Low Frequency of Human Papillomavirus in Strictly Site-Coded Oral Squamous Cell Carcinomas, Using the Latest NHI/SEER-ICD Systems: A Pilot Observational Study and Critical Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Observational Study
2.1.1. Entry Criteria
- (i).
- age ≥ 18 years;
- (ii).
- ability to provide informed consent;
- (iii).
- suspected OSCC, located strictly in the oral cavity, having applied the 2021 NIH/SEER ICD-0-3.2 topographical classification codes (from C02.0 to C02.3 for the tongue, C03.0 and C03.1 for the gum, C04.0 and C04.1 for the floor of the mouth, C05.0 and C05.1 for palate, and C06.0, C06.1 and C06.2 for cheek mucosa, mouth vestibule and retromolar area); the codes referring to not otherwise specified (NOS) oral sites have not been considered [8,10];
- (iv).
- no previous diagnosis of cancer in the head or neck regions.
2.1.2. Data Collection and Clinical Examination
2.1.3. Sample Collection
2.1.4. Histological Examination and p16 Immunohistochemistry
2.1.5. DNA Extraction and HPV DNA Detection
2.2. Critical Review
2.2.1. Focused Questions
- ▪
- How many studies, published in the last decade and regarding the frequency of HPV in OSCC, report a standardized coding of oral sites?
- ▪
- Are there differences in the frequency of HPV status when the ‘not otherwise specified (NOS) tongue’ are excluded?
- ▪
- Are there discordant results regarding the frequency of HPV between PCR DNA (considered the gold standard) and p16 IHC?
2.2.2. Search Criteria
2.2.3. Comparison Data Criteria
2.3. Statistical Analysis
3. Results
3.1. Observational Study
3.2. Critical Review
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fakhry, C.; Lacchetti, C.; Rooper, L.M.; Jordan, R.C.; Rischin, D.; Sturgis, E.M.; Bell, D.; Lingen, M.W.; Harichand-Herdt, S.; Thibo, J.; et al. Human papillomavirus testing in head and neck carcinomas: ASCO clinical practice guideline endorsement of the college of American pathologists guideline. J. Clin. Oncol. 2018, 36, 3152–3161. [Google Scholar] [CrossRef]
- Schache, A.G.; Liloglou, T.; Risk, J.M.; Filia, A.; Jones, T.M.; Sheard, J.; Woolgar, J.A.; Helliwell, T.R.; Triantafyllou, A.; Robinson, M.; et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: Sensitivity, specificity, and prognostic discrimination. Clin. Cancer Res. 2011, 17, 6262–6271. [Google Scholar] [CrossRef] [Green Version]
- Bishop, J.A.; Ma, X.J.; Wang, H.; Luo, Y.; Illei, P.B.; Begum, S.; Taube, J.M.; Koch, W.M.; Westra, W.H. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am. J. Surg. Pathol. 2012, 36, 1874–1882. [Google Scholar] [CrossRef] [Green Version]
- Campisi, G.; Panzarella, V. Human Papillomavirus Infection: A Risk Factor for Oral and Oropharyngeal Cancers. In Textbook of Oral Cancer: Prevention, Diagnosis and Management; Warnakulasuriya, S., Greenspan, J.S., Eds.; Springer: Cham, Switzerland, 2020; pp. 31–45. [Google Scholar]
- Lydiatt, W.M.; Patel, S.G.; O’Sullivan, B.; Brandwein, M.S.; Ridge, J.A.; Migliacci, J.C.; Loomis, A.M.; Shah, J.P. Head and neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Mirghani, H.; Casiraghi, O.; Amen, F.; He, M.; Ma, X.J.; Saulnier, P.; Lacroix, L.; Drusch, F.; Lakdhar, A.B.; Saint Guily, J.L.; et al. Diagnosis of HPV-driven head and neck cancer with a single test in routine clinical practice. Mod. Pathol. 2015, 28, 1518–1527. [Google Scholar] [CrossRef] [Green Version]
- Syrjänen, S. Oral manifestations of human papillomavirus infections. Eur. J. Oral Sci. 2018, 126, 49–66. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. International Classification of Diseases for Oncology, 3rd ed.; First Revision; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Ndiaye, C.; Mena, M.; Alemany, L.; Arbyn, M.; Castellsagué, X.; Laporte, L.; Bosch, F.X.; de Sanjosé, S.; Trottier, H. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: A systematic review and meta-analysis. Lancet Oncol. 2014, 15, 1319–1331. [Google Scholar] [CrossRef]
- National Cancer Institute. Surveillance, Epidemiology, and E.R.P.-(NIH/SEER). Head and Neck Equivalent Terms and Definitions C000-C148, C300-C339, C410, C411, C442, C479 Excludes lymphoma and leukemia M9590–M9992 and Kaposi sarcoma M9140. 2020; pp. 1–50. Available online: https://seer.cancer.gov/tools/solidtumor/Head_Neck_STM.pdf (accessed on 31 December 2020).
- Donà, M.G.; Spriano, G.; Pichi, B.; Rollo, F.; Laquintana, V.; Covello, R.; Pellini, R.; Giuliani, M.; Pescarmona, E.; Benevolo, M. Human papillomavirus infection and p16 overexpression in oropharyngeal squamous cell carcinoma: A case series from 2010 to 2014. Future Microbiol. 2015, 10, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M. HPV testing of head and neck cancer in clinical practice. Recent Results Cancer Res. 2017, 206, 101–111. [Google Scholar] [PubMed]
- Wasylyk, B.; Abecassis, J.; Jung, A.C. Identification of clinically relevant HPV-related HNSCC: In p16 should we trust? Oral Oncol. 2013, 49, e33-7. [Google Scholar] [CrossRef]
- Harris, S.L.; Thorne, L.B.; Seaman, W.T.; Neil Hayes, D.; Couch, M.E.; Kimple, R.J. Association of p16INK4a overexpression with improved outcomes in young patients with squamous cell cancers of the oral tongue. Head Neck 2011, 33, 1622–1627. [Google Scholar] [CrossRef]
- Laco, J.; Vosmikova, H.; Novakova, V.; Celakovsky, P.; Dolezalova, H.; Tucek, L.; Nekvindova, J.; Vosmik, M.; Cermakova, E.; Ryska, A. The role of high-risk human papillomavirus infection in oral and oropharyngeal squamous cell carcinoma in non-smoking and non-drinking patients: A clinicopathological and molecular study of 46 cases. Virchows Arch. 2011, 458, 179–187. [Google Scholar] [CrossRef]
- Blumberg, J.; Monjane, L.; Prasad, M.; Carrilho, C.; Judson, B.L. Investigation of the presence of HPV related oropharyngeal and oral tongue squamous cell carcinoma in Mozambique. Cancer Epidemiol. 2015, 39, 1000–1005. [Google Scholar] [CrossRef]
- Emmett, S.; Jenkins, G.; Boros, S.; Whiteman, D.C.; Panizza, B.; Antonsson, A. Low prevalence of human papillomavirus in oral cavity squamous cell carcinoma in Queensland, Australia. ANZ J. Surg. 2017, 87, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Vidal Loustau, A.C.; Dulguerov, N.; Curvoisier, D.; McKee, T.; Lombardi, T. Low prevalence of HPV-induced oral squamous cell carcinoma in Geneva, Switzerland. Oral Dis. 2019, 25, 1283–1290. [Google Scholar] [CrossRef]
- Duray, A.; Descamps, G.; Decaestecker, C.; Remmelink, M.; Sirtaine, N.; Lechien, J.; Ernoux-Neufcoeur, P.; Bletard, N.; Somja, J.; Depuydt, C.E.; et al. Human papillomavirus DNA strongly correlates with a poorer prognosis in oral cavity carcinoma. Laryngoscope 2012, 122, 1558–1565. [Google Scholar] [CrossRef] [PubMed]
- Elango, K.J.; Suresh, A.; Erode, E.M.; Subhadradevi, L.; Ravindran, H.K.; Iyer, S.K.; Iyer, S.K.R.; Kuriakose, M.A. Role of human papilloma virus in oral tongue squamous cell carcinoma. Asian Pac. J. Cancer Prev. 2011, 12, 889–896. [Google Scholar]
- Duncan, L.D.; Winkler, M.; Carlson, E.R.; Heidel, R.E.; Kang, E.; Webb, D. P16 immunohistochemistry can be used to detect human papillomavirus in oral cavity squamous cell carcinoma. J. Oral Maxillofac. Surg. 2013, 71, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Ndiaye, C.; Alemany, L.; Diop, Y.; Ndiaye, N.; Diémé, M.J.; Tous, S.; Klaustermeier, J.E.; Alejo, M.; Castellsagué, X.; Bosch, F.X.; et al. The role of human papillomavirus in head and neck cancer in Senegal. Infect. Agent. Cancer 2013, 8, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rushatamukayanunt, P.; Morita, K.I.; Matsukawa, S.; Harada, H.; Shimamoto, H.; Tomioka, H.; Omura, K. Lack of association between high-risk human papillomaviruses and oral squamous cell carcinoma in young Japanese patients. Asian Pac. J. Cancer Prev. 2014, 15, 4135–4141. [Google Scholar] [CrossRef] [Green Version]
- Krüger, M.; Pabst, A.M.; Walter, C.; Sagheb, K.; Günther, C.; Blatt, S.; Weise, K.; Al-Nawas, B.; Ziebart, T. The prevalence of human papilloma virus (HPV) infections in oral squamous cell carcinomas: A retrospective analysis of 88 patients and literature overview. J. Cranio Maxillofac. Surg. 2014, 42, 1506–1514. [Google Scholar] [CrossRef]
- Singh, V.; Husain, N.; Akhtar, N.; Kumar, V.; Tewari, S.; Mishra, S.; Misra, S.; Khan, M.Y. Do human papilloma viruses play any role in oral squamous cell carcinoma in North Indians. Asian Pac. J. Cancer Prev. 2015, 16, 7077–7084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes, M.; Rojas-Alcayaga, G.; Pennacchiotti, G.; Carrillo, D.; Muñoz, J.P.; Peña, N.; Montes, R.; Lobos, N.; Aguayo, F. Human papillomavirus infection in oral squamous cell carcinomas from Chilean patients. Exp. Mol. Pathol. 2015, 99, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Campisi, G.; Giovannelli, L.; Calvino, F.; Matranga, D.; Colella, G.; Di Liberto, C.; Capra, G.; Leao, J.C.; Lo Muzio, L.; Capogreco, M.; et al. HPV infection in relation to OSCC histological grading and TNM stage. Evaluation by traditional statistics and fuzzy logic model. Oral Oncol. 2006, 42, 638–645. [Google Scholar] [CrossRef]
- Qureishi, A.; Mawby, T.; Fraser, L.; Shah, K.A.; Møller, H.; Winter, S. Current and future techniques for human papilloma virus (HPV) testing in oropharyngeal squamous cell carcinoma. Eur. Arch. Oto Rhino Laryngol. 2017, 274, 2675–2683. [Google Scholar] [CrossRef]
- Bradley, K.T.; Budnick, S.D.; Logani, S. Immunohistochemical detection of p16INK4a in dysplastic lesions of the oral cavity. Mod. Pathol. 2006, 19, 1310–1316. [Google Scholar] [CrossRef]
- Chung, C.H.; Zhang, Q.; Kong, C.S.; Harris, J.; Fertig, E.J.; Harari, P.M.; Wang, D.; Redmond, K.P.; Shenouda, G.; Trotti, A.; et al. p16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J. Clin. Oncol. 2014, 32, 3930–3938. [Google Scholar] [CrossRef] [Green Version]
- Hendawi, N.; Niklander, S.; Allsobrook, O.; Khurram, S.A.; Bolt, R.; Doorbar, J.; Speight, P.M.; Hunter, K.D. Human papillomavirus (HPV) can establish productive infection in dysplastic oral mucosa, but HPV status is poorly predicted by histological features and p16 expression. Histopathology 2020, 76, 592–602. [Google Scholar] [CrossRef]
- Taberna, M.; Mena, M.; Tous, S.; Pavón, M.A.; Oliva, M.; León, X.; Garcia, J.; Guix, M.; Hijano, R.; Bonfill, T.; et al. HPV-relatedness definitions for classifying HPV-related oropharyngeal cancer patient do impact on TNM classification and patients’ survival. PLoS ONE 2018, 13, e0194107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| OSCC (No. 40) | ||
|---|---|---|
| N | (%) | |
| Sex | ||
| Female | 17 | (42.5) |
| Male | 23 | (57.5) |
| Family history of cancer | ||
| Negative | 40 | (100) |
| Positive | 0 | (0) |
| Tobacco consumption (packs/year) | ||
| Non-smokers (0) | 19 | (47.5) |
| Light-smokers (<25) | 3 | (7.5) |
| Moderate/heavy-smokers (>25) | 18 | (45) |
| Alcohol consumption (DU/week) | ||
| Non-drinkers (0 or 1) | 37 | (92.5) |
| Moderate-drinkers (<16) | 0 | (0) |
| Heavy-drinkers (>16) | 3 | (7.5) |
| Mechanical trauma | ||
| Sharp cusps | 12 | (30) |
| Incongruous prosthesis | 3 | (7.5) |
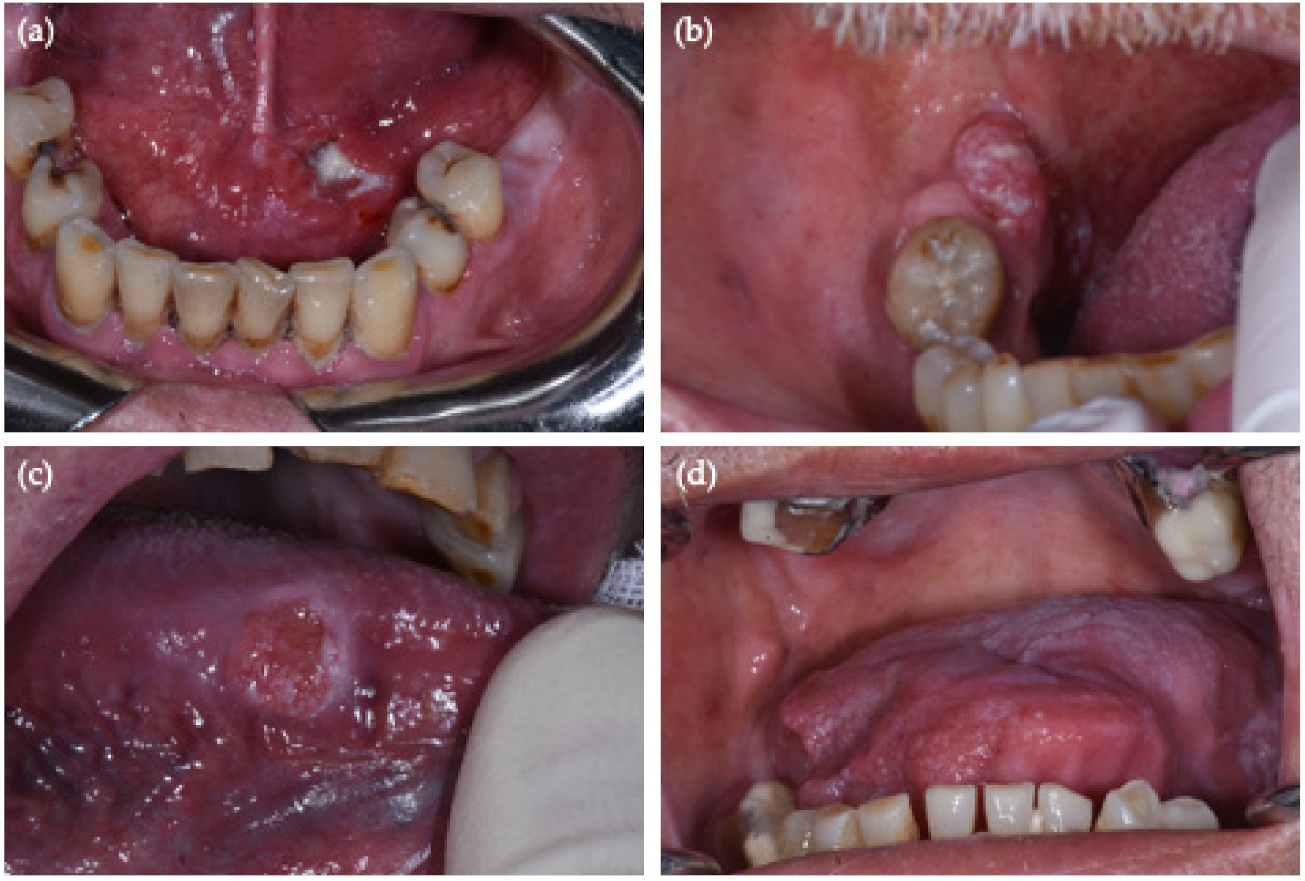
| OSCC Site (By New NIH/SEER ICD-0-3.2 System) | No. (%, 95% CI) | No. HPV-Positive OSCC (%, 95% CI) | Sex/Age | PCR HPV-DNA | p16 IHC |
|---|---|---|---|---|---|
| Border of tongue (C02.1) | 13 (32.5%, 95% CI = [19–49%]) | 2 (15%, 95% CI = [2–45%]) | M/65 | HR-HPV 31 + 68 | − |
| M/65 | HR-HPV 66 | − | |||
| Overlapping lesions of tongue/no base of tongue (C02.8 = C02.1 + C02.2) | 4 (10%, 95% CI = [3–24%]) | 0 | − | − | − |
| Gum (C03.1) | 2 (5%, 95% CI = [0.6–17%]) | 0 | − | − | − |
| Anterior floor of mouth (C04.0) | 4 (10%, 95% CI = [3–24%]) | 1 (25%, 95% CI = [0.6–80%]) | M/44 | HR-HPV 51 | + |
| Hard palate (C05.0) | 3 (7.5%, 95% CI = [2–20%]) | 0 | − | − | − |
| Buccal mucosa (C06.0) | 7 (17.5%, 95% CI = [7–33%]) | 0 | − | − | − |
| Retromolar area (C06.2) | 7 (17.5%, 95% CI = [7–33%]) | 1 (14%, 95% CI = [0.3–58%]) | M/61 | HR-HPV 67 | + |
| HPV Test Results | No./Total OSCC (%, 95% CI) | HPV-Positive OSCC Sites (by 2021 NIH/SEER ICD-0-3.2 System) |
|---|---|---|
| PCR DNA (+) | 4/40 (10%, 95% CI = [2–24%]) | Retromolar area (C06.2) Anterior floor of mouth (C04.0) n.2 Border of the tongue (C02.1) |
| PCR DNA (−) | 36/40 (90%, 95% CI = [76–97%]) | — |
| p16 IHC (+) | 2/40 (5%, 95% CI = [0.6–17%]) | Retromolar area (C06.2) Anterior floor of mouth (C04.0) |
| p16 IHC (−) | 38/40 (95%, 95% CI = [83–99%]) | — |
| PCR DNA (+) p16 IHC (+) | 2/40 (5%, 95% CI = [0.6–17%]) | Retromolar area (C06.2) Anterior floor of mouth (C04.0) |
| PCR DNA (−) p16 IHC (−) | 36/40 (90%, 95% CI = [76–97%]) | — |
| PCR DNA (+) p16 IHC (−) | 2/40 (5%, 95% CI = [0.6–17%]) | n.2 Border of the tongue (C02.1) |
| PCR DNA (−) p16 IHC (+) | 0/40 (0%, 95% CI = [0–9%]) | — |
| Authors (Year) | No.OSCCs Cases (Tissue Sample) | No. HPV-DNA PCR Positive Cases (%, 95% CI) | 2021 NIH/SEER ICD-0-3.2 Site-Coded HPV-DNA PCR Positive Cases | No. p16 Positive Cases/No. HPV-DNA PCR Positive Cases (Sensitivity %, 95% CI) | No. p16 Negative/No. HPV-DNA PCR Positive Cases (Specificity%, 95% CI) | No. p16 Positive Cases/No. HPV-DNA PCR Negative Cases (%, 95% CI) |
|---|---|---|---|---|---|---|
| Harris et al. (2010) [14] | 25 (PE) | 2/25 (8%, 95% CI = [0.9–26%]) | 2 tongue, NOS (C02.9) | 0/2 (0%, 95% CI = [0–84%]) | 12/23 (52%, 95% CI = [31–73%]) | 11/23 (48%, 95% CI = [27–69%]) |
| Laco et al. (2011) [15] | 24 (FFPE) | 3/24 (12,5%, 95% CI = [2–32%]) | 1 cheek mucosa (C06.0) 1 gum, NOS (C03.9) 1 anterior 2/3 of tongue, NOS (C02.3) | not reported | not reported | not reported |
| Elango et al. (2011) [20] | 60 (PE) | 29/60 (48%, 95% CI = [35–62%]) | 29 tongue, NOS (C02.9) | not reported | not reported | not reported |
| Duray et al. (2012) [19] | 147 (FFPE) | 65/147 (44%, 95% CI = [36–53%]) | 13 tongue, NOS (C02.9) 12 floor of the mouth, NOS (C04.9) 2 gum, NOS (C03.9) 1 cheek mucosa (C06.0) | 42/65 (65%, 95% CI = [52–76%]) | 46/82 (56%, 95% CI = [45–67%]) | 36/82 (44%, 95% CI = [33-55%]) |
| Duncan et al. (2013) [21] | 81 (FFPE) | 7/81 (8.6%, 95% CI = [3–17%]) | 13 tongue, NOS (C02.9) 2 gum, NOS (C03.9) 12 floor of the mouth, NOS (C04.9) 1 cheek mucosa (C06.0) 1 retromolar area (C06.2) 2 lip, NOS (C00.9) 1 palate, NOS (C05.9) 33 mouth, NOS (C06.9) | 7/7 (100%, 95% CI = [59–100%]) | 67/74 (91%, 95% CI = [81–96%] | 7/74 (9,4%, 95% CI = [4–19%]) |
| Ndiaye et al. (2013) [22] | 41 (PE) | 1/41 (2.4%, 95% CI = [0.06–13%]) | 1 gum, NOS (C03.9) | 0/1 (0%, 95% CI = [0–98%]) | 40/40 (100%, 95% CI = [91–100%]) | 0/40 (0%, 95% CI = [0–88%]) |
| Rushatamukayanunt et al. (2014) [23] | 275 (FFPE) | 69/275 (25.1%, 95% CI = [20–31%]) | 29 floor of the mouth, NOS (C04.9) 16 tongue, NOS (C02.9) 2 upper gum (C03.0) 13 lower gum (C03.1) 4 cheek mucosa (C06.0) 5 lip, NOS (C00.9) | not reported | not reported | not reported |
| Kruger et Al (2014) [24] | 88 (FF) | 5/88 (6%, 95% CI = [1.9–13%]) | 2 tongue, NOS (C02.9) 1 upper gum (C03.0) 1 lower gum (C03.1) 1 cheek mucosa (C06.0) | not reported | not reported | not reported |
| Singh et al. (2015) [25] | 250 (FFPE) | 23/250 (9.2%, 95% CI = [6–13%]) | 12 cheek mucosa (C06.0) 4 lower gum (C03.1) 1 retromolar area (C06.2) 6 tongue, NOS (C02.9) | 9/23 (39.1%, 95% CI = [20–61%]) | 220/227 (97%, 95% CI = [94–99%]) | 7/227 (3.1%, 95% CI = [1–6%]) |
| Reyes et al. (2015) [26] | 80 (FFPE) | 9/80 (11%, 95% CI = [5–20%]) | 3 tongue, NOS (C02.9) | 2/9 (22%, 95% CI = [3–60%]) | not reported | not reported |
| Blumberg et al. (2015) [16] | 29 (FFPE) | 0 (0%) | --- | 0 | 27/29 (93%, 95% CI = [77–99%]) | 2/29 (6.9%, 95% CI = [0.8–23%]) |
| Emmett et al. (2017) [17] | 63 (FFPE) | 5/63 (8%, 95% CI = [3–18%]) | 4 tongue (excluding base of tongue), NOS (C02.9) 1 floor of the mouth, NOS (C04.9) | 1/5 (20%, 95% CI = [0.5–72%]) | 57/58 (98%, 95% CI = [91–100%]) | 1/58 (1.7%, 95% CI = [0.04–9%]) |
| Vidal Loustao et al. (2019) [18] | 152 (FFPE) | 5/152 (3.3%, 95% CI = [1–7%]) | * floor of the mouth, NOS (C04.9) * anterior 2/3 of tongue, NOS (C02.3) | 1/5 (20%, 95% CI = [0.5–72%]) | 136/147 (93%, 95% CI = [87–96%]) | 11/147 (7.5%, 95% CI = [4–13%]) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panzarella, V.; Campisi, G.; Giardina, Y.; Maniscalco, L.; Capra, G.; Rodolico, V.; Di Fede, O.; Mauceri, R. Low Frequency of Human Papillomavirus in Strictly Site-Coded Oral Squamous Cell Carcinomas, Using the Latest NHI/SEER-ICD Systems: A Pilot Observational Study and Critical Review. Cancers 2021, 13, 4595. https://doi.org/10.3390/cancers13184595
Panzarella V, Campisi G, Giardina Y, Maniscalco L, Capra G, Rodolico V, Di Fede O, Mauceri R. Low Frequency of Human Papillomavirus in Strictly Site-Coded Oral Squamous Cell Carcinomas, Using the Latest NHI/SEER-ICD Systems: A Pilot Observational Study and Critical Review. Cancers. 2021; 13(18):4595. https://doi.org/10.3390/cancers13184595
Chicago/Turabian StylePanzarella, Vera, Giuseppina Campisi, Ylenia Giardina, Laura Maniscalco, Giuseppina Capra, Vito Rodolico, Olga Di Fede, and Rodolfo Mauceri. 2021. "Low Frequency of Human Papillomavirus in Strictly Site-Coded Oral Squamous Cell Carcinomas, Using the Latest NHI/SEER-ICD Systems: A Pilot Observational Study and Critical Review" Cancers 13, no. 18: 4595. https://doi.org/10.3390/cancers13184595
APA StylePanzarella, V., Campisi, G., Giardina, Y., Maniscalco, L., Capra, G., Rodolico, V., Di Fede, O., & Mauceri, R. (2021). Low Frequency of Human Papillomavirus in Strictly Site-Coded Oral Squamous Cell Carcinomas, Using the Latest NHI/SEER-ICD Systems: A Pilot Observational Study and Critical Review. Cancers, 13(18), 4595. https://doi.org/10.3390/cancers13184595











