Defining the Role of GLI/Hedgehog Signaling in Chemoresistance: Implications in Therapeutic Approaches
Abstract
Simple Summary
Abstract
1. Introduction
2. Hedgehog Signaling
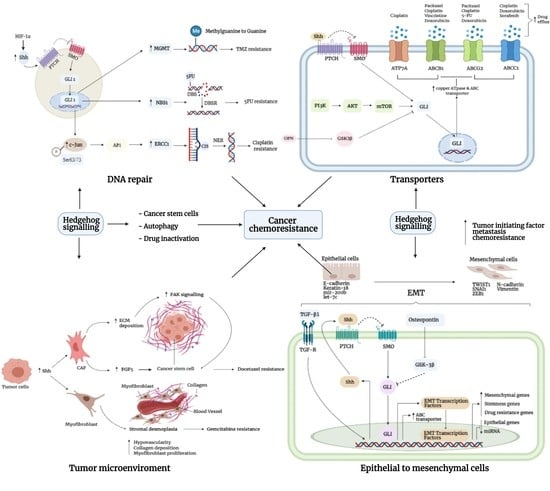
3. Mechanism of Chemoresistance
3.1. DNA Repair
3.2. Autophagy
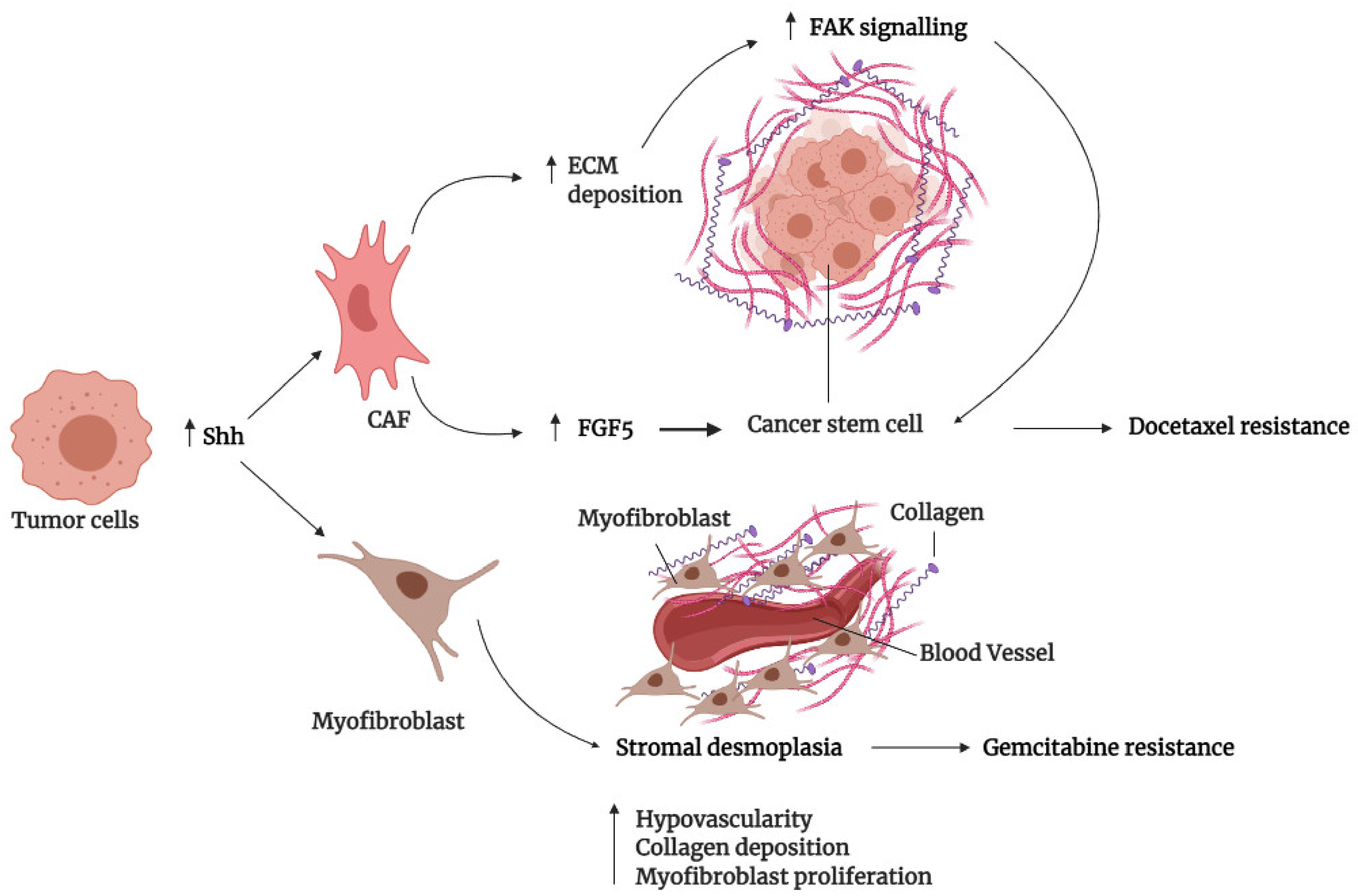
3.3. Tumor Microenvironment
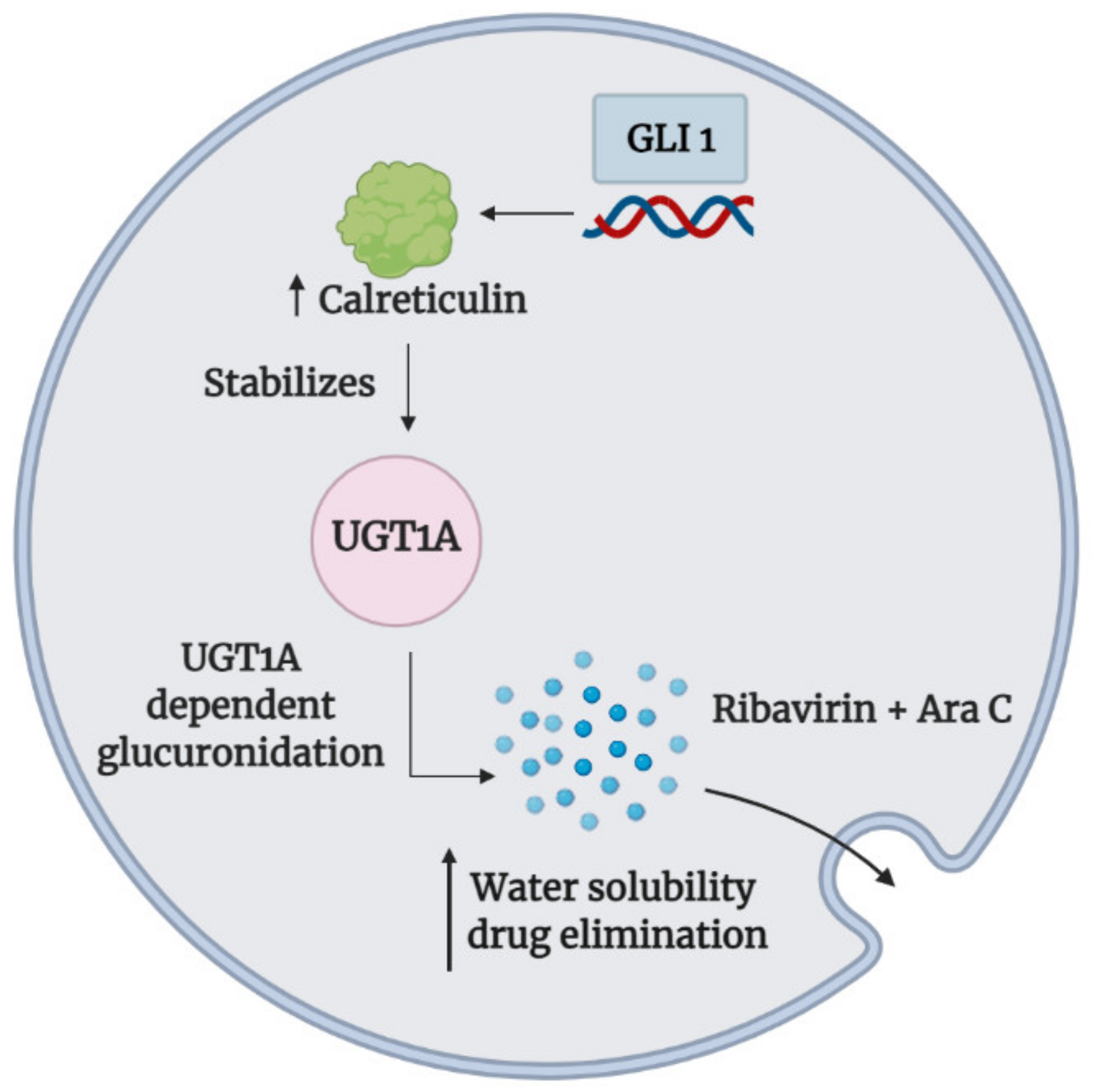
3.4. Drug Inactivation
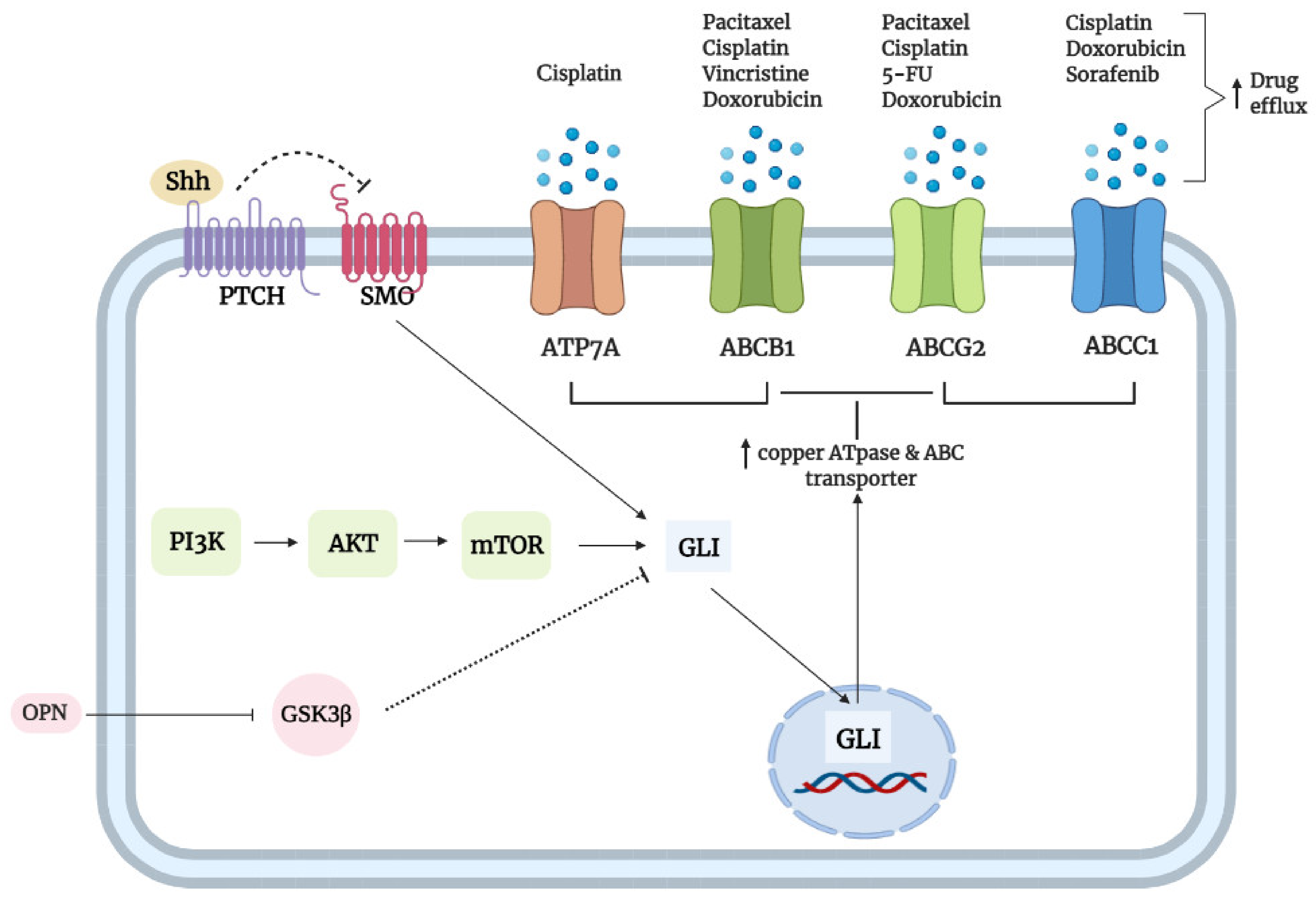
3.5. Transporters
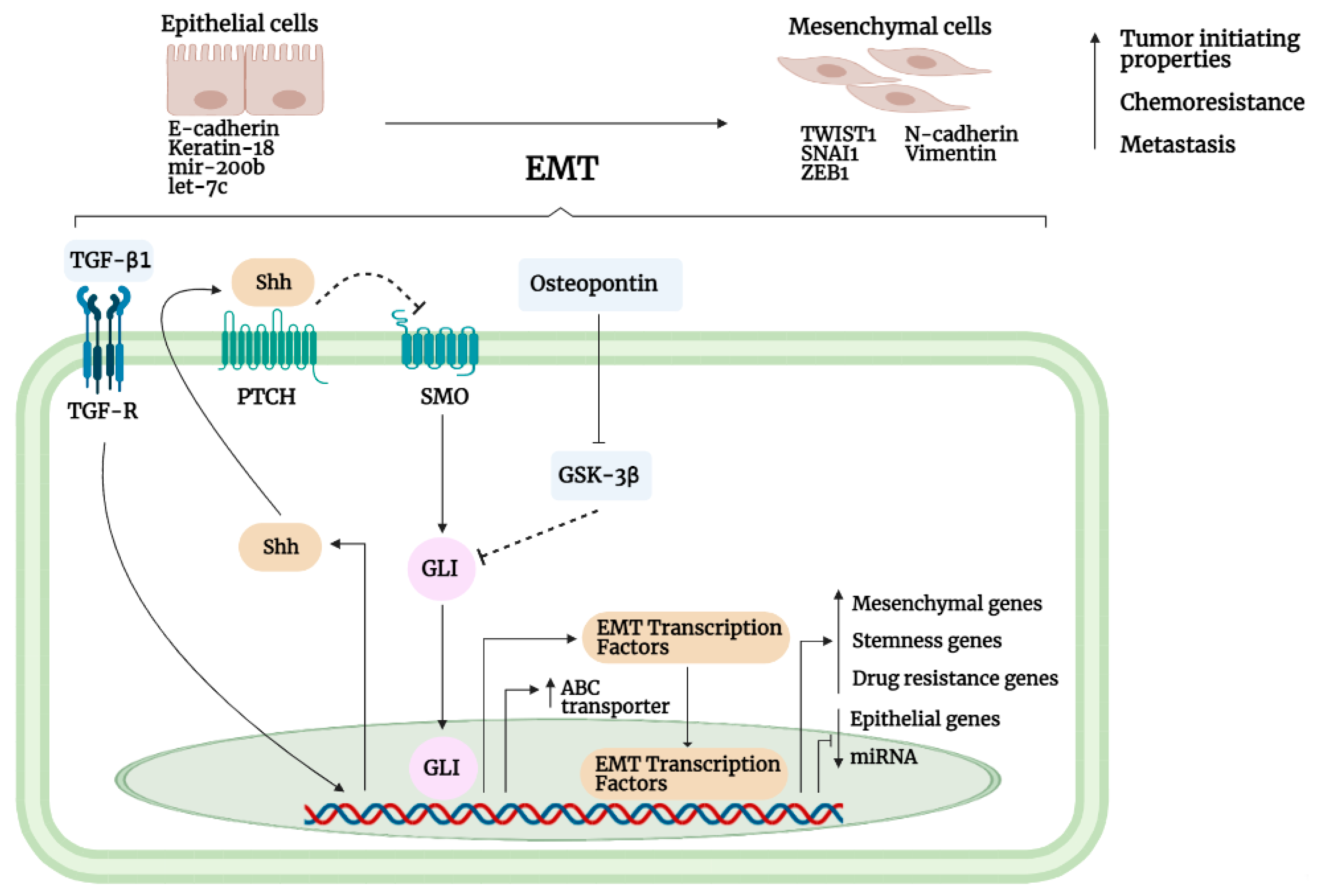
3.6. Epithelial-to-Mesenchymal Interaction
3.7. Cancer Stem Cells
3.8. Section Summary
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- André, T.; Boni, C.; Mounedji-Boudiaf, L.; Navarro, M.; Tabernero, J.; Hickish, T.; Topham, C.; Zaninelli, M.; Clingan, P.; Bridgewater, J.; et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N. Engl. J. Med. 2004, 350, 2343–2351. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.M.; Moulder-Thompson, S.L. Neoadjuvant treatment of breast cancer. Ann. Oncol. 2012, 23, 231. [Google Scholar] [CrossRef] [PubMed]
- D’Alterio, C.; Scala, S.; Sozzi, G.; Roz, L.; Bertolini, G. Paradoxical effects of chemotherapy on tumor relapse and metastasis promotion. Semin. Cancer Biol. 2020, 60, 351–361. [Google Scholar] [CrossRef]
- Madden, E.C.; Gorman, A.M.; Logue, S.E.; Samali, A. Tumour cell secretome in chemoresistance and tumour recurrence. Trends Cancer 2020, 6, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.M.; Mao, Y.; Zeng, J.; Holla, V.; Johnson, A.; Brusco, L.; Chen, K.; Mendelsohn, J.; Routbort, M.J.; Mills, G.B.; et al. Implementation of biomarker-driven cancer therapy: Existing tools and remaining gaps. Discov. Med. 2014, 17, 101–114. [Google Scholar]
- Liu, D. Cancer biomarkers for targeted therapy. Biomark. Res. 2019, 7, 101–114. [Google Scholar] [CrossRef]
- Mokhtari, R.B.; Homayouni, T.S.; Baluch, N.; Morgatskaya, E.; Kumar, S.; Das, B.; Yeger, H. Combination therapy in combating cancer. Oncotarget 2017, 8, 38022–38043. [Google Scholar] [CrossRef]
- Kukcinaviciute, E.; Jonusiene, V.; Sasnauskiene, A.; Dabkeviciene, D.; Eidenaite, E.; Laurinavicius, A. Significance of notch and Wnt signaling for chemoresistance of colorectal cancer cells HCT. J. Cell. Biochem. 2018, 119, 5913–5920. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Tao, F.; Zhang, X.; Zhang, Y.; Sun, X.; Wu, D. Role of Wnt/ β-Catenin signaling in the chemoresistance modulation of colorectal cancer. Biomed Res. Int. 2020, 2020, 9390878. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Cang, W.; Li, Q.; Liao, X.; Zhan, M.; Deng, H.; Li, S.; Jin, W.; Pang, Z.; Qiu, X.; et al. Erlotinib overcomes paclitaxel-resistant cancer stem cells by blocking the EGFR-CREB/GRβ-IL-6 axis in MUC1-positive cervical cancer. Oncogenesis 2019, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Hopper-Borge, E.A.; Nasto, R.E.; Ratushny, V.; Weiner, L.M.; Golemis, E.A.; Astsaturov, I. Mechanisms of tumor resistance to EGFR-targeted therapies. Expert Opin. Ther. Targets 2009, 13, 339–362. [Google Scholar] [CrossRef] [PubMed]
- West, K.A.; Castillo, S.S.; Dennis, P.A. Activation of the PI3K/Akt pathway and chemotherapeutic resistance. Drug Resist. Updat. 2002, 5, 234–248. [Google Scholar] [CrossRef]
- Deng, J.; Bai, X.; Feng, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Inhibition of PI3K/Akt/MTOR signaling pathway alleviates ovarian cancer chemoresistance through reversing epithelial-mesenchymal transition and decreasing cancer stem cell marker expression. BMC Cancer 2019, 19, 618. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shen, S.; Guo, J.; Chen, H.; Yu Greenblatt, D.; Kleeff, J.; Liao, Q.; Chen, G.; Friess, H.; Sing Leung, P. Mitogen-activated protein kinases and chemoresistance in pancreatic cancer cells. J. Surg. Res. 2006, 136, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Salaroglio, I.C.; Mungo, E.; Gazzano, E.; Kopecka, J.; Riganti, C. ERK is a pivotal player of chemo-immune-resistance in cancer. Int. J. Mol. Sci. 2019, 20, 2505. [Google Scholar] [CrossRef]
- Godwin, P.; Baird, A.M.; Heavey, S.; Barr, M.P.; O’Byrne, K.J.; Gately, K. Targeting nuclear factor-kappa B to overcome resistance to chemotherapy. Front. Oncol. 2013, 3, 120. [Google Scholar] [CrossRef]
- Li, Q.; Yang, G.; Feng, M.; Zheng, S.; Cao, Z.; Qiu, J.; You, L.; Zheng, L.; Hu, Y.; Zhang, T.; et al. NF-ΚB in pancreatic cancer: Its key role in chemoresistance. Cancer Lett. 2018, 421, 127–134. [Google Scholar] [CrossRef]
- Skoda, A.M.; Simovic, D.; Karin, V.; Kardum, V.; Vranic, S.; Serman, L. The role of the hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci. 2018, 18, 8–20. [Google Scholar] [CrossRef]
- Sari, I.N.; Phi, L.T.H.; Jun, N.; Wijaya, Y.T.; Lee, S.; Kwon, H.Y. Hedgehog signaling in cancer: A prospective therapeutic target for eradicating cancer stem cells. Cells 2018, 7, 208. [Google Scholar] [CrossRef]
- Lupi, O. Correlations between the sonic hedgehog pathway and basal cell carcinoma. Int. J. Dermatol. 2007, 46, 1113–1117. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, A.; Levesque, M.P.; Dummer, R.; Kabashima, K. Hedgehog signaling in basal cell carcinoma. J. Dermatol. Sci. 2015, 78, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, A.; Chaudhary, S.C.; Rana, M.; Elmets, C.A.; Athar, M. Basal cell carcinoma pathogenesis and therapy involving hedgehog signaling and beyond. Mol. Carcinog. 2017, 56, 2543–2557. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Yang, J.Y. Targeting the hedgehog pathway in pediatric medulloblastoma. Cancers 2015, 7, 2110–2123. [Google Scholar] [CrossRef]
- Yoon, J.W.; Gilbertson, R.; Iannaccone, S.; Iannaccone, P.; Walterhouse, D. Defining a role for sonic hedgehog pathway activation in desmoplastic medulloblastoma by identifying GLI1 target genes. Int. J. Cancer 2009, 124, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, G.; Pea, F.; Moscarella, E.; Argenziano, G. Sonidegib for the treatment of advanced basal cell carcinoma. Front. Oncol. 2020, 10, 582866. [Google Scholar] [CrossRef]
- Sandhiya, S.; Melvin, G.; Kumar, S.S.; Dkhar, S.A. The dawn of hedgehog inhibitors: Vismodegib. J. Pharmacol. Pharmacother. 2013, 4, 4–7. [Google Scholar] [CrossRef]
- Catenacci, D.V.T.; Junttila, M.R.; Karrison, T.; Bahary, N.; Horiba, M.N.; Nattam, S.R.; Marsh, R.; Wallace, J.; Kozloff, M.; Rajdev, L.; et al. Randomized phase Ib/II study of gemcitabine plus placebo or vismodegib, a hedgehog pathway inhibitor, in patients with metastatic pancreatic cancer. J. Clin. Oncol. 2015, 33, 4284–4292. [Google Scholar] [CrossRef]
- Xie, H.; Paradise, B.D.; Ma, W.W.; Fernandez-Zapico, M.E. Recent advances in the clinical targeting of hedgehog/GLI signaling in cancer. Cells 2019, 8, 394. [Google Scholar] [CrossRef]
- Li, Y.; Song, Q.; Day, B.W. Phase I and phase II Sonidegib and Vismodegib clinical trials for the treatment of paediatric and adult MB patients: A systemic review and meta-analysis. Acta Neuropathol. Commun. 2019, 7, 123. [Google Scholar] [CrossRef]
- Gould, S.E.; Low, J.A.; Marsters, J.C.; Robarge, K.; Rubin, L.L.; De Sauvage, F.J.; Sutherlin, D.P.; Wong, H.; Yauch, R.L. Discovery and preclinical development of Vismodegib. Expert Opin. Drug Discov. 2014, 9, 969–984. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, K.; Ruela-de-Sousa, R.; Fuhler, G.; Aberson, H.; Ferreira, C.; Peppelenbosch, M.; Spek, C. Hedgehog signaling maintains chemoresistance in myeloid leukemic cells. Oncogene 2010, 29, 6314–6322. [Google Scholar] [CrossRef] [PubMed]
- Nüsslein-volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Le, H.; Kleinerman, R.; Lerman, O.Z.; Brown, D.; Galiano, R.; Gurtner, G.C.; Warren, S.M.; Levine, J.P.; Saadeh, P.B. Hedgehog signaling is essential for normal wound healing. Wound Repair Regen. 2008, 16, 768–773. [Google Scholar] [CrossRef]
- Carballo, G.B.; Honorato, J.R.; De Lopes, G.P.F.; Spohr, T.C.L.D.S.E. A highlight on sonic hedgehog pathway. Cell Commun. Signal. 2018, 16, 11. [Google Scholar] [CrossRef]
- Merchant, J.L. Hedgehog signalling in gut development, physiology and cancer. J. Physiol. 2012, 590, 421–432. [Google Scholar] [CrossRef]
- Perler, F.B. Protein splicing of inteins and hedgehog autoproteolysis: Structure, function, and evolution. Cell 1998, 92, 1–4. [Google Scholar] [CrossRef]
- Chen, X.; Tukachinsky, H.; Huang, C.H.; Jao, C.; Chu, Y.R.; Tang, H.Y.; Mueller, B.; Schulman, S.; Rapoport, T.A.; Salic, A. Processing and turnover of the hedgehog protein in the endoplasmic reticulum. J. Cell Biol. 2011, 192, 825–838. [Google Scholar] [CrossRef]
- Buglino, J.A.; Resh, M.D. Palmitoylation of hedgehog proteins. Vitam. Horm. 2012, 88, 229–252. [Google Scholar]
- Tukachinsky, H.; Kuzmickas, R.P.; Jao, C.Y.; Liu, J.; Salic, A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2012, 2, 308–320. [Google Scholar] [CrossRef]
- Chan, J.A.; Balasubramanian, S.; Witt, R.M.; Nazemi, K.J.; Choi, Y.; Pazyra-Murphy, M.F.; Walsh, C.O.; Thompson, M.; Segal, R.A. Proteoglycan interactions with sonic hedgehog specify mitogenic responses. Nat. Neurosci. 2009, 12, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Christ, A.; Herzog, K.; Willnow, T.E. LRP2, an auxiliary receptor that controls sonic hedgehog signaling in development and disease. Dev. Dyn. 2016, 245, 569–579. [Google Scholar] [CrossRef]
- Petrov, K.; Wierbowski, B.M.; Salic, A. Sending and receiving hedgehog signals. Annu. Rev. Cell Dev. Biol. 2017, 33, 145–168. [Google Scholar] [CrossRef] [PubMed]
- Marigo, V.; Davey, R.A.; Zuo, Y.; Cunningham, J.M.; Tabin, C.J. Biochemical evidence that patched is the hedgehog receptor. Nature 1996, 384, 176–179. [Google Scholar] [CrossRef]
- Mastronardi, F.G.; Dimitroulakos, J.; Kamel-Reid, S.; Manoukian, A.S. Co-localization of patched and activated sonic hedgehog to lysosomes in neurons. Neuroreport 2000, 11, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, J.J.; Whalen, E.J.; Liu, R.; Xiao, K.; Kim, J.; Chen, M.; Wang, J.; Chen, W.; Lefkowitz, R.J. β-arrestin-mediated localization of smoothened to the primary cilium. Science 2008, 320, 1777–1781. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ren, X.R.; Nelson, C.D.; Barak, L.S.; Chen, J.K.; Beachy, P.A.; De Sauvage, F.; Lefkowitz, R.J. Activity-dependent internalization of smoothened mediated by β-arrestin 2 and GRK. Science 2004, 306, 2257–2260. [Google Scholar] [CrossRef] [PubMed]
- Bangs, F.; Anderson, K.V. Primary cilia and mammalian hedgehog signaling. Cold Spring Harb. Perspect. Biol. 2017, 9, a028175. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Tong, C.; Wang, B.; Luo, L.; Jiang, J. Hedgehog signalling activity of smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature 2004, 432, 1045–1050. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, J. Decoding the phosphorylation code in hedgehog signal transduction. Cell Res. 2013, 23, 186–200. [Google Scholar] [CrossRef]
- Ma, G.; Li, S.; Han, Y.; Li, S.; Yue, T.; Wang, B.; Correspondence, J.J. Regulation of smoothened trafficking and hedgehog signaling by the SUMO pathway. Dev. Cell 2016, 39, 438–451. [Google Scholar] [CrossRef]
- Han, Y.; Shi, Q.; Jiang, J. Multisite interaction with sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc. Natl. Acad. Sci. USA 2015, 112, 6383–6388. [Google Scholar] [CrossRef]
- Shafique, S.; Rashid, S. Structural basis of ΒTrCP1-associated GLI3 processing. Sci. Rep. 2019, 9, 6865. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhang, L.; Zhang, Q.; Tong, C.; Wang, B.; Hou, F.; Amanai, K.; Jiang, J. Phosphorylation by double-time/CKIε and CKIα targets cubitus interruptus for slimb/β-TRCP-mediated proteolytic processing. Dev. Cell 2005, 9, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Tukachinsky, H.; Lopez, L.V.; Salic, A. A mechanism for vertebrate hedgehog signaling: Recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 2010, 191, 415–428. [Google Scholar] [CrossRef]
- Humke, E.W.; Dorn, K.V.; Milenkovic, L.; Scott, M.P.; Rohatgi, R. The output of hedgehog signaling is controlled by the dynamic association between suppressor of fused and the gli proteins. Genes Dev. 2010, 24, 670–682. [Google Scholar] [CrossRef]
- Katoh, Y.; Katoh, M. Hedgehog target genes: Mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med. 2009, 9, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, C.R.; Szczepny, A.; Watkins, D.N.; Cain, J.E. Hedgehog signaling in the maintenance of cancer stem cells. Cancers 2015, 7, 1554–1585. [Google Scholar] [CrossRef]
- Niewiadomski, P.; Niedziółka, S.M.; Markiewicz, Ł.; Uśpieński, T.; Baran, B.; Chojnowska, K. Gli proteins: Regulation in development and cancer. Cells 2019, 8, 147. [Google Scholar] [CrossRef]
- De Reyniès, A.; Javelaud, D.; Elarouci, N.; Marsaud, V.; Gilbert, C.; Mauviel, A. Large-scale pan-cancer analysis reveals broad prognostic association between TGF-β ligands, not hedgehog, and GLI1/2 expression in tumors. Sci. Rep. 2020, 10, 14491. [Google Scholar] [CrossRef]
- Nolan-Stevaux, O.; Lau, J.; Truitt, M.L.; Chu, G.C.; Hebrok, M.; Fernández-Zapico, M.E.; Hanahan, D. GLI1 Is regulated through smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009, 23, 24–36. [Google Scholar] [CrossRef]
- Eberl, M.; Klingler, S.; Mangelberger, D.; Loipetzberger, A.; Damhofer, H.; Zoidl, K.; Schnidar, H.; Hache, H.; Bauer, H.C.; Solca, F.; et al. Hedgehog-EGFR cooperation response genes determine the oncogenic phenotype of basal cell carcinoma and tumour-initiating pancreatic cancer cells. EMBO Mol. Med. 2012, 4, 218–233. [Google Scholar] [CrossRef]
- Po, A.; Silvano, M.; Miele, E.; Capalbo, C.; Eramo, A.; Salvati, V.; Todaro, M.; Besharat, Z.M.; Catanzaro, G.; Cucchi, D.; et al. Noncanonical GLI1 signaling promotes stemness features and in vivo growth in lung adenocarcinoma. Oncogene 2017, 36, 4641–4652. [Google Scholar] [CrossRef]
- Varnat, F.; Siegl-Cachedenier, I.; Malerba, M.; Gervaz, P.; Ruiz, I.; Altaba, A. Loss of WNT-TCF Addiction and enhancement of HH-GLI1 signalling define the metastatic transition of human colon carcinomas. EMBO Mol. Med. 2010, 2, 440–457. [Google Scholar] [CrossRef]
- Noubissi, F.K.; Goswami, S.; Sanek, N.A.; Kawakami, K.; Minamoto, T.; Moser, A.; Grinblat, Y.; Spiegelman, V.S. Wnt signaling stimulates transcriptional outcome of the hedgehog pathway by stabilizing GLI1 MRNA. Cancer Res. 2009, 69, 8572–8578. [Google Scholar] [CrossRef]
- Zubčić, V.; Rinčić, N.; Kurtović, M.; Trnski, D.; Musani, V.; Ozretić, P.; Levanat, S.; Leović, D.; Sabol, M. GANT61 and lithium chloride inhibit the growth of head and neck cancer cell lines through the regulation of GLI3 processing by GSK3β. Int. J. Mol. Sci. 2020, 21, 6410. [Google Scholar] [CrossRef]
- Ozretić, P.; Trnski, D.; Musani, V.; Maurac, I.; Kalafatić, D.; Orešković, S.; Levanat, S.; Sabol, M. Non-canonical hedgehog signaling activation in ovarian borderline tumors and ovarian carcinomas. Int. J. Oncol. 2017, 51, 1869–1877. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Matsubara, S.; Ding, Q.; Tsukasa, K.; Yoshimitsu, M.; Kosai, K.; Takao, S. Efficient elimination of pancreatic cancer stem cells by hedgehog/GLI Inhibitor GANT61 in combination with MTOR inhibition. Mol. Cancer 2016, 15, 49. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Robins, P.; Lindahl, T.; Barnes, D.E. 5-Fluorouracil incorporated into DNA is excised by the Smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007, 67, 940–945. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.; Hanna, A.; Samant, R.S.; Shevde, L.A. The impact of hedgehog signaling pathway on DNA repair mechanisms in human cancer. Cancers 2015, 7, 1333–1348. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Y.; Che, J.; Sun, K.K.; Shen, X.J.; Yang, D.; Zhong, N.; Zhao, H. Cyclopamine increases the radiosensitivity of human pancreatic cancer cells by regulating the DNA repair signal pathway through an epidermal growth factor receptor-dependent pathway. Mol. Med. Rep. 2013, 8, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cai, J.; Zhao, S.; Yao, K.; Sun, Y.; Li, Y.; Chen, L.; Li, R.; Zhai, X.; Zhang, J.; et al. GANT61, a GLI inhibitor, sensitizes glioma cells to the temozolomide treatment. J. Exp. Clin. Cancer Res. 2016, 35, 184. [Google Scholar] [CrossRef]
- Wang, K.; Chen, D.; Qian, Z.; Cui, D.; Gao, L.; Lou, M. Hedgehog/Gli1 Signaling pathway regulates MGMT expression and chemoresistance to temozolomide in human glioblastoma. Cancer Cell Int. 2017, 17, 117. [Google Scholar] [CrossRef]
- Wei, M.; Ma, R.; Huang, S.; Liao, Y.; Ding, Y.; Li, Z.; Guo, Q.; Tan, R.; Zhang, L.; Zhao, L. Oroxylin A increases the sensitivity of temozolomide on glioma cells by hypoxia-inducible factor 1α/hedgehog pathway under hypoxia. J. Cell. Physiol. 2019, 234, 17392–17404. [Google Scholar] [CrossRef]
- Lee, S.Y. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016, 3, 198–210. [Google Scholar] [CrossRef]
- Zhu, H.; Lo, H.-W. The human glioma-associated oncogene homolog 1 (GLI1) family of transcription factors in gene regulation and diseases. Curr. Genom. 2010, 11, 238–245. [Google Scholar] [CrossRef]
- Alnaim, L. Individualization of 5-fluorouracil in the treatment of colorectal cancer. SRX Pharmacol. 2010, 2010, 1–12. [Google Scholar] [CrossRef][Green Version]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Abubakar, A.; Lim, S.H.; Ng, W.; Lee, M.; de Souza, P.; Shin, J.S.; et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) complex in rectal cancer correlates with poor response to neoadjuvant radiotherapy and prognosis. BMC Cancer 2018, 18, 869. [Google Scholar] [CrossRef]
- Zhang, L.; Song, R.; Gu, D.; Zhang, X.; Yu, B.; Liu, B.; Xie, J. The role of GLI1 for 5-Fu resistance in colorectal cancer. Cell Biosci. 2017, 7, 17. [Google Scholar] [CrossRef]
- Zhang, R.; Ma, J.; Avery, J.T.; Sambandam, V.; Nguyen, T.H.; Xu, B.; Suto, M.J.; Boohaker, R.J. GLI1 Inhibitor SRI-38832 attenuates chemotherapeutic resistance by downregulating NBS1 transcription in BRAFV600E colorectal cancer. Front. Oncol. 2020, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, L.; Tsang, B.; Gardner, K.; Bostick-Bruton, F.; Reed, E. Phorbol ester exposure activates an AP-1-mediated increase in ERCC-1 messenger RNA expression in human ovarian tumor cells. Cell. Mol. Life Sci. 1999, 55, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Gardner, K.; Zhang, L.; Tsang, B.; Bostick-Brutont, F.; Reed, E. Cisplatin induction of ERCC-1 MRNA expression in A2780/CP70 human ovarian cancer cells. J. Biol. Chem. 1998, 273, 23419–23425. [Google Scholar] [CrossRef] [PubMed]
- Kudo, K.; Gavin, E.; Das, S.; Amable, L.; Shevde, L.A.; Reed, E. Inhibition of Gli1 results in altered C-jun activation, inhibition of cisplatin-induced upregulation of ERCC1, XPD and XRCC1, and inhibition of platinum-DNA adduct repair. Oncogene 2012, 31, 4718–4724. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhao, Z.; Yang, Y.; O’Connell, D.; Zhang, X.; Oh, S.; Ma, B.; Lee, J.H.; Zhang, T.; Varghese, B.; et al. Truncating mutation in the autophagy gene UVRAG confers oncogenic properties and chemosensitivity in colorectal cancers. Nat. Commun. 2015, 6, 7839. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Jeong, E.G.; Ahn, C.H.; Kim, S.S.; Lee, S.H.; Yoo, N.J. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum. Pathol. 2008, 39, 1059–1063. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Coppola, D.; Matsushita, N.; Cualing, H.D.; Sun, M.; Sato, Y.; Liang, C.; Jung, J.U.; Cheng, J.Q.; Mulé, J.J.; et al. Bif-1 interacts with beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007, 9, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.W.; Lee, S.H. The roles of autophagy in cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef]
- Degenhardt, K.; Mathew, R.; Beaudoin, B.; Bray, K.; Anderson, D.; Chen, G.; Mukherjee, C.; Shi, Y.; Gélinas, C.; Fan, Y.; et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006, 10, 51–64. [Google Scholar] [CrossRef]
- Sun, T.; Liu, H.; Ming, L. Multiple roles of autophagy in the sorafenib resistance of hepatocellular carcinoma. Cell. Physiol. Biochem. 2017, 44, 716–727. [Google Scholar] [CrossRef]
- Saxena, R.; Klochkova, A.; Murray, M.G.; Kabir, M.F.; Samad, S.; Beccari, T.; Gang, J.; Patel, K.; Hamilton, K.E.; Whelan, K.A. Roles for autophagy in esophageal carcinogenesis: Implications for improving patient outcomes. Cancers 2019, 11, 1697. [Google Scholar] [CrossRef]
- Wang, J.; Wu, G.S. Role of autophagy in cisplatin resistance in ovarian cancer cells. J. Biol. Chem. 2014, 289, 17163–17173. [Google Scholar] [CrossRef] [PubMed]
- Drullion, C.; Trégoat, C.; Lagarde, V.; Tan, S.; Gioia, R.; Priault, M.; Djavaheri-Mergny, M.; Brisson, A.; Auberger, P.; Mahon, F.X.; et al. Apoptosis and autophagy have opposite roles on imatinib-induced K562 leukemia cell senescence. Cell Death Dis. 2012, 3, e373. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, S.; Pirtoli, L.; Tini, P.; Cevenini, G.; Calderaro, F.; Toscano, M.; Miracco, C.; Comincini, S. Different involvement of autophagy in human malignant glioma cell lines undergoing irradiation and temozolomide combined treatments. J. Cell. Biochem. 2012, 113, 2308–2318. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Deng, L.; Chen, Q.; Wang, Y.; Xu, R.; Shi, C.; Shao, J.; Hu, G.; Gao, M.; Rao, H.; et al. Inhibition of hedgehog signaling pathway impedes cancer cell proliferation by promotion of autophagy. Eur. J. Cell Biol. 2015, 94, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Lo Ré, A.E.; Fernández-Barrena, M.G.; Almada, L.L.; Mills, L.D.; Elsawa, S.F.; Lund, G.; Ropolo, A.; Molejon, M.I.; Vaccaro, M.I.; Fernandez-Zapico, M.E. Novel AKT1-GLI3-VMP1 pathway mediates KRAS oncogene-induced autophagy in cancer cells. J. Biol. Chem. 2012, 287, 25325–25334. [Google Scholar] [CrossRef]
- Amantini, C.; Morelli, M.B.; Nabissi, M.; Cardinali, C.; Santoni, M.; Gismondi, A.; Santoni, G. Capsaicin triggers autophagic cell survival which drives epithelial mesenchymal transition and chemoresistance in bladder cancer cells in an hedgehog-dependent manner. Oncotarget 2016, 7, 50180–50194. [Google Scholar] [CrossRef]
- Honorato, J.R.; Hauser-Davis, R.A.; Saggioro, E.M.; Correia, F.V.; Sales-Junior, S.F.; Soares, L.O.S.; Lima, L.D.R.; Moura-Neto, V.; Lopes, G.P.D.F.; Spohr, T.C.L.D.S. Role of sonic hedgehog signaling in cell cycle, oxidative stress, and autophagy of temozolomide resistant glioblastoma. J. Cell. Physiol. 2020, 235, 3798–3814. [Google Scholar] [CrossRef]
- Jin, M.Z.; Jin, W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef]
- Lei, J.; Ma, J.; Ma, Q.; Li, X.; Liu, H.; Xu, Q.; Duan, W.; Sun, Q.; Xu, J.; Wu, Z.; et al. Hedgehog signaling regulates hypoxia induced epithelial to mesenchymal transition and invasion in pancreatic cancer cells via a ligand-independent manner. Mol. Cancer 2013, 12, 66. [Google Scholar] [CrossRef]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfried, E.; Schwarz, S.; Rothe, G.; Hoves, S.; et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef]
- Ward, C.; Meehan, J.; Gray, M.E.; Murray, A.F.; Argyle, D.J.; Kunkler, I.H.; Langdon, S.P. The impact of tumour PH on cancer progression: Strategies for clinical intervention. Explor. Target. Antitumor Ther. 2020, 1, 71–100. [Google Scholar] [CrossRef]
- Senthebane, D.A.; Rowe, A.; Thomford, N.E.; Shipanga, H.; Munro, D.; Al Mazeedi, M.A.M.; Almazyadi, H.A.M.; Kallmeyer, K.; Dandara, C.; Pepper, M.S.; et al. The role of tumor microenvironment in chemoresistance: To survive, keep your enemies closer. Int. J. Mol. Sci. 2017, 18, 1586. [Google Scholar] [CrossRef] [PubMed]
- Hui, M.; Cazet, A.; Jessica, Y.; Cooper, C.; McFarland, A.; Nair, R.; O’Toole, S.; Swarbrick, A. Targeting the hedgehog signalling pathway in triple negative breast cancer. Ann. Oncol. 2015, 26, iii31. [Google Scholar] [CrossRef]
- Hui, M.N.; Cazet, A.; Elsworth, B.; Roden, D.; Cox, T.; Yang, J.; McFarland, A.; Deng, N.; Chan, C.-L.; O’Toole, S.; et al. Targeting the hedgehog signalling pathway in triple negative breast cancer. J. Clin. Oncol. 2018, 36, e24216. [Google Scholar] [CrossRef]
- Cazet, A.S.; Hui, M.N.; Elsworth, B.L.; Wu, S.Z.; Roden, D.; Chan, C.L.; Skhinas, J.N.; Collot, R.; Yang, J.; Harvey, K.; et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun. 2018, 9, 2897. [Google Scholar] [CrossRef] [PubMed]
- Feig, C.; Gopinathan, A.; Neesse, A.; Chan, D.S.; Cook, N.; Tuveson, D.A. The pancreas cancer microenvironment. Clin. Cancer Res. 2012, 18, 4266–4276. [Google Scholar] [CrossRef]
- Thayer, S.P.; Di Magliano, M.P.; Heiser, P.W.; Nielsen, C.M.; Roberts, D.J.; Lauwers, G.Y.; Qi, Y.P.; Gysin, S.; Fernández-del Castillo, C.; Yajnik, V.; et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 2003, 425, 851–856. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Wang, L.; Multani, A.S.; Combs, C.; Deramaudt, T.B.; Hruban, R.H.; Rustgi, A.K.; Chang, S.; Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005, 7, 469–483. [Google Scholar] [CrossRef]
- Tian, H.; Callahan, C.A.; Dupree, K.J.; Darbonne, W.C.; Ahn, C.P.; Scales, S.J.; De Sauvage, F.J. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 4254–4259. [Google Scholar] [CrossRef]
- Olive, K.P.; Jacobetz, M.A.; Davidson, C.J.; Gopinathan, A.; McIntyre, D.; Honess, D.; Madhu, B.; Goldgraben, M.A.; Caldwell, M.E.; Allard, D.; et al. Inhibition of hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009, 324, 1457–1461. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Tu, Y.; Hu, M. Challenges and opportunities with predicting in vivo phase II metabolism via glucuronidation from in vitro data. Curr. Pharmacol. Rep. 2016, 2, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Allain, E.P.; Rouleau, M.; Lévesque, E.; Guillemette, C. Emerging roles for UDP-glucuronosyltransferases in drug resistance and cancer progression. Br. J. Cancer 2020, 122, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Zahreddine, H.A.; Culjkovic-Kraljacic, B.; Assouline, S.; Gendron, P.; Romeo, A.A.; Morris, S.J.; Cormack, G.; Jaquith, J.B.; Cerchietti, L.; Cocolakis, E.; et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature 2014, 511, 90–93. [Google Scholar] [CrossRef]
- Kentsis, A.; Volpon, L.; Topisirovic, I.; Soll, C.E.; Culjkovic, B.; Shao, L.; Borden, K.L.B. Further evidence that ribavirin interacts with EIF4E. RNA 2005, 11, 1762–1766. [Google Scholar] [CrossRef] [PubMed]
- Kentsis, A.; Topisirovic, I.; Culjkovic, B.; Shao, L.; Borden, K.L.B. Ribavirin suppresses ElF4E-mediated oncogenic transformation by physical mimicry of the 7-Methyl Guanosine MRNA Cap. Proc. Natl. Acad. Sci. USA 2004, 101, 18105–18110. [Google Scholar] [CrossRef] [PubMed]
- Zahreddine, H.A.; Culjkovic-Kraljacic, B.; Gasiorek, J.; Duchaine, J.; Borden, K.L.B. GLI1-Inducible glucuronidation targets a broad spectrum of drugs. ACS Chem. Biol. 2019, 14, 348–355. [Google Scholar] [CrossRef]
- Ford, R.C.; Beis, K. Learning the ABCs one at a time: Structure and mechanism of ABC transporters. Biochem. Soc. Trans. 2019, 47, 23–36. [Google Scholar] [CrossRef]
- Pan, S.-T.; Li, Z.-L.; He, Z.-X.; Qiu, J.-X.; Zhou, S.-F. Molecular mechanisms for tumour resistance to chemotherapy. Clin. Exp. Pharmacol. Physiol. 2016, 43, 723–737. [Google Scholar] [CrossRef]
- Begicevic, R.R.; Falasca, M. ABC Transporters in cancer stem cells: Beyond chemoresistance. Int. J. Mol. Sci. 2017, 18, 2362. [Google Scholar] [CrossRef]
- Santisteban, M. ABC Transporters as molecular effectors of pancreatic oncogenic pathways: The hedgehog-GLI model. J. Gastrointest. Cancer 2010, 41, 153–158. [Google Scholar] [CrossRef]
- Steg, A.D.; Katre, A.A.; Bevis, K.S.; Ziebarth, A.; Dobbin, Z.C.; Shah, M.M.; Alvarez, R.D.; Landen, C.N. Smoothened antagonists reverse taxane resistance in ovarian cancer. Mol. Cancer Ther. 2012, 11, 1587–1597. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, L.; Cheng, M.; Wang, Q.; Hu, X.; Chen, Q. The hedgehog signaling pathway promotes chemotherapy resistance via multidrug resistance protein 1 in ovarian cancer. Oncol. Rep. 2020, 44, 2610–2620. [Google Scholar] [CrossRef]
- Chen, Y.; Bieber, M.M.; Teng, N.N.H. Hedgehog signaling regulates drug sensitivity by targeting ABC transporters ABCB1 and ABCG2 in epithelial ovarian cancer. Mol. Carcinog. 2014, 53, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Fang, T.; Duan, Z.; Xiang, D.; Wang, Y.; Zhang, M.; Zhai, F.; Cui, X.; Yang, L. Dihydroartemisinin sensitizes esophageal squamous cell carcinoma to cisplatin by inhibiting sonic hedgehog signaling. Front. Cell Dev. Biol. 2020, 8596788. [Google Scholar] [CrossRef]
- Das, S.; Samant, R.S.; Shevde, L.A. Nonclassical activation of hedgehog signaling enhances multidrug resistance and makes cancer cells refractory to smoothened-targeting hedgehog inhibition. J. Biol. Chem. 2013, 288, 11824–11833. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Gu, D.; Zhang, X.; Liu, B.; Xie, J. The role of GLI2-ABCG2 signaling axis for 5Fu resistance in gastric cancer. J. Genet. Genomics 2017, 44, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Gu, D.; Zhang, X.; Li, J.; Liu, B.; Xie, J. GLI1-mediated regulation of side population is responsible for drug resistance in gastric cancer. Oncotarget 2017, 8, 27412–27427. [Google Scholar] [CrossRef]
- Yao, Y.; Zhou, D.; Shi, D.; Zhang, H.; Zhan, S.; Shao, X.; Sun, K.; Sun, L.; Wu, G.; Tian, K.; et al. GLI1 Overexpression promotes gastric cancer cell proliferation and migration and induces drug resistance by combining with the AKT-MTOR pathway. Biomed. Pharmacother. 2019, 111, 993–1004. [Google Scholar] [CrossRef]
- Xu, M.; Gong, A.; Yang, H.; George, S.K.; Jiao, Z.; Huang, H.; Jiang, X.; Zhang, Y. Sonic hedgehog-glioma associated oncogene homolog 1 signaling enhances drug resistance in CD44+/Musashi-1+ gastric cancer stem cells. Cancer Lett. 2015, 369, 124–133. [Google Scholar] [CrossRef]
- Huang, F.T.; Zhuan-Sun, Y.X.; Zhuang, Y.Y.; Wei, S.L.; Tang, J.; Chen, W.B.; Zhang, S.N. Inhibition of hedgehog signaling depresses self-renewal of pancreatic cancer stem cells and reverses chemoresistance. Int. J. Oncol. 2012, 41, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhou, X.T.; Zou, H.Y.; Wu, J. Hedgehog signaling pathway affects the sensitivity of hepatoma cells to drug therapy through the ABCC1 transporter. Lab. Investig. 2017, 97, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lingala, S.; Khoobyari, S.; Nolta, J.; Zern, M.A.; Wu, J. Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J. Hepatol. 2011, 55, 838–845. [Google Scholar] [CrossRef]
- Zhou, X.T.; Ding, J.; Li, H.Y.; Zuo, J.L.; Ge, S.Y.; Jia, H.L.; Wu, J. Hedgehog signalling mediates drug resistance through targeting TAP1 in hepatocellular carcinoma. J. Cell. Mol. Med. 2020, 24, 4298–4311. [Google Scholar] [CrossRef]
- Amable, L.; Fain, J.; Gavin, E.; Reed, E. Gli1 Contributes to cellular resistance to cisplatin through altered cellular accumulation of the drug. Oncol. Rep. 2014, 32, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Kim, A.Y.; Kwak, J.H.; Je, N.K.; Lee, Y.; Jung, Y.S. Epithelial-mesenchymal transition is associated with acquired resistance to 5-fluorocuracil in HT-29 colon cancer cells. Toxicol. Res. 2015, 31, 151–156. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Zarrabi, A.; Hushmandi, K.; Kalantari, M.; Mohammadinejad, R.; Javaheri, T.; Sethi, G. Association of the Epithelial–Mesenchymal Transition (EMT) with cisplatin resistance. Int. J. Mol. Sci. 2020, 21, 4002. [Google Scholar] [CrossRef]
- Liu, Y.-R.; Liang, L.; Zhao, J.M.; Zhang, Y.; Zhang, M.; Zhong, W.-L.; Zhang, Q.; Wei, J.-J.; Li, M.; Yuan, J.; et al. Twist1 confers multidrug resistance in colon cancer through upregulation of ATP-binding cassette transporters. Oncotarget 2017, 8, 52901–52912. [Google Scholar] [CrossRef]
- Jiang, Z.S.; Sun, Y.Z.; Wang, S.M.; Ruan, J.S. Epithelial-mesenchymal transition: Potential regulator of ABC transporters in tumor progression. J. Cancer 2017, 8, 2319–2327. [Google Scholar] [CrossRef]
- Liu, Y.; Du, F.; Zhao, Q.; Jin, J.; Ma, X.; Li, H. Acquisition of 5-fluorouracil resistance induces epithelial-mesenchymal transitions through the hedgehog signaling pathway in HCT-8 colon cancer cells. Oncol. Lett. 2015, 9, 2675–2679. [Google Scholar] [CrossRef]
- Maitah, M.Y.; Ali, S.; Ahmad, A.; Gadgeel, S.; Sarkar, F.H. Up-regulation of sonic hedgehog contributes to TGF-Β1-induced epithelial to mesenchymal transition in NSCLC Cells. PLoS ONE 2011, 6, e16068. [Google Scholar] [CrossRef]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; Hausen, A.Z.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting MicroRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef]
- Hojo, N.; Huisken, A.L.; Wang, H.; Chirshev, E.; Kim, N.S.; Nguyen, S.M.; Campos, H.; Glackin, C.A.; Ioffe, Y.J.; Unternaehrer, J.J. Snail knockdown reverses stemness and inhibits tumour growth in ovarian cancer. Sci. Rep. 2018, 8, 8704. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.H.; Lan, H.Y.; Huang, C.H.; Tai, S.K.; Tzeng, C.H.; Kao, S.Y.; Wu, K.J.; Hung, M.C.; Yang, M.H. RAC1 Activation mediates twist1-induced cancer cell migration. Nat. Cell Biol. 2012, 14, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Maitah, M.Y.; Ginnebaugh, K.R.; Li, Y.; Bao, B.; Gadgeel, S.M.; Sarkar, F.H. Inhibition of hedgehog signaling sensitizes NSCLC cells to standard therapies through modulation of EMT-regulating MiRNAs. J. Hematol. Oncol. 2013, 6, 77. [Google Scholar] [CrossRef]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.C.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef]
- Singh, A.; Settleman, J. EMT, Cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Wang, J.; Liu, Y.; Peng, Y.; Tan, W. GPCR-like signaling mediated by smoothened contributes to acquired chemoresistance through activating gli. Mol. Cancer 2014, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Peng, Y.; Liu, Y.; Xin, H.; Zhan, X.; Tan, W. Twist1 and snail link hedgehog signaling to tumor-initiating cell-like properties and acquired chemoresistance independently of ABC transporters. Stem Cells 2015, 33, 1063–1074. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Izadpanah, M.H.; Abbaszadegan, M.R.; Fahim, Y.; Forghanifard, M.M. Ectopic expression of TWIST1 upregulates the stemness marker OCT4 in the esophageal squamous cell carcinoma cell line KYSE. Cell. Mol. Biol. Lett. 2017, 22, 33. [Google Scholar] [CrossRef]
- Zhou, W.; Lv, R.; Qi, W.; Wu, D.; Xu, Y.; Liu, W.; Mou, Y.; Wang, L. Snail contributes to the maintenance of stem cell-like phenotype cells in human pancreatic cancer. PLoS ONE 2014, 9, 87409. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ma, L.; Zhang, Z.; Liu, X.; Gao, H.; Zhuang, Y.; Yang, P.; Kornmann, M.; Tian, X.; Yang, Y. hedgehog signaling regulates epithelial-mesenchymal transition in pancreatic cancer stem-like cells. J. Cancer 2016, 7, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Atashzar, M.R.; Baharlou, R.; Karami, J.; Abdollahi, H.; Rezaei, R.; Pourramezan, F.; Zoljalali Moghaddam, S.H. Cancer stem cells: A review from origin to therapeutic implications. J. Cell. Physiol. 2020, 235, 790–803. [Google Scholar] [CrossRef]
- Kim, W.T.; Ryu, C.J. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017, 50, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.; Park, D.J.; Schmidt, B.; Thomas, N.J.; Lee, H.J.; Kim, T.S.; Janjigian, Y.Y.; Cohen, D.J.; Yoon, S.S. CD44 Expression denotes a subpopulation of gastric cancer cells in which hedgehog signaling promotes chemotherapy resistance. Clin. Cancer Res. 2014, 20, 3974–3988. [Google Scholar] [CrossRef]
- Song, Z.; Yue, W.; Wei, B.; Wang, N.; Li, T.; Guan, L.; Shi, S.; Zeng, Q.; Pei, X.; Chen, L. Sonic hedgehog pathway is essential for maintenance of cancer stem-like cells in human gastric cancer. PLoS ONE 2011, 6, e17687. [Google Scholar] [CrossRef]
- Wang, D.; Nagle, P.W.; Wang, H.H.; Smit, J.K.; Faber, H.; Baanstra, M.; Karrenbeld, A.; Chiu, R.K.; Plukker, J.T.M.; Coppes, R.P. Hedgehog pathway as a potential intervention target in esophageal cancer. Cancers 2019, 11, 821. [Google Scholar] [CrossRef]
- Kobune, M.; Ito, Y.; Kawano, Y.; Sasaki, K.; Uchida, H.; Nakamura, K.; Dehari, H.; Chiba, H.; Takimoto, R.; Matsunaga, T.; et al. Indian hedgehog gene transfer augments hematopoietic support of human stromal cells including NOD/SCID-B2m-/- repopulating cells. Blood 2004, 104, 1002–1009. [Google Scholar] [CrossRef]
- Kobune, M.; Takimoto, R.; Murase, K.; Iyama, S.; Sato, T.; Kikuchi, S.; Kawano, Y.; Miyanishi, K.; Sato, Y.; Niitsu, Y.; et al. Drug resistance is dramatically restored by hedgehog inhibitors in CD34+ leukemic cells. Cancer Sci. 2009, 100, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Long, B.; Wang, L.-X.; Zheng, F.-M.; Lai, S.-P.; Xu, D.-R.; Hu, Y.; Lin, D.-J.; Zhang, X.-Z.; Dong, L.; Long, Z.-J.; et al. Targeting GLI1 suppresses cell growth and enhances chemosensitivity in CD34+ enriched acute myeloid leukemia progenitor cells. Cell. Physiol. Biochem. 2016, 38, 1288–1302. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; An, Y.; Wei, J.S.; Ji, Z.L.; Lu, Z.P.; Wu, J.L.; Jiang, K.R.; Chen, P.; Xu, Z.K.; Miao, Y. Cyclopamine reverts acquired chemoresistance and down-regulates cancer stem cell markers in pancreatic cancer cell lines. Swiss Med. Wkly. 2011, 141, w13208. [Google Scholar] [CrossRef]
- Ren, Y.; Deng, R.; Cai, R.; Lu, X.; Luo, Y.; Wang, Z.; Zhu, Y.; Yin, M.; Ding, Y.; Lin, J. TUSC3 Induces drug resistance and cellular stemness via hedgehog signaling pathway in colorectal cancer. Carcinogenesis 2020, 41, 1755–1766. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, Q.; Guo, K.; Qin, W.; Liao, W.; Wang, S.; Ding, Y.; Lin, J. TUSC3 promotes colorectal cancer progression and Epithelial-Mesenchymal Transition (EMT) through WNT/β-catenin and MAPK signalling. J. Pathol. 2016, 239, 60–71. [Google Scholar] [CrossRef]
- Varnat, F.; Duquet, A.; Malerba, M.; Zbinden, M.; Mas, C.; Gervaz, P.; Ruiz, I.; Altaba, A. human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol. Med. 2009, 1, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.A.; Chen, Y.F.; Bao, Y.; Mahara, S.; Yatim, S.M.J.M.; Oguz, G.; Lee, P.L.; Feng, M.; Cai, Y.; Tan, E.Y.; et al. Hypoxic tumor microenvironment activates GLI2 via HIF-1α and TGF-Β2 to promote chemoresistance in colorectal cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E5990–E5999. [Google Scholar] [CrossRef] [PubMed]

| Molecular Target | Drug | Status | Cancer Target | Drug Combination | Mechanism of Chemoresistance | Treatment Outcome | Citation |
|---|---|---|---|---|---|---|---|
| Shh/Ihh | 5E1 | Preclinical study | Gastric | Oxaliplatin and Mitomycin | Hh signaling enhanced self-renewing capacity of tumorspheres, which promotes oxaliplatin and mitomycin resistance | Hh signaling inhibition enhanced cell death of CD44+ HGC-7 tumorsphere cells and primary gastric tumor samples when co-treated with oxaliplatin and mitomycin | [158] |
| SMO | Sonidegib/Erismodegib (NVP-LDE-225) | Preclinical study and phase I clinical trail EDALINE | Breast | Docetaxel | M6-Hh cells activate CAFs via paracrine Hh signaling to induce ECM remodeling and consequently chemoresistant CSCs phenotype in adjacent epithelial cells | SMO inhibition reduced collagen fibrillar deposition and consequently reversed the CSC phenotype of M6-Hh cells, which sensitizes M6-Hh tumors, patient-derived xenografts, and metastatic TNBC patients to docetaxel | [105,106] |
| Preclinical study | Ovarian | Paclitaxel | The upregulation of SMO, GLI1, and GLI2 modulate ABCB1 expression to promote paclitaxel resistance | SMO inhibition sensitized taxane resistant ALDH-negative and positive A2780 and SKOV3 cell lines to paclitaxel by downregulating ABCB1 expression | [122] | ||
| Vismodegib (GDC-0449) | Preclinical study | Myeloid Leukemia | Ribavirin, cytarabine, methotrexate, venetoclax, 5-fluorouracil, sunitinib, and idarubicin | High levels of GLI1 mediated the UGT1A-dependent glucuronidation of several cancer therapeutics via calreticulin-dependent UGT1A protein stabilization to enhance drug elimination | Inhibition of GLI1 reduced UGT1A dependent glucuronidation of cancer therapeutics to promote drug accumulation (e.g., eIF4E-ribavirin complex) in primary AML specimens and acquired resistant cell lines | [114,117] | |
| Colon | 5-fluorouracil | Elevated GLI1 levels induced EMT phenotype, which was associated with 5-fluorouracil resistance | Inhibition of SMO reversed EMT signals and 5-fluorouracil resistance in colon cancer HCT-8 cell line | [141] | |||
| Lung | Cisplatin | Elevated Hh signaling contributed to TGF-β1 induced EMT and cisplatin resistance via the downregulation of miR-200b and let-7c | The inhibition of Hh signaling sensitized A549 EMT cells to cisplatin through the restoration of miR-200b and let-7c expression and reversal of EMT genes | [146] | |||
| Esophageal | Carboplatin | High levels of Shh and PTCH1 were associated with increased CSC traits and therapeutic resistance | Inhibition of Hh pathway via SMO decreased CD44+ CSC phenotype and sphere-forming potential in esophageal cancer OE21 cell line, which restored carboplatin sensitivity | [159] | |||
| Preclinical study and phase II clinical trial | Gastric | 5-fluorouracil, cisplatin, oxaliplatin, and leucovorin | Increased Hh signaling enhanced CSC traits in gastric cancer (GC) cells to promote 5-fluorouracil and cisplatin resistance | SMO inhibition sensitized CD44+ AGS, MKN45, and N87 spheroid cells and mice xenografts as well as GC patients with high CD44 median scores to the combination treatment of 5-fluorouracil and cisplatin and 5-fluorouracil, oxaliplatin, and leucovorin, respectively | [157] | ||
| Saridegib (IPI-926) | Preclinical study | Pancreatic | Gemcitabine | Hh pathway activation enhanced stromal desmoplasia and hypovascularity to reduced gemcitabine delivery | SMO inhibition depletes stromal desmoplasia and increases MVD to enhance gemcitabine delivery and consequently intracellular dFdCTP accumulation in KPC mice tissues | [111] | |
| Cyclopamine | Preclinical study | Pancreatic | Gemcitabine and 5-fluorouracil | Hh signaling induced gemcitabine resistance through CSC induction, as well as ABCB1 and ABCG2-mediated drug efflux | Inhibition of SMO restored gemcitabine sensitivity in acquired gemcitabine-resistant SW1990 and CFPAC cell lines expressing CD44 and CD133, as well as restored gemcitabine and 5-fluorouracil sensitivity in PANC-1 tumorspheres by inhibiting their self-renewing capacity and ABCB1 and ABCG2 expression | [131,163] | |
| Glioma | Temozolomide | GLI1 upregulates MGMT at the promoter level to promote TMZ resistance | SMO inhibition restricts Hh/GLI signaling to downregulate MGMT expression and consequently improved TMZ toxicity in GBM U251 and U87 cell lines and xenografts | [75] | |||
| Myeloid Leukemia | Cytarabine | Ihh and its downstream effector (GLI1 and GLI2) were enriched in CD34+ subpopulations, which was associated with cytarabine resistance | Inhibition of Ihh autocrine Hh signaling via SMO induced apoptosis and sensitized cytokine responsive CD34+ Kasumi-1, Kasumi-3, and TF-1 cell lines to cytarabine treatment | [161] | |||
| GLI1/2 | GANT-58 | Preclinical study | Myeloid leukemia and cervical | Doxorubicin and vincristine | GLI1 transcriptionally upregulates TWIST1 and SNAI1 expression to promote tumor-initiating properties and consequently chemoresistance | GLI1 inhibition reduced TWIST1 and SNAI1 levels, which restored the chemosensitivity of multidrug-resistant chronic myelogenous leukemia K562 and human cervical epidermoid carcinoma KB sublines to doxorubicin and vincristine | [150] |
| GANT-61 | Preclinical study | Myeloid Leukemia | Vincristine | High GLI1 levels upregulate ABCB1 to promote vincristine resistance | GLI1 inhibition reversed vincristine resistance of Lucena-1 cell line by interfering with ABCB1 expression | [32] | |
| Cytarabine | GLI1 enrichment in CD34+ cells enhanced colony-forming capacity and cytarabine resistance | GLI1 inhibition induced a significant reduction in cell viability of CD34+ compared to CD34- primary AML cells when treated with cytarabine | [162] | ||||
| Ovarian | Cisplatin | GLI2 transcriptionally upregulates ABCB1 to promotes cisplatin resistance | GLI2 inhibition reduced ABCB1 levels, which enhanced cisplatin-induced DNA damage in cisplatin-resistant OC SK-OV-3 cell line | [123] | |||
| Glioma | Temozolomide | High levels of GLI and MGMT expression in glioblastoma cells lead to TMZ resistance | Inhibition of GLI with GANT-61 reduced the level of MGMT, which restored the sensitivity to temozolomide in GBM U251 and U87 cell lines | [73] | |||
| Hepatoma | Cisplatin, doxorubicin, and sorafenib | The binding of GLI1/2 to the GLI1-binding consensus sequence within the ABCB2 promoter initiated its transcription to confer drug resistance | Inhibition of GLI1/2 downregulated the ABCB2 expression in Huh-7 DN (CD133-/EpCAM-) and trans (CD133−/EpCAM− transwell-selected) hepatoma subpopulations, which enhanced their chemotherapeutic drug sensitivity | [132] | |||
| Colorectal | 5-fluorouracil and cisplatin | Hh signaling has SMO as a mediator of TUSC3-induced CSC phenotype and drug resistance | Inhibition of Hh signaling at the GLI level inhibited the expression of CD133 and ABCC1 and decreased the number of TUSC3-overexpressing CACO2 and RKO2 tumorspheres, which was associated with increased 5-fluorouracil and cisplatin sensitivity | [164] | |||
| SIR-38832 | Preclinical study | Colorectal | 5-fluorouracil | GLI1 transcriptionally upregulates NBS1 and consequently MRN complex function to reduce 5-fluorouracil induced DNA damage | Inhibition of GLI1 activity reduced total NBS1 levels and impaired MRN complex function to increase 5-fluorouracil induced DNA damage in HT29 cell lines and xenografts | [81] | |
| Not specified | Dihydroartemisinin | Preclinical study | Esophageal | Cisplatin | Hh activation upregulates ABCB1 levels to reduce cisplatin enrichment | Hh inhibition reduced ABCB1 levels and consequently enhanced cisplatin accumulation in the TE-1 cell line | [125] |
| HIF-1α | Oroxylin A | Preclinical study | Glioma | TMZ | HIF-1α activates Shh/GLI1/MGMT signaling to promote TMZ resistance | HIF-1α degradation inhibits Hh pathway activation and increases SUFU expression, thus reducing MGMT levels and restoring TMZ sensitivity in glioma U251 and C6 cell lines and xenografts | [75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, J.Y.; Sugumar, V.; Alshanon, A.F.; Wong, W.F.; Fung, S.Y.; Looi, C.Y. Defining the Role of GLI/Hedgehog Signaling in Chemoresistance: Implications in Therapeutic Approaches. Cancers 2021, 13, 4746. https://doi.org/10.3390/cancers13194746
Chai JY, Sugumar V, Alshanon AF, Wong WF, Fung SY, Looi CY. Defining the Role of GLI/Hedgehog Signaling in Chemoresistance: Implications in Therapeutic Approaches. Cancers. 2021; 13(19):4746. https://doi.org/10.3390/cancers13194746
Chicago/Turabian StyleChai, Jian Yi, Vaisnevee Sugumar, Ahmed F. Alshanon, Won Fen Wong, Shin Yee Fung, and Chung Yeng Looi. 2021. "Defining the Role of GLI/Hedgehog Signaling in Chemoresistance: Implications in Therapeutic Approaches" Cancers 13, no. 19: 4746. https://doi.org/10.3390/cancers13194746
APA StyleChai, J. Y., Sugumar, V., Alshanon, A. F., Wong, W. F., Fung, S. Y., & Looi, C. Y. (2021). Defining the Role of GLI/Hedgehog Signaling in Chemoresistance: Implications in Therapeutic Approaches. Cancers, 13(19), 4746. https://doi.org/10.3390/cancers13194746