MAPK15 Controls Hedgehog Signaling in Medulloblastoma Cells by Regulating Primary Ciliogenesis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Plasmids and Lentiviral Vectors
2.3. Luciferase Reporter Assay
2.4. Medullo-Spheres
2.5. Western Blotting
2.6. Immunofluorescence
2.7. Transient Knock-Down of Endogenous MAPK15
2.8. Cell Count
2.9. Statistical Analysis
3. Results
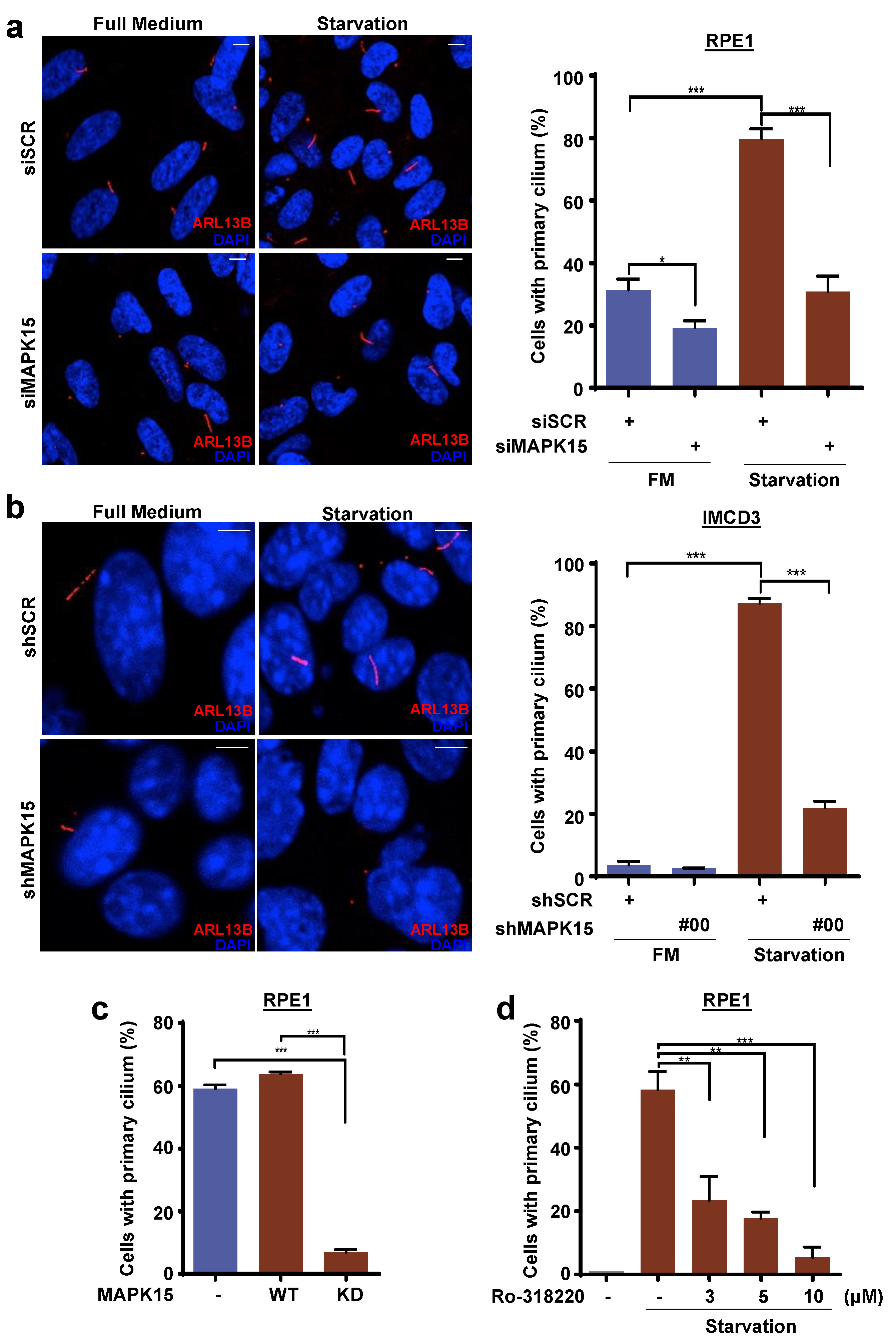
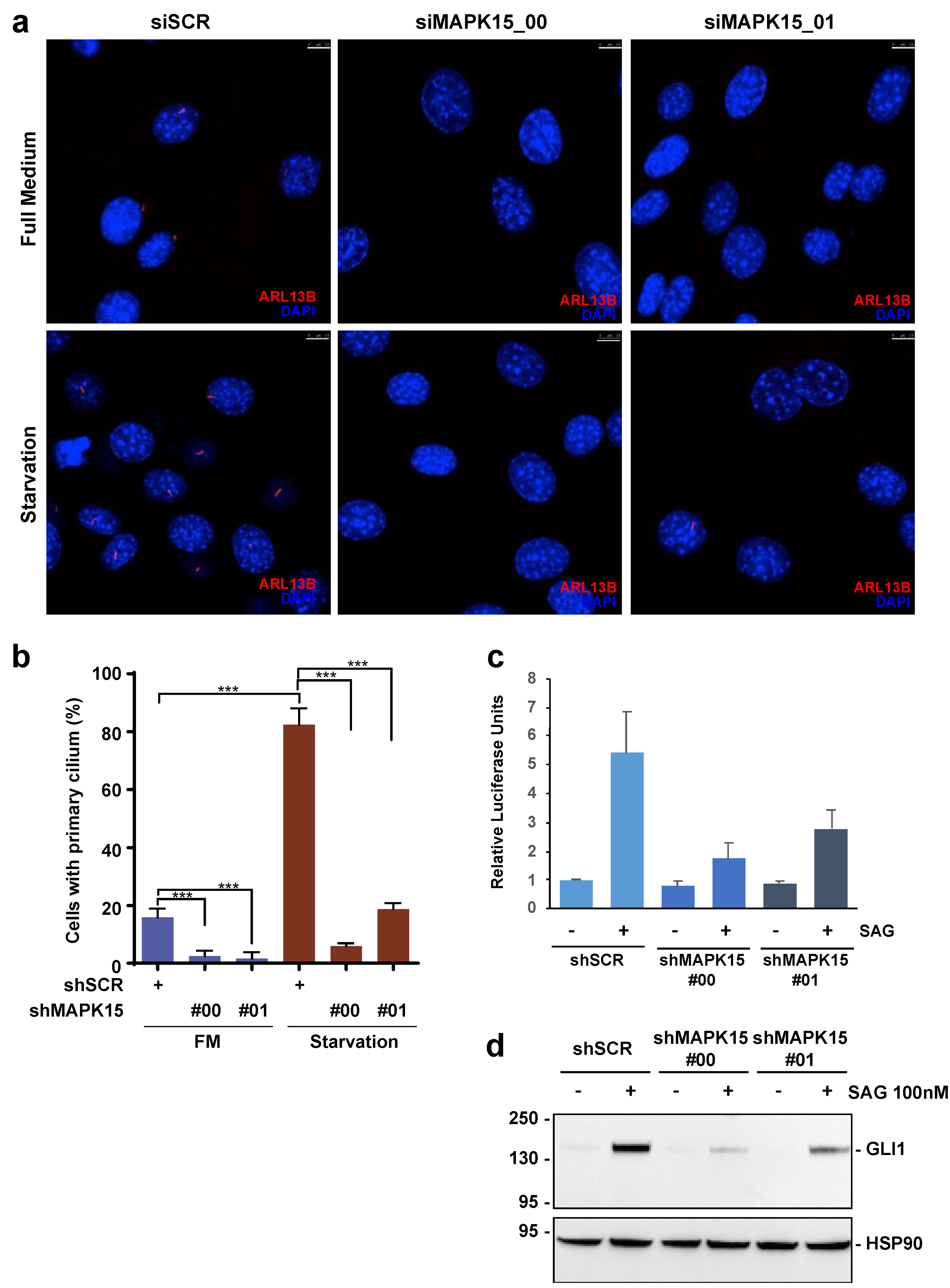
3.1. MAPK15 Controls Primary Ciliogenesis in A Kinase-Dependent Fashion
3.2. MAPK15 Regulates the HH Pathway in NIH3T3 Cells
3.3. MAPK15 Controls Hedgehog Signaling in Human Medulloblastoma Cells
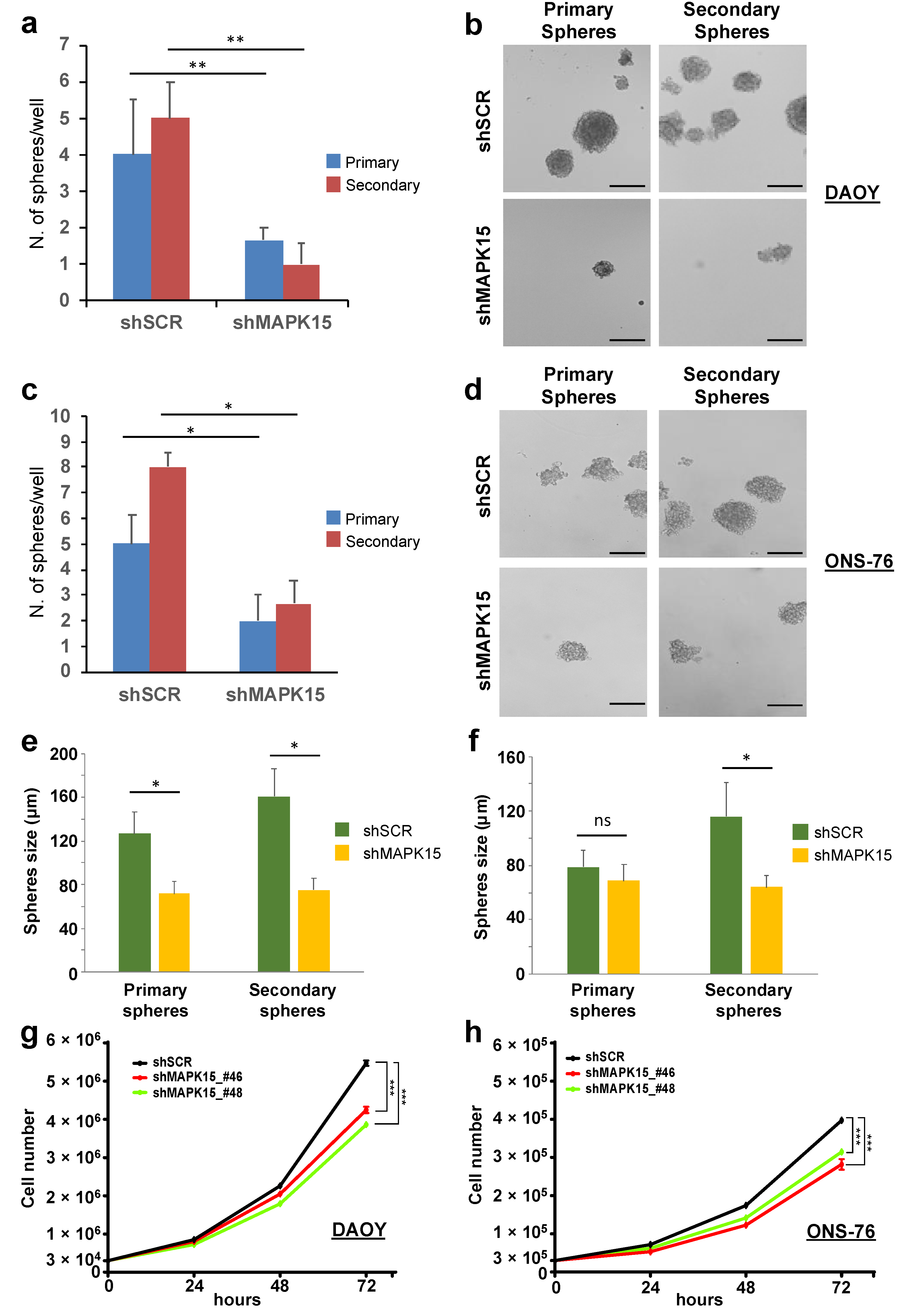
3.4. MAPK15 Regulates Self-Renewal In Vitro of Medulloblastoma Stem Cell-Like Cells
3.5. MAPK15 Effects on the Hedgehog Pathway Depends on the Ability of the Kinase of Affecting Ciliary Structures
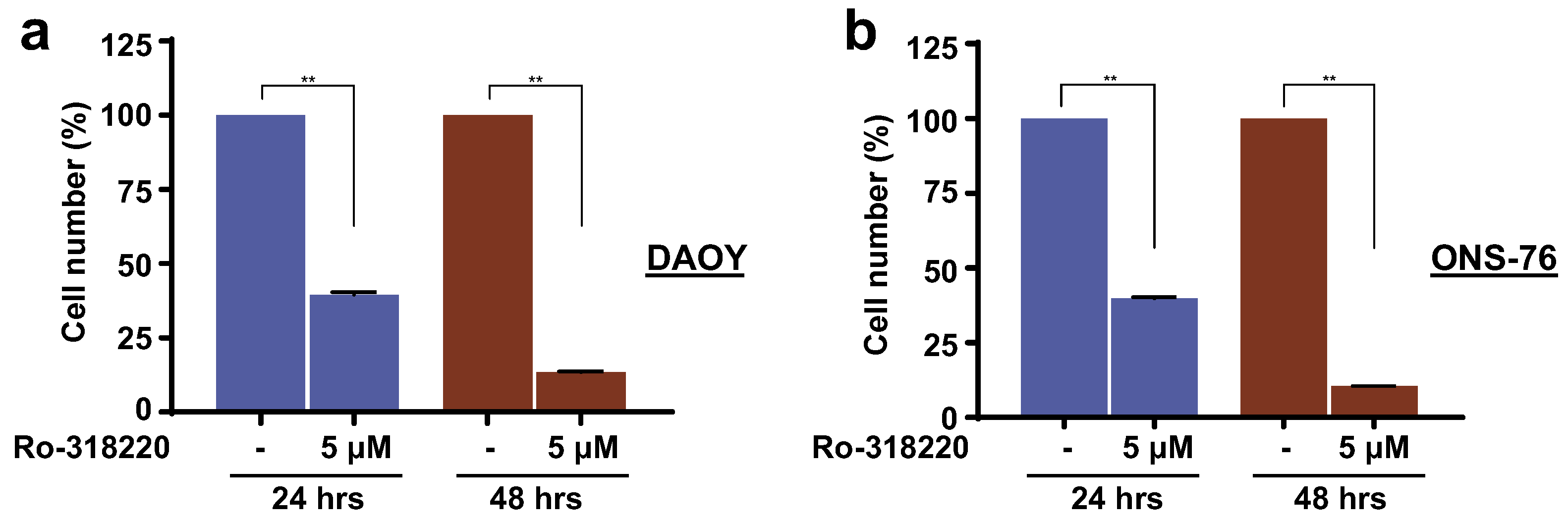
3.6. Inhibition of MAPK15 Kinase Activity by Ro-318220 Prevents Cell Proliferation of SHH-Driven Medulloblastoma Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kool, M.; Korshunov, A.; Remke, M.; Jones, D.T.; Schlanstein, M.; Northcott, P.A.; Cho, Y.J.; Koster, J.; Schouten-van Meeteren, A.; van Vuurden, D.; et al. Molecular subgroups of medulloblastoma: An international meta-analysis of transcrip-tome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012, 123, 473–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Northcott, P.A.; Jones, D.T.W.; Kool, M.; Robinson, G.; Gilbertson, R.J.; Cho, Y.-J.; Pomeroy, S.L.; Korshunov, A.; Lichter, P.; Taylor, M.D.; et al. Medulloblastomics: The end of the beginning. Nat. Rev. Cancer 2012, 12, 818–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Northcott, P.A.; Buchhalter, I.; Morrissy, S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Gröbner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.; et al. The whole-genome landscape of medulloblastoma subtypes. Nat. Cell Biol. 2017, 547, 311–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, V.; Kumar, V.; McGuire, T.; Coulter, D.W.; Sharp, J.G.; Mahato, R.I. Challenges and Recent Advances in Medulloblastoma Therapy. Trends Pharmacol. Sci. 2017, 38, 1061–1084. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.K.; Kuo, W.-L.; Hershenson, M.B.; Rosner, M.R. Extracellular Signal-Regulated Kinase 7 (ERK7), a Novel ERK with a C-Terminal Domain That Regulates Its Activity, Its Cellular Localization, and Cell Growth. Mol. Cell. Biol. 1999, 19, 1301–1312. [Google Scholar] [CrossRef] [Green Version]
- Colecchia, D.; Strambi, A.; Sanzone, S.; Iavarone, C.; Rossi, M.; Dall’Armi, C.; Piccioni, F.; Di Pianella, A.V.; Chiariello, M. MAPK15/ERK8 stimulates autophagy by interacting with LC3 and GABARAP proteins. Autophagy 2012, 8, 1724–1740. [Google Scholar] [CrossRef] [Green Version]
- Kazatskaya, A.; Kuhns, S.; Lambacher, N.J.; Kennedy, J.E.; Brear, A.G.; McManus, G.J.; Sengupta, P.; Blacque, O.E. Primary Cilium Formation and Ciliary Protein Trafficking Is Regulated by the Atypical MAP Kinase MAPK15 in Caeno-rhabditis elegans and Human Cells. Genetics 2017, 207, 1423–1440. [Google Scholar] [CrossRef] [Green Version]
- Miyatake, K.; Kusakabe, M.; Takahashi, C.; Nishida, E. ERK7 regulates ciliogenesis by phosphorylating the actin regu-lator CapZIP in cooperation with Dishevelled. Nat. Commun. 2015, 6, 6666. [Google Scholar] [CrossRef] [Green Version]
- Klevernic, I.V.; Martin, N.M.; Cohen, P. Regulation of the activity and expression of ERK8 by DNA damage. FEBS Lett. 2009, 583, 680–684. [Google Scholar] [CrossRef] [Green Version]
- Rossi, M.; Colecchia, D.; Ilardi, G.; Acunzo, M.; Nigita, G.; Sasdelli, F.; Celetti, A.; Strambi, A.; Staibano, S.; Croce, C.M.; et al. MAPK15 upregulation promotes cell proliferation and prevents DNA damage in male germ cell tumors. Oncotarget 2016, 7, 20981–20998. [Google Scholar] [CrossRef]
- Zacharogianni, M.; Kondylis, V.; Tang, Y.; Farhan, H.; Xanthakis, D.; Fuchs, F.; Boutros, M.; Rabouille, C. ERK7 is a neg-ative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EM-BO J. 2011, 30, 3684–3700. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.M.; Zhu, F.; Cho, Y.Y.; Carper, A.; Peng, C.; Zheng, D.; Yao, K.; Lau, A.T.; Zykova, T.A.; Kim, H.G.; et al. Extracellu-lar signal-regulated kinase 8-mediated c-Jun phosphorylation increases tumorigenesis of human colon cancer. Cancer Res. 2010, 70, 3218–3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colecchia, D.; Rossi, M.; Sasdelli, F.; Sanzone, S.; Strambi, A.; Chiariello, M. MAPK15 mediates BCR-ABL1-induced au-tophagy and regulates oncogene-dependent cell proliferation and tumor formation. Autophagy 2015, 11, 1790–1802. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Marshall, W.F. Ciliogenesis: Building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 2011, 12, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Eguether, T.; Hahne, M. Mixed signals from the cell’s antennae: Primary cilia in cancer. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Dahmane, N.; Altaba, A.R. Sonic hedgehog regulates the growth and patterning of the cerebellum. Dev. 1999, 126, 3089–3100. [Google Scholar] [CrossRef]
- Ericson, J.; Muhr, J.; Placzek, M.; Lints, T.; Jessell, T.M.; Edlund, T. Sonic hedgehog induces the differentiation of ventral forebrain neurons: A common signal for ventral patterning within the neural tube. Cell 1995, 81, 747–756. [Google Scholar] [CrossRef] [Green Version]
- Palma, V.; Lim, D.; Dahmane, N.; Sánchez, P.; Brionne, T.C.; Herzberg, C.D.; Gitton, Y.; Carleton, A.; Alvarez-Buylla, A.; Altaba, A.R. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Dev. 2005, 132, 335–344. [Google Scholar] [CrossRef] [Green Version]
- Petrova, R.; Joyner, A.L. Roles for Hedgehog signaling in adult organ homeostasis and repair. Dev. 2014, 141, 3445–3457. [Google Scholar] [CrossRef] [Green Version]
- Pietrobono, S.; Gagliardi, S.; Stecca, B. Non-canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Tran-scription Factors Beyond Smoothened. Front Genet. 2019, 10, 556. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.-G.; Kim, H.J.; Dlugosz, A.A.; Ellison, D.W.; Gilbertson, R.J.; Alvarez-Buylla, A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat. Med. 2009, 15, 1062–1065. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.Y.; Seol, A.D.; So, P.-L.; Ermilov, A.N.; Bichakjian, C.K.; Epstein, E.H.; Dlugosz, A.A.; Reiter, J.F. Primary cilia can both mediate and suppress Hedgehog pathway–dependent tumorigenesis. Nat. Med. 2009, 15, 1055–1061. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, H.; Hui, C.; Nakafuku, M.; Kondoh, H. A binding site for Gli proteins is essential for HNF-3beta floor plate en-hancer activity in transgenics and can respond to Shh in vitro. Development 1997, 124, 1313–1322. [Google Scholar] [CrossRef]
- Pietrobono, S.; Gaudio, E.; Gagliardi, S.; Zitani, M.; Carrassa, L.; Migliorini, F.; Petricci, E.; Manetti, F.; Makukhin, N.; Bond, A.G.; et al. Targeting non-canonical activation of GLI1 by the SOX2-BRD4 transcriptional complex improves the efficacy of HEDGEHOG pathway inhibition in melanoma. Oncogene 2021, 40, 1–16. [Google Scholar] [CrossRef]
- Pietrobono, S.; Santini, R.; Gagliardi, S.; Dapporto, F.; Colecchia, D.; Chiariello, M.; Leone, C.; Valoti, M.; Manetti, F.; Petricci, E.; et al. Targeted inhibition of Hedgehog-GLI signaling by novel acylguanidine derivatives inhibits melanoma cell growth by inducing replication stress and mitotic catastrophe. Cell Death Dis. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Pietrobono, S.; Anichini, G.; Sala, C.; Manetti, F.; Almada, L.L.; Pepe, S.; Carr, R.M.; Paradise, B.D.; Sarkaria, J.N.; Davila, J.I.; et al. ST3GAL1 is a target of the SOX2-GLI1 transcriptional complex and promotes melanoma metastasis through AXL. Nat. Commun. 2020, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Bernatik, O.; Paclikova, P.; Kotrbova, A.; Bryja, V.; Cajanek, L. Primary Cilia Formation Does Not Rely on WNT/β-Catenin Signaling. Front. Cell Dev. Biol. 2021, 9, 623753. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Fisher, R.L.; Bowser, S.S.; Pentecost, B.T.; Sui, H. Three-dimensional architecture of epithelial primary cilia. Proc. Natl. Acad. Sci. USA 2019, 116, 9370–9379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheway, G.; Nazlamova, L.; Hancock, J.T. Signaling through the Primary Cilium. Front. Cell Dev. Biol. 2018, 6, 8. [Google Scholar] [CrossRef]
- Schneider, L.; Clement, C.A.; Teilmann, S.C.; Pazour, G.J.; Hoffmann, E.K.; Satir, P.; Christensen, S.T. PDGFRalpha signaling is regulated through the primary cilium in fibroblasts. Curr. Biol. 2005, 15, 1861–1866. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Taipale, J.; Young, K.E.; Maiti, T.; Beachy, P.A. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA 2002, 99, 14071–14076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bay, S.N.; Long, A.; Caspary, T. Disruption of the ciliary GTPase Arl13b suppresses Sonic hedgehog overactivation and inhibits medulloblastoma formation. Proc. Natl. Acad. Sci. USA 2018, 115, 1570–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Lopez, J.; Kumar, R.; Smith, K.S.; Northcott, P.A. Deconstructing Sonic Hedgehog Medulloblastoma: Molecular Subtypes, Drivers, and Beyond. Trends Genet. 2021, 37, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Zanini, C.; Ercole, E.; Mandili, G.; Salaroli, R.; Poli, A.; Renna, C.; Papa, V.; Cenacchi, G.; Forni, M. Medullospheres from DAOY, UW228 and ONS-76 Cells: Increased Stem Cell Population and Proteomic Modifications. PLoS ONE 2013, 8, e63748. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, D.P.; Coyle, B.; Walker, D.A.; Grabowska, A.M. In vitro models of medulloblastoma: Choosing the right tool for the job. J. Biotechnol. 2016, 236, 10–25. [Google Scholar] [CrossRef]
- Bahmad, H.F.; Poppiti, R.J. Medulloblastoma cancer stem cells: Molecular signatures and therapeutic targets. J. Clin. Pathol. 2020, 73, 243–249. [Google Scholar] [CrossRef]
- Manoranjan, B.; Venugopal, C.; McFarlane, N.; Doble, B.; Dunn, S.E.; Scheinemann, K.; Singh, S.K. Medulloblastoma stem cells: Where development and cancer cross pathways. Pediatr. Res. 2012, 71, 516–522. [Google Scholar] [CrossRef] [Green Version]
- Roessler, E.; Ermilov, A.N.; Grange, D.K.; Wang, A.; Grachtchouk, M.; Dlugosz, A.A.; Muenke, M. A previously uni-dentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum. Mol. Genet. 2005, 14, 2181–2188. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Murone, M.; Luoh, S.M.; Ryan, A.; Gu, Q.; Zhang, C.; Bonifas, J.M.; Lam, C.W.; Hynes, M.; Goddard, A.; et al. Acti-vating Smoothened mutations in sporadic basal-cell carcinoma. Nature 1998, 391, 90–92. [Google Scholar] [CrossRef]
- Dennler, S.; André, J.; Alexaki, I.; Li, A.; Magnaldo, T.; ten Dijke, P.; Wang, X.J.; Verrecchia, F.; Mauviel, A. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007, 67, 6981–6986. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietrobono, S.; Franci, L.; Imperatore, F.; Zanini, C.; Stecca, B.; Chiariello, M. MAPK15 Controls Hedgehog Signaling in Medulloblastoma Cells by Regulating Primary Ciliogenesis. Cancers 2021, 13, 4903. https://doi.org/10.3390/cancers13194903
Pietrobono S, Franci L, Imperatore F, Zanini C, Stecca B, Chiariello M. MAPK15 Controls Hedgehog Signaling in Medulloblastoma Cells by Regulating Primary Ciliogenesis. Cancers. 2021; 13(19):4903. https://doi.org/10.3390/cancers13194903
Chicago/Turabian StylePietrobono, Silvia, Lorenzo Franci, Francesco Imperatore, Cristina Zanini, Barbara Stecca, and Mario Chiariello. 2021. "MAPK15 Controls Hedgehog Signaling in Medulloblastoma Cells by Regulating Primary Ciliogenesis" Cancers 13, no. 19: 4903. https://doi.org/10.3390/cancers13194903
APA StylePietrobono, S., Franci, L., Imperatore, F., Zanini, C., Stecca, B., & Chiariello, M. (2021). MAPK15 Controls Hedgehog Signaling in Medulloblastoma Cells by Regulating Primary Ciliogenesis. Cancers, 13(19), 4903. https://doi.org/10.3390/cancers13194903






