. Lnc-C/EBPβ Negatively Regulates the Suppressive Function of Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2018, 6, 1352–1363. [Google Scholar] [CrossRef] [PubMed]
- Alkhateeb, T.; Kumbhare, A.; Bah, I.; Youssef, D.; Yao, Z.Q.; McCall, C.E.; El Gazzar, M. S100A9 Maintains Myeloid-Derived Suppressor Cells in Chronic Sepsis by Inducing MiR-21 and MiR-181b. Mol. Immunol. 2019, 112, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Corzo, C.A.; Luetteke, N.; Yu, B.; Nagaraj, S.; Bui, M.M.; Ortiz, M.; Nacken, W.; Sorg, C.; Vogl, T.; et al. Inhibition of Dendritic Cell Differentiation and Accumulation of Myeloid-Derived Suppressor Cells in Cancer Is Regulated by S100A9 Protein. J. Exp. Med. 2008, 205, 2235–2249. [Google Scholar] [CrossRef]
- Mazzoni, C.J.; Gomes, C.A.; Souza, N.A.; de Queiroz, R.G.; Justiniano, S.C.B.; Ward, R.D.; Kyriacou, C.P.; Peixoto, A.A. Molecular Evolution of the Period Gene in Sandflies. J. Mol. Evol. 2002, 55, 553–562. [Google Scholar] [CrossRef]
- Steggerda, S.M.; Bennett, M.K.; Chen, J.; Emberley, E.; Huang, T.; Janes, J.R.; Li, W.; MacKinnon, A.L.; Makkouk, A.; Marguier, G.; et al. Inhibition of Arginase by CB-1158 Blocks Myeloid Cell-Mediated Immune Suppression in the Tumor Microenvironment. J. Immunother. Cancer 2017, 5, 101. [Google Scholar] [CrossRef]
- Srivastava, M.K.; Sinha, P.; Clements, V.K.; Rodriguez, P.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cells Inhibit T-Cell Activation by Depleting Cystine and Cysteine. Cancer Res. 2010, 70, 68–77. [Google Scholar] [CrossRef]
- Yu, J.; Du, W.; Yan, F.; Wang, Y.; Li, H.; Cao, S.; Yu, W.; Shen, C.; Liu, J.; Ren, X. Myeloid-Derived Suppressor Cells Suppress Antitumor Immune Responses through IDO Expression and Correlate with Lymph Node Metastasis in Patients with Breast Cancer. J. Immunol. 2013, 190, 3783–3797. [Google Scholar] [CrossRef]
- Baumann, T.; Dunkel, A.; Schmid, C.; Schmitt, S.; Hiltensperger, M.; Lohr, K.; Laketa, V.; Donakonda, S.; Ahting, U.; Lorenz-Depiereux, B.; et al. Regulatory Myeloid Cells Paralyze T Cells through Cell-Cell Transfer of the Metabolite Methylglyoxal. Nat. Immunol. 2020, 21, 555–566. [Google Scholar] [CrossRef]
- Molon, B.; Ugel, S.; Del Pozzo, F.; Soldani, C.; Zilio, S.; Avella, D.; De Palma, A.; Mauri, P.; Monegal, A.; Rescigno, M.; et al. Chemokine Nitration Prevents Intratumoral Infiltration of Antigen-Specific T Cells. J. Exp. Med. 2011, 208, 1949–1962. [Google Scholar] [CrossRef] [PubMed]
- Hanson, E.M.; Clements, V.K.; Sinha, P.; Ilkovitch, D.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cells down-Regulate L-Selectin Expression on CD4+ and CD8+ T Cells. J. Immunol. 2009, 183, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yu, S.; Kappes, J.; Wang, J.; Grizzle, W.E.; Zinn, K.R.; Zhang, H.-G. Expansion of Spleen Myeloid Suppressor Cells Represses NK Cell Cytotoxicity in Tumor-Bearing Host. Blood 2007, 109, 4336–4342. [Google Scholar] [CrossRef] [PubMed]
- Elkabets, M.; Ribeiro, V.S.G.; Dinarello, C.A.; Ostrand-Rosenberg, S.; Di Santo, J.P.; Apte, R.N.; Vosshenrich, C.A.J. IL-1β Regulates a Novel Myeloid-Derived Suppressor Cell Subset That Impairs NK Cell Development and Function. Eur. J. Immunol. 2010, 40, 3347–3357. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, H.; Mabuchi, S.; Yokoi, E.; Komura, N.; Kozasa, K.; Matsumoto, Y.; Kawano, M.; Takahashi, R.; Sasano, T.; Shimura, K.; et al. Prostaglandin E2 Produced by Myeloid-Derived Suppressive Cells Induces Cancer Stem Cells in Uterine Cervical Cancer. Oncotarget 2018, 9, 36317–36330. [Google Scholar] [CrossRef] [PubMed]
- Komura, N.; Mabuchi, S.; Shimura, K.; Yokoi, E.; Kozasa, K.; Kuroda, H.; Takahashi, R.; Sasano, T.; Kawano, M.; Matsumoto, Y.; et al. The Role of Myeloid-Derived Suppressor Cells in Increasing Cancer Stem-like Cells and Promoting PD-L1 Expression in Epithelial Ovarian Cancer. Cancer Immunol. Immunother. 2020, 69, 2477–2499. [Google Scholar] [CrossRef]
- Pan, P.-Y.; Ma, G.; Weber, K.J.; Ozao-Choy, J.; Wang, G.; Yin, B.; Divino, C.M.; Chen, S.-H. Immune Stimulatory Receptor CD40 Is Required for T-Cell Suppression and T Regulatory Cell Activation Mediated by Myeloid-Derived Suppressor Cells in Cancer. Cancer Res. 2010, 70, 99–108. [Google Scholar] [CrossRef]
- Huang, B.; Pan, P.-Y.; Li, Q.; Sato, A.I.; Levy, D.E.; Bromberg, J.; Divino, C.M.; Chen, S.-H. Gr-1+CD115+ Immature Myeloid Suppressor Cells Mediate the Development of Tumor-Induced T Regulatory Cells and T-Cell Anergy in Tumor-Bearing Host. Cancer Res. 2006, 66, 1123–1131. [Google Scholar] [CrossRef]
- Kim, K.; Skora, A.D.; Li, Z.; Liu, Q.; Tam, A.J.; Blosser, R.L.; Diaz, L.A.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B.; et al. Eradication of Metastatic Mouse Cancers Resistant to Immune Checkpoint Blockade by Suppression of Myeloid-Derived Cells. Proc. Natl. Acad. Sci. USA 2014, 111, 11774–11779. [Google Scholar] [CrossRef]
- Romano, A.; Parrinello, N.L.; La Cava, P.; Tibullo, D.; Giallongo, C.; Camiolo, G.; Puglisi, F.; Parisi, M.; Pirosa, M.C.; Martino, E.; et al. PMN-MDSC and Arginase Are Increased in Myeloma and May Contribute to Resistance to Therapy. Expert Rev. Mol. Diagn. 2018, 18, 675–683. [Google Scholar] [CrossRef]
- Maaninka, K.; Lappalainen, J.; Kovanen, P.T. Human Mast Cells Arise from a Common Circulating Progenitor. J. Allergy Clin. Immunol. 2013, 132, 463–469.e3. [Google Scholar] [CrossRef] [PubMed]
- Schmetzer, O.; Valentin, P.; Church, M.K.; Maurer, M.; Siebenhaar, F. Murine and Human Mast Cell Progenitors. Eur. J. Pharmacol. 2016, 778, 2–10. [Google Scholar] [CrossRef]
- Gurish, M.F.; Boyce, J.A. Mast Cell Growth, Differentiation, and Death. Clin. Rev. Allergy Immunol. 2002, 22, 107–118. [Google Scholar] [CrossRef]
- Gentek, R.; Ghigo, C.; Hoeffel, G.; Bulle, M.J.; Msallam, R.; Gautier, G.; Launay, P.; Chen, J.; Ginhoux, F.; Bajénoff, M. Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity 2018, 48, 1160–1171.e5. [Google Scholar] [CrossRef] [PubMed]
- Lundequist, A.; Pejler, G. Biological Implications of Preformed Mast Cell Mediators. Cell Mol. Life Sci. 2011, 68, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Wernersson, S.; Pejler, G. Mast Cell Secretory Granules: Armed for Battle. Nat. Rev. Immunol. 2014, 14, 478–494. [Google Scholar] [CrossRef]
- Espinosa, E.; Valitutti, S. New Roles and Controls of Mast Cells. Curr. Opin. Immunol. 2018, 50, 39–47. [Google Scholar] [CrossRef]
- Komi, D.E.A.; Rambasek, T.; Wöhrl, S. Mastocytosis: From a Molecular Point of View. Clin. Rev. Allergy Immunol. 2018, 54, 397–411. [Google Scholar] [CrossRef]
- Gaudenzio, N.; Laurent, C.; Valitutti, S.; Espinosa, E. Human Mast Cells Drive Memory CD4+ T Cells toward an Inflammatory IL-22+ Phenotype. J. Allergy Clin. Immunol. 2013, 131, 1400–1407.e11. [Google Scholar] [CrossRef]
- Galli, S.J.; Nakae, S.; Tsai, M. Mast Cells in the Development of Adaptive Immune Responses. Nat. Immunol. 2005, 6, 135–142. [Google Scholar] [CrossRef]
- Gilfillan, A.M.; Tkaczyk, C. Integrated Signalling Pathways for Mast-Cell Activation. Nat. Rev. Immunol. 2006, 6, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Recent Advances in Mast Cell Activation and Regulation–PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/32226609/ (accessed on 20 November 2020).
- Agier, J.; Pastwińska, J.; Brzezińska-Błaszczyk, E. An Overview of Mast Cell Pattern Recognition Receptors. Inflamm. Res. 2018, 67, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Saluja, R.; Zoltowska, A.; Ketelaar, M.E.; Nilsson, G. IL-33 and Thymic Stromal Lymphopoietin in Mast Cell Functions. Eur. J. Pharmacol. 2016, 778, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Joulia, R.; Mailhol, C.; Valitutti, S.; Didier, A.; Espinosa, E. Direct Monitoring of Basophil Degranulation by Using Avidin-Based Probes. J. Allergy Clin. Immunol. 2017, 140, 1159–1162.e6. [Google Scholar] [CrossRef] [PubMed]
- Rönnberg, E.; Ghaib, A.; Ceriol, C.; Enoksson, M.; Arock, M.; Säfholm, J.; Ekoff, M.; Nilsson, G. Divergent Effects of Acute and Prolonged Interleukin 33 Exposure on Mast Cell IgE-Mediated Functions. Front. Immunol. 2019, 10, 1361. [Google Scholar] [CrossRef]
- Jönsson, F.; Daëron, M. Mast Cells and Company. Front. Immunol. 2012, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- McNeil, B.D.; Pundir, P.; Meeker, S.; Han, L.; Undem, B.J.; Kulka, M.; Dong, X. Identification of a Mast-Cell-Specific Receptor Crucial for Pseudo-Allergic Drug Reactions. Nature 2015, 519, 237–241. [Google Scholar] [CrossRef]
- Pundir, P.; Liu, R.; Vasavda, C.; Serhan, N.; Limjunyawong, N.; Yee, R.; Zhan, Y.; Dong, X.; Wu, X.; Zhang, Y.; et al. A Connective Tissue Mast-Cell-Specific Receptor Detects Bacterial Quorum-Sensing Molecules and Mediates Antibacterial Immunity. Cell Host Microbe 2019, 26, 114–122.e8. [Google Scholar] [CrossRef]
- Corbière, A.; Loste, A.; Gaudenzio, N. MRGPRX2 Sensing of Cationic Compounds-A Bridge between Nociception and Skin Diseases? Exp. Dermatol. 2020. [Google Scholar] [CrossRef]
- Lu, L.; Kulka, M.; Unsworth, L.D. Peptide-Mediated Mast Cell Activation: Ligand Similarities for Receptor Recognition and Protease-Induced Regulation. J. Leukoc. Biol. 2017, 102, 237–251. [Google Scholar] [CrossRef]
- Abraham, S.N.; St John, A.L. Mast Cell-Orchestrated Immunity to Pathogens. Nat. Rev. Immunol. 2010, 10, 440–452. [Google Scholar] [CrossRef]
- Arizono, N.; Matsuda, S.; Hattori, T.; Kojima, Y.; Maeda, T.; Galli, S.J. Anatomical Variation in Mast Cell Nerve Associations in the Rat Small Intestine, Heart, Lung, and Skin. Similarities of Distances between Neural Processes and Mast Cells, Eosinophils, or Plasma Cells in the Jejunal Lamina Propria. Lab. Investig. 1990, 62, 626–634. [Google Scholar] [PubMed]
- Barbara, G.; Stanghellini, V.; De Giorgio, R.; Cremon, C.; Cottrell, G.S.; Santini, D.; Pasquinelli, G.; Morselli-Labate, A.M.; Grady, E.F.; Bunnett, N.W.; et al. Activated Mast Cells in Proximity to Colonic Nerves Correlate with Abdominal Pain in Irritable Bowel Syndrome. Gastroenterology 2004, 126, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Serhan, N.; Basso, L.; Sibilano, R.; Petitfils, C.; Meixiong, J.; Bonnart, C.; Reber, L.L.; Marichal, T.; Starkl, P.; Cenac, N.; et al. House Dust Mites Activate Nociceptor-Mast Cell Clusters to Drive Type 2 Skin Inflammation. Nat. Immunol. 2019, 20, 1435–1443. [Google Scholar] [CrossRef]
- Suzuki, J.; Isobe, M.; Izawa, A.; Takahashi, W.; Yamazaki, S.; Okubo, Y.; Amano, J.; Sekiguchi, M. Differential Th1 and Th2 Cell Regulation of Murine Cardiac Allograft Acceptance by Blocking Cell Adhesion of ICAM-1/LFA-1 and VCAM-1/VLA-4. Transpl. Immunol. 1999, 7, 65–72. [Google Scholar] [CrossRef]
- Siiskonen, H.; Harvima, I. Mast Cells and Sensory Nerves Contribute to Neurogenic Inflammation and Pruritus in Chronic Skin Inflammation. Front. Cell Neurosci. 2019, 13, 422. [Google Scholar] [CrossRef] [PubMed]
- Stelekati, E.; Bahri, R.; D’Orlando, O.; Orinska, Z.; Mittrücker, H.-W.; Langenhaun, R.; Glatzel, M.; Bollinger, A.; Paus, R.; Bulfone-Paus, S. Mast Cell-Mediated Antigen Presentation Regulates CD8+ T Cell Effector Functions. Immunity 2009, 31, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Lotfi-Emran, S.; Ward, B.R.; Le, Q.T.; Pozez, A.L.; Manjili, M.H.; Woodfolk, J.A.; Schwartz, L.B. Human Mast Cells Present Antigen to Autologous CD4+ T Cells. J. Allergy Clin. Immunol. 2018, 141, 311–321.e10. [Google Scholar] [CrossRef]
- Gaudenzio, N.; Espagnolle, N.; Mars, L.T.; Liblau, R.; Valitutti, S.; Espinosa, E. Cell-Cell Cooperation at the T Helper Cell/Mast Cell Immunological Synapse. Blood 2009, 114, 4979–4988. [Google Scholar] [CrossRef]
- Nakae, S.; Suto, H.; Iikura, M.; Kakurai, M.; Sedgwick, J.D.; Tsai, M.; Galli, S.J. Mast Cells Enhance T Cell Activation: Importance of Mast Cell Costimulatory Molecules and Secreted TNF. J. Immunol. 2006, 176, 2238–2248. [Google Scholar] [CrossRef]
- Shefler, I.; Salamon, P.; Levi-Schaffer, F.; Mor, A.; Hershko, A.Y.; Mekori, Y.A. MicroRNA-4443 Regulates Mast Cell Activation by T Cell-Derived Microvesicles. J. Allergy Clin. Immunol. 2018, 141, 2132–2141.e4. [Google Scholar] [CrossRef] [PubMed]
- Shefler, I.; Salamon, P.; Reshef, T.; Mor, A.; Mekori, Y.A. T Cell-Induced Mast Cell Activation: A Role for Microparticles Released from Activated T Cells. J. Immunol. 2010, 185, 4206–4212. [Google Scholar] [CrossRef] [PubMed]
- Bradding, P.; Pejler, G. The Controversial Role of Mast Cells in Fibrosis. Immunol. Rev. 2018, 282, 198–231. [Google Scholar] [CrossRef] [PubMed]
- Ud-Din, S.; Wilgus, T.A.; Bayat, A. Mast Cells in Skin Scarring: A Review of Animal and Human Research. Front. Immunol. 2020, 11, 552205. [Google Scholar] [CrossRef]
- Pejler, G.; Rönnberg, E.; Waern, I.; Wernersson, S. Mast Cell Proteases: Multifaceted Regulators of Inflammatory Disease. Blood 2010, 115, 4981–4990. [Google Scholar] [CrossRef]
- Vangansewinkel, T.; Lemmens, S.; Geurts, N.; Quanten, K.; Dooley, D.; Pejler, G.; Hendrix, S. Mouse Mast Cell Protease 4 Suppresses Scar Formation after Traumatic Spinal Cord Injury. Sci. Rep. 2019, 9, 3715. [Google Scholar] [CrossRef]
- Galli, S.J.; Gaudenzio, N.; Tsai, M. Mast Cells in Inflammation and Disease: Recent Progress and Ongoing Concerns. Annu. Rev. Immunol. 2020, 38, 49–77. [Google Scholar] [CrossRef]
- Varricchi, G.; Rossi, F.W.; Galdiero, M.R.; Granata, F.; Criscuolo, G.; Spadaro, G.; de Paulis, A.; Marone, G. Physiological Roles of Mast Cells: Collegium Internationale Allergologicum Update 2019. Int. Arch. Allergy Immunol. 2019, 179, 247–261. [Google Scholar] [CrossRef]
- Varricchi, G.; Galdiero, M.R.; Loffredo, S.; Marone, G.; Iannone, R.; Marone, G.; Granata, F. Are Mast Cells MASTers in Cancer? Front. Immunol. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Pittoni, P.; Tripodo, C.; Piconese, S.; Mauri, G.; Parenza, M.; Rigoni, A.; Sangaletti, S.; Colombo, M.P. Mast Cell Targeting Hampers Prostate Adenocarcinoma Development but Promotes the Occurrence of Highly Malignant Neuroendocrine Cancers. Cancer Res. 2011, 71, 5987–5997. [Google Scholar] [CrossRef]
- Carlini, M.J.; Dalurzo, M.C.L.; Lastiri, J.M.; Smith, D.E.; Vasallo, B.C.; Puricelli, L.I.; Lauría de Cidre, L.S. Mast Cell Phenotypes and Microvessels in Non-Small Cell Lung Cancer and Its Prognostic Significance. Hum. Pathol. 2010, 41, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Siiskonen, H.; Poukka, M.; Bykachev, A.; Tyynelä-Korhonen, K.; Sironen, R.; Pasonen-Seppänen, S.; Harvima, I.T. Low Numbers of Tryptase+ and Chymase+ Mast Cells Associated with Reduced Survival and Advanced Tumor Stage in Melanoma. Melanoma Res. 2015, 25, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Welsh, T.J.; Green, R.H.; Richardson, D.; Waller, D.A.; O’Byrne, K.J.; Bradding, P. Macrophage and Mast-Cell Invasion of Tumor Cell Islets Confers a Marked Survival Advantage in Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2005, 23, 8959–8967. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Rudolfsson, S.; Hammarsten, P.; Halin, S.; Pietras, K.; Jones, J.; Stattin, P.; Egevad, L.; Granfors, T.; Wikström, P.; et al. Mast Cells Are Novel Independent Prognostic Markers in Prostate Cancer and Represent a Target for Therapy. Am. J. Pathol. 2010, 177, 1031–1041. [Google Scholar] [CrossRef]
- Fleischmann, A.; Schlomm, T.; Köllermann, J.; Sekulic, N.; Huland, H.; Mirlacher, M.; Sauter, G.; Simon, R.; Erbersdobler, A. Immunological Microenvironment in Prostate Cancer: High Mast Cell Densities Are Associated with Favorable Tumor Characteristics and Good Prognosis. Prostate 2009, 69, 976–981. [Google Scholar] [CrossRef]
- Ribatti, D.; Vacca, A.; Ria, R.; Marzullo, A.; Nico, B.; Filotico, R.; Roncali, L.; Dammacco, F. Neovascularisation, Expression of Fibroblast Growth Factor-2, and Mast Cells with Tryptase Activity Increase Simultaneously with Pathological Progression in Human Malignant Melanoma. Eur. J. Cancer 2003, 39, 666–674. [Google Scholar] [CrossRef]
- Takebayashi, S.; Jimi, S.; Kawamoto, N. Cutaneous Malignant Melanoma: Correlation between Neovascularization and Peritumor Accumulation of Mast Cells Overexpressing Vascular Endothelial Growth Factor. Hum. Pathol. 2000, 31, 955–960. [Google Scholar] [CrossRef]
- Imada, A.; Shijubo, N.; Kojima, H.; Abe, S. Mast Cells Correlate Angiogenesis and Poor Outcome in Stage I Lung Adenocarcinoma. Eur. Respir. J. 2000, 15, 1087–1093. [Google Scholar] [CrossRef]
- Esposito, I.; Menicagli, M.; Funel, N.; Bergmann, F.; Boggi, U.; Mosca, F.; Bevilacqua, G.; Campani, D. Inflammatory Cells Contribute to the Generation of an Angiogenic Phenotype in Pancreatic Ductal Adenocarcinoma. J. Clin. Pathol. 2004, 57, 630–636. [Google Scholar] [CrossRef]
- Gorzalczany, Y.; Merimsky, O.; Sagi-Eisenberg, R. Mast Cells Are Directly Activated by Cancer Cell–Derived Extracellular Vesicles by a CD73- and Adenosine-Dependent Mechanism. Transl. Oncol. 2019, 12, 1549–1556. [Google Scholar] [CrossRef]
- Visciano, C.; Liotti, F.; Prevete, N.; Cali’, G.; Franco, R.; Collina, F.; De Paulis, A.; Marone, G.; Santoro, M.; Melillo, R.M. Mast Cells Induce Epithelial-to-Mesenchymal Transition and Stem Cell Features in Human Thyroid Cancer Cells through an IL-8-Akt-Slug Pathway. Oncogene 2015, 34, 5175–5186. [Google Scholar] [CrossRef] [PubMed]
- Öhrvik, H.; Grujic, M.; Waern, I.; Gustafson, A.M.; Ernst, N.; Roers, A.; Hartmann, K.; Pejler, G. Mast Cells Promote Melanoma Colonization of Lungs. Oncotarget 2016, 7, 68990–69001. [Google Scholar] [CrossRef] [PubMed]
- Giannou, A.D.; Marazioti, A.; Spella, M.; Kanellakis, N.I.; Apostolopoulou, H.; Psallidas, I.; Prijovich, Z.M.; Vreka, M.; Zazara, D.E.; Lilis, I.; et al. Mast Cells Mediate Malignant Pleural Effusion Formation. J. Clin. Investig. 2015, 125, 2317–2334. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Lei, Z.; Zhang, G.M.; Li, D.; Song, C.; Li, B.; Liu, Y.; Yuan, Y.; Unkeless, J.; Xiong, H.; et al. SCF-Mediated Mast Cell Infiltration and Activation Exacerbate the Inflammation and Immunosuppression in Tumor Microenvironment. Blood 2008, 112, 1269–1279. [Google Scholar] [CrossRef]
- Reddy, S.M.; Reuben, A.; Barua, S.; Jiang, H.; Zhang, S.; Wang, L.; Gopalakrishnan, V.; Hudgens, C.W.; Tetzlaff, M.T.; Reuben, J.M.; et al. Poor Response to Neoadjuvant Chemotherapy Correlates with Mast Cell Infiltration in Inflammatory Breast Cancer. Cancer Immunol. Res. 2019, 7, 1025–1035. [Google Scholar] [CrossRef]
- Kaesler, S.; Wölbing, F.; Kempf, W.E.; Skabytska, Y.; Köberle, M.; Volz, T.; Sinnberg, T.; Amaral, T.; Möckel, S.; Yazdi, A.; et al. Targeting Tumor-Resident Mast Cells for Effective Anti-Melanoma Immune Responses. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Zelenay, S.; Van Der Veen, A.G.; Böttcher, J.P.; Snelgrove, K.J.; Rogers, N.; Acton, S.E.; Chakravarty, P.; Girotti, M.R.; Marais, R.; Quezada, S.A.; et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell 2015, 162, 1257–1270. [Google Scholar] [CrossRef]
- Terabe, M.; Park, J.M.; Berzofsky, J.A. Role of IL-13 in Regulation of Anti-Tumor Immunity and Tumor Growth. Cancer Immunol. Immunother. 2004, 53, 79–85. [Google Scholar] [CrossRef]
- Kennedy, L.; Hodges, K.; Meng, F.; Alpini, G.; Francis, H. Histamine and Histamine Receptor Regulation of Gastrointestinal Cancers. Transll. Gastrointest. Cancer 2012, 1, 215–227. [Google Scholar]
- Balkwill, F. Tumour Necrosis Factor and Cancer. Nat. Rev. Cancer 2009, 9, 361–371. [Google Scholar] [CrossRef]
- Arinobu, Y.; Iwasaki, H.; Gurish, M.F.; Mizuno, S.; Shigematsu, H.; Ozawa, H.; Tenen, D.G.; Austen, K.F.; Akashi, K. Developmental Checkpoints of the Basophil/Mast Cell Lineages in Adult Murine Hematopoiesis. Proc. Natl. Acad. Sci. USA 2005, 102, 18105–18110. [Google Scholar] [CrossRef] [PubMed]
- Kopf, M.; Brombacher, F.; Hodgkin, P.D.; Ramsay, A.J.; Milbourne, E.A.; Dai, W.J.; Ovington, K.S.; Behm, C.A.; Köhler, G.; Young, I.G.; et al. IL-5-Deficient Mice Have a Developmental Defect in CD5+ B-1 Cells and Lack Eosinophilia but Have Normal Antibody and Cytotoxic T Cell Responses. Immunity 1996, 4, 15–24. [Google Scholar] [CrossRef]
- Schroeder, J.T.; Chichester, K.L.; Bieneman, A.P. Human Basophils Secrete IL-3: Evidence of Autocrine Priming for Phenotypic and Functional Responses in Allergic Disease. J. Immunol. 2009, 182, 2432–2438. [Google Scholar] [CrossRef] [PubMed]
- Gould, H.J.; Sutton, B.J.; Beavil, A.J.; Beavil, R.L.; McCloskey, N.; Coker, H.A.; Fear, D.; Smurthwaite, L. The Biology of IGE and the Basis of Allergic Disease. Annu. Rev. Immunol. 2003, 21, 579–628. [Google Scholar] [CrossRef] [PubMed]
- Knol, E.F.; Gibbs, B.F. Basophil Stimulation and Signaling Pathways. Methods Mol. Biol. 2014, 1192, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Yuk, C.M.; Park, H.J.; Kwon, B.-I.; Lah, S.J.; Chang, J.; Kim, J.-Y.; Lee, K.-M.; Park, S.-H.; Hong, S.; Lee, S.-H. Basophil-Derived IL-6 Regulates TH17 Cell Differentiation and CD4 T Cell Immunity. Sci. Rep. 2017, 7, 41744. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.W. Eosinophil Activation Status in Separate Compartments and Association with Asthma. Front. Med. 2017, 4, 75. [Google Scholar] [CrossRef]
- Johnston, L.K.; Hsu, C.-L.; Krier-Burris, R.A.; Chhiba, K.D.; Chien, K.B.; McKenzie, A.; Berdnikovs, S.; Bryce, P.J. IL-33 Precedes IL-5 in Regulating Eosinophil Commitment and Is Required for Eosinophil Homeostasis. J. Immunol. 2016, 197, 3445–3453. [Google Scholar] [CrossRef]
- Zimmermann, N.; Hogan, S.P.; Mishra, A.; Brandt, E.B.; Bodette, T.R.; Pope, S.M.; Finkelman, F.D.; Rothenberg, M.E. Murine Eotaxin-2: A Constitutive Eosinophil Chemokine Induced by Allergen Challenge and IL-4 Overexpression. J. Immunol. 2000, 165, 5839–5846. [Google Scholar] [CrossRef]
- Bandeira-Melo, C.; Sugiyama, K.; Woods, L.J.; Weller, P.F. Cutting Edge: Eotaxin Elicits Rapid Vesicular Transport-Mediated Release of Preformed IL-4 from Human Eosinophils. J. Immunol. 2001, 166, 4813–4817. [Google Scholar] [CrossRef]
- Simon, D.; Hoesli, S.; Roth, N.; Staedler, S.; Yousefi, S.; Simon, H.-U. Eosinophil Extracellular DNA Traps in Skin Diseases. J. Allergy Clin. Immunol. 2011, 127, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Munitz, A.; Levi-Schaffer, F. Inhibitory Receptors on Eosinophils: A Direct Hit to a Possible Achilles Heel? J. Allergy Clin. Immunol. 2007, 119, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, Y.-G.; Gao, J.-L.; Lyu, G.-Y.; Su, J.; Zhang, Q.I.; Ji, X.; Yan, J.-Z.; Qiu, Q.-L.; Zhang, Y.-L.; et al. Low Local Blood Perfusion, High White Blood Cell and High Platelet Count Are Associated with Primary Tumor Growth and Lung Metastasis in a 4T1 Mouse Breast Cancer Metastasis Model. Oncol. Lett. 2015, 10, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Prizment, A.E.; Vierkant, R.A.; Smyrk, T.C.; Tillmans, L.S.; Lee, J.J.; Sriramarao, P.; Nelson, H.H.; Lynch, C.F.; Thibodeau, S.N.; Church, T.R.; et al. Tumor Eosinophil Infiltration and Improved Survival of Colorectal Cancer Patients: Iowa Women’s Health Study. Mod. Pathol. 2016, 29, 516–527. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, N.; Qian, L.; Wang, X.; Fan, P.; Kuai, J.; Lin, S.; Liu, C.; Jiang, W.; Qin, S.; et al. CTLA4 Blockade Promotes Vessel Normalization in Breast Tumors via the Accumulation of Eosinophils. Int. J. Cancer 2020, 146, 1730–1740. [Google Scholar] [CrossRef]
- Lucarini, V.; Ziccheddu, G.; Macchia, I.; La Sorsa, V.; Peschiaroli, F.; Buccione, C.; Sistigu, A.; Sanchez, M.; Andreone, S.; D’Urso, M.T.; et al. IL-33 Restricts Tumor Growth and Inhibits Pulmonary Metastasis in Melanoma-Bearing Mice through Eosinophils. Oncoimmunology 2017, 6, e1317420. [Google Scholar] [CrossRef]
- Rosner, S.; Kwong, E.; Shoushtari, A.N.; Friedman, C.F.; Betof, A.S.; Brady, M.S.; Coit, D.G.; Callahan, M.K.; Wolchok, J.D.; Chapman, P.B.; et al. Peripheral Blood Clinical Laboratory Variables Associated with Outcomes Following Combination Nivolumab and Ipilimumab Immunotherapy in Melanoma. Cancer Med. 2018, 7, 690–697. [Google Scholar] [CrossRef]
- Bax, H.J.; Chauhan, J.; Stavraka, C.; Khiabany, A.; Nakamura, M.; Pellizzari, G.; Ilieva, K.M.; Lombardi, S.; Gould, H.J.; Corrigan, C.J.; et al. Basophils from Cancer Patients Respond to Immune Stimuli and Predict Clinical Outcome. Cells 2020, 9, 1631. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, X.; Wang, G.; Zhou, Y.; Luo, M.; Wang, S.; Hong, C. The Impacts of Pretreatment Circulating Eosinophils and Basophils on Prognosis of Stage I–III Colorectal Cancer. Asia Pac. J. Clin. Oncol. 2018, 14, e243–e251. [Google Scholar] [CrossRef]
- Cohen, M.; Giladi, A.; Gorki, A.-D.; Solodkin, D.G.; Zada, M.; Hladik, A.; Miklosi, A.; Salame, T.-M.; Halpern, K.B.; David, E.; et al. Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell 2018, 175, 1031–1044.e18. [Google Scholar] [CrossRef]
- De Monte, L.; Wörmann, S.; Brunetto, E.; Heltai, S.; Magliacane, G.; Reni, M.; Paganoni, A.M.; Recalde, H.; Mondino, A.; Falconi, M.; et al. Basophil Recruitment into Tumor-Draining Lymph Nodes Correlates with Th2 Inflammation and Reduced Survival in Pancreatic Cancer Patients. Cancer Res. 2016, 76, 1792–1803. [Google Scholar] [CrossRef] [PubMed]
- Crawford, G.; Hayes, M.D.; Seoane, R.C.; Ward, S.; Dalessandri, T.; Lai, C.; Healy, E.; Kipling, D.; Proby, C.; Moyes, C.; et al. Epithelial Damage and Tissue Γδ T Cells Promote a Unique Tumor-Protective IgE Response. Nat. Immunol. 2018, 19, 859–870. [Google Scholar] [CrossRef] [PubMed]
- de Paulis, A.; Prevete, N.; Fiorentino, I.; Rossi, F.W.; Staibano, S.; Montuori, N.; Ragno, P.; Longobardi, A.; Liccardo, B.; Genovese, A.; et al. Expression and Functions of the Vascular Endothelial Growth Factors and Their Receptors in Human Basophils. J. Immunol. 2006, 177, 7322–7331. [Google Scholar] [CrossRef] [PubMed]
- Prevete, N.; Staiano, R.I.; Granata, F.; Detoraki, A.; Necchi, V.; Ricci, V.; Triggiani, M.; De Paulis, A.; Marone, G.; Genovese, A. Expression and Function of Angiopoietins and Their Tie Receptors in Human Basophils and Mast Cells. J. Biol. Regul. Homeost. Agents 2013, 27, 827–839. [Google Scholar]
- Cerny-Reiterer, S.; Ghanim, V.; Hoermann, G.; Aichberger, K.J.; Herrmann, H.; Muellauer, L.; Repa, A.; Sillaber, C.; Walls, A.F.; Mayerhofer, M.; et al. Identification of Basophils as a Major Source of Hepatocyte Growth Factor in Chronic Myeloid Leukemia: A Novel Mechanism of BCR-ABL1-Independent Disease Progression. Neoplasia 2012, 14, 572–584. [Google Scholar] [CrossRef]
- MacGlashan, D.W.; Peters, S.P.; Warner, J.; Lichtenstein, L.M. Characteristics of Human Basophil Sulfidopeptide Leukotriene Release: Releasability Defined as the Ability of the Basophil to Respond to Dimeric Cross-Links. J. Immunol. 1986, 136, 2231–2239. [Google Scholar]
- Duah, E.; Teegala, L.R.; Kondeti, V.; Adapala, R.K.; Keshamouni, V.G.; Kanaoka, Y.; Austen, K.F.; Thodeti, C.K.; Paruchuri, S. Cysteinyl Leukotriene 2 Receptor Promotes Endothelial Permeability, Tumor Angiogenesis, and Metastasis. Proc. Natl. Acad. Sci. USA 2019, 116, 199–204. [Google Scholar] [CrossRef]
- Horiuchi, T.; Weller, P.F. Expression of Vascular Endothelial Growth Factor by Human Eosinophils: Upregulation by Granulocyte Macrophage Colony-Stimulating Factor and Interleukin-5. Am. J. Respir. Cell Mol. Biol. 1997, 17, 70–77. [Google Scholar] [CrossRef]
- Ben Efraim, A.H.N.; Eliashar, R.; Levi-Schaffer, F. Hypoxia Modulates Human Eosinophil Function. Clin. Mol. Allergy 2010, 8, 10. [Google Scholar] [CrossRef]
- Puxeddu, I.; Berkman, N.; Ribatti, D.; Bader, R.; Haitchi, H.M.; Davies, D.E.; Howarth, P.H.; Levi-Schaffer, F. Osteopontin Is Expressed and Functional in Human Eosinophils. Allergy 2010, 65, 168–174. [Google Scholar] [CrossRef]
- Cools-Lartigue, J.; Spicer, J.; McDonald, B.; Gowing, S.; Chow, S.; Giannias, B.; Bourdeau, F.; Kubes, P.; Ferri, L. Neutrophil Extracellular Traps Sequester Circulating Tumor Cells and Promote Metastasis. J. Clin. Investig. 2013, 123, 3446–3458. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domagala, M.; Laplagne, C.; Leveque, E.; Laurent, C.; Fournié, J.-J.; Espinosa, E.; Poupot, M. Cancer Cells Resistance Shaping by Tumor Infiltrating Myeloid Cells. Cancers 2021, 13, 165. https://doi.org/10.3390/cancers13020165
Domagala M, Laplagne C, Leveque E, Laurent C, Fournié J-J, Espinosa E, Poupot M. Cancer Cells Resistance Shaping by Tumor Infiltrating Myeloid Cells. Cancers. 2021; 13(2):165. https://doi.org/10.3390/cancers13020165
Chicago/Turabian StyleDomagala, Marcin, Chloé Laplagne, Edouard Leveque, Camille Laurent, Jean-Jacques Fournié, Eric Espinosa, and Mary Poupot. 2021. "Cancer Cells Resistance Shaping by Tumor Infiltrating Myeloid Cells" Cancers 13, no. 2: 165. https://doi.org/10.3390/cancers13020165
APA StyleDomagala, M., Laplagne, C., Leveque, E., Laurent, C., Fournié, J.-J., Espinosa, E., & Poupot, M. (2021). Cancer Cells Resistance Shaping by Tumor Infiltrating Myeloid Cells. Cancers, 13(2), 165. https://doi.org/10.3390/cancers13020165





