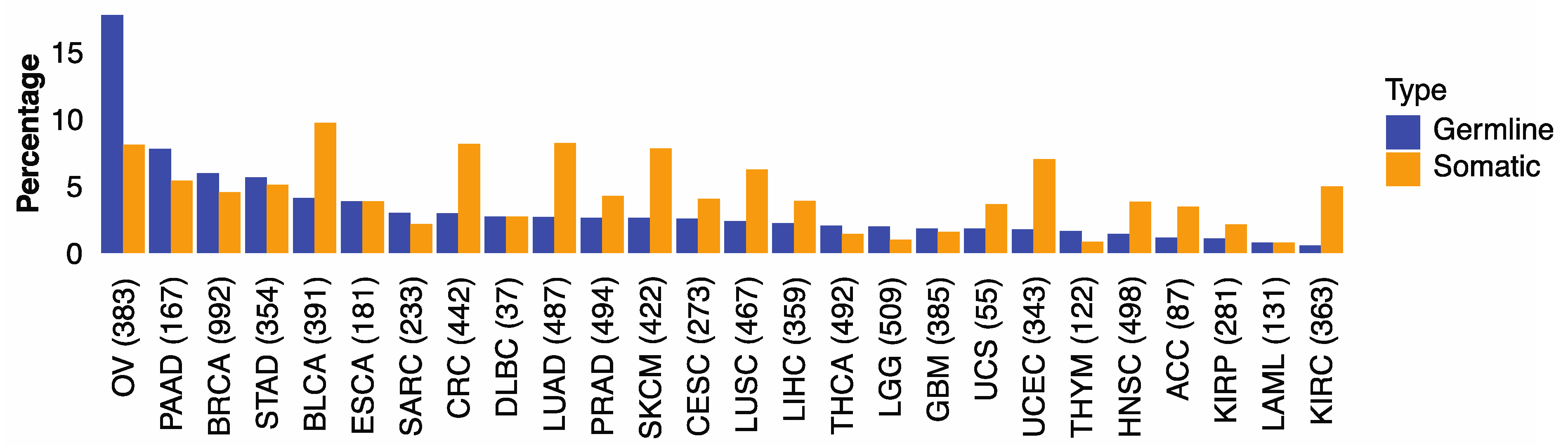Simple Summary
Tumors with homologous recombination deficiency (HRD) respond to Poly-ADP ribose polymerase inhibitor (PARPi) therapy in breast, ovarian, prostate, and pancreatic cancers. However, in addition to a handful of known pathogenic variants including those affecting BRCA1/2, it remains unclear what other genomic events can cause HRD. Our study systematically examined the germline and somatic genome of over 9000 cancers. We identified alterations associated with HRD, including mutations in ATM and ATR genes; specific deletions in stomach, bladder, and lung cancer; and BRCA-wild type breast, ovarian, and pancreatic cancers. These findings suggest a potentially broader utility for PARPi for cancers harboring a wide range of genomic alterations.
Abstract
Germline BRCA1/2 mutations associated with HRD are clinical biomarkers for sensitivity to poly-ADP ribose polymerase inhibitors (PARPi) treatment in breast, ovarian, pancreatic, and prostate cancers. However, it remains unclear whether other mutations may also lead to HRD and PARPi sensitivity across a broader range of cancer types. Our goal was to determine the germline or somatic alterations associated with the HRD phenotype that might therefore confer PARPi sensitivity. Using germline and somatic genomic data from over 9000 tumors representing 32 cancer types, we examined associations between HRD scores and pathogenic germline variants, somatic driver mutations, and copy number deletions in 30 candidate genes involved in homologous recombination. We identified several germline and somatic mutations (e.g., BRCA1/2, PALB2, ATM, and ATR mutations) associated with HRD phenotype in ovarian, breast, pancreatic, stomach, bladder, and lung cancer. The co-occurrence of germline BRCA1 variants and somatic TP53 mutations was significantly associated with increasing HRD in breast cancer. Notably, we also identified multiple somatic copy number deletions associated with HRD. Our study suggests that multiple cancer types include tumor subsets that show HRD phenotype and should be considered in the future clinical studies of PARPi and synthetic lethality strategies exploiting HRD, which can be caused by a large number of genomic alterations.
1. Introduction
Tumor cells with homologous recombination deficiency (HRD) are vulnerable to the inhibition of the DNA damage repair mechanism mediated by poly-ADP ribose polymerase (PARP). PARP inhibitors (PARPi) leverage this synthetic lethality and are US FDA approved to treat germline BRCA1 or BRCA2-mutant breast, ovarian, pancreatic, and prostate cancers [1,2,3,4,5]. Clinical trials have also demonstrated that somatic BRCA1/2 and germline PALB2 mutations, as well as HRD determined by the myChoice® CDx HRD assay (Myriad Genetics Inc., Salt Lake, UT, USA) can serve as biomarkers of PARPi response [2,6,7]. Notably, pre-clinical studies that investigated PARPi sensitivity showed a broader range of activity than its current clinical use across cancer types including gastrointestinal and genitourinary cancers [8,9,10]. These observations suggest that genomic alterations in genes involved in homologous recombination (HR) other than BRCA1/2 could also cause HRD [2,7] and implicate new treatment opportunities.
Many genes are involved in the homologous recombination (e.g., BRCA1/2, PALB2) and DNA damage sensing (e.g., ATM, ATR, CHEK2) and therefore regulate HR, which we refer to as “HR genes” herein. The function of HR genes can be disrupted through inherited germline variants, acquired somatic mutations, epigenetic silencing, and somatic copy number variations. Approximately 5% of all cancers represented in The Cancer Genome Atlas (TCGA) carry germline pathogenic variants in HR genes [11,12], and cancers also frequently harbor somatic mutations or show copy-number deletions [13]. These genetic alterations are expected to disrupt DNA repair function. However, their relative contribution to shaping HRD and their potential as biomarkers for PARPi sensitivity remain to be determined.
Here, we systematically determined the associations between HRD phenotypes and pathogenic germline variants, somatic driver mutations, and somatic copy number deletions of 30 HR genes across 32 cancer types. Using the HRD phenotype score captured by whole-exome sequencing and SNP array data from over 9000 cases in TCGA cohort, we identified cancer type specific associations between germline and somatic mutations in each HR gene with the HRD phenotype. We further utilized a structural equation modeling method to estimate the combined germline or somatic contributions and described how germline variants may collaborate with specific somatic drivers. We also identified novel somatic copy number deletions that are associated with the HRD phenotype. Altogether, these findings provide a catalog of genomic alterations that may lead to the HRD phenotype and serve potential biomarkers for PARPi treatment.
2. Materials and Methods
2.1. Patients
We obtained germline variants and somatic mutations from 10,389 and 10,295 cases, respectively including 10,080 samples with both types of data. We excluded the 342 hypermutators and 226 microsatellite instability (MSI) high tumors from the 10,080 cases, resulting in 9512 cases that were included in our mutation analyses (Figure S1 and Table S1).
We collected gene-level and arm-level copy number variation (CNV) events from 10,713 and 10,399 cases, including 10,149 samples with both types of date. We excluded 561 cases without HRD score and 252 cases without somatic mutation data from 10,149 cases, resulting in 9336 cases included our CNV analyses (Figure S1 and Table S1).
2.2. Genomic Datasets of TCGA Patients
TCGA HRD score: We obtained the pre-calculated HRD scores from Knijnenburg et al. [13]. The HRD score is constructed from three types of CNV [13]: (i) binary deep deletions from PanCanAtlas GISTIC2.0 analysis corresponding to GISTIC calls of “−2” which indicate loss of more than half of baseline ploidy [14]; (ii) CNV burden scores generated by TCGA PanCanAtlas Aneuploidy study using ABSOLUTE and SNP-array data [15]; and (iii) segment loss-of-heterozygosity (LOH) scores that capture the total number of segments with LOH [15,16]. The Knijnenburg et al. HRD score is calculated as the sum of the number of loss-of-heterozygosity in the genomic region of BRCA1/2 [15,16], telomeric allelic imbalance [17], and large-scale state transitions across the entire genome region [18]. The resulting HRD score is conceptually similar to the clinically available MyChoice DCx assay.
TCGA germline cancer predisposing variants: we obtained 853 germlines pathogenic/likely pathogenic variants that were identified from 10,389 patients. Germline variants were downloaded from The Genomic Data Commons database (GDC, https://gdc.cancer.gov/about-data/publications/PanCanAtlas-Germline-AWG (accessed on 11 August 2021)) [12,19].
TCGA somatic mutations: all somatic mutations of 10,295 cancers were obtained from the Multi-Center Mutation Calling in Multiple Cancers (MC3) dataset [20]. Tumor mutation burden (TMB) was calculated as the total number of all somatic mutations in all human genes. The pre-calculated functional prediction results of all somatic mutations and the designation of “likely driver mutation” for 299 genes were taken from TCGA PanCanAtlas Driver study which used [21]. The likely somatic driver mutations included truncating mutations, frameshift and in-frame shift indels, nonsense, splice site, and translation start site mutations, or missense mutations predicted as deleterious.
TCGA somatic copy number variation data: we obtained arm-level and gene-level CNV data from the PanCanAtlas Aneuploidy study (https://gdc.cancer.gov/about-data/publications/pancanatlas, accessed date 11 August 2021) [15]. The acute myeloid leukemia (LAML) data is not available, thus LAML was excluded from the CNV analysis. The arm-level and gene-level events indicate that the copy number gain/loss effect an entire chromosome arm or a specific genomic region that encodes gene. CNV was assessed with Affymetrix SNP 6.0 arrays (Santa Clara, CA, USA) [15] and gene-level CNV values were generated by GISTIC [14]. GISTIC calls of “−2” and “2” which indicate a loss or gain of more than half of baseline ploidy were assigned as deep deletions or amplification, respectively. Chromosome arm-level events were determined using the ABSOLUTE algorithm [16]. Only deep deletions (−2) were considered for association analysis in our study.
Hypermutators: we designated 344 TCGA cases hypermutated based on Bailey et al. [21]. The hypermutators were defined as samples with mutation burden greater than 1.5 times the interquartile range above the third quartile in their respective cancer types, and the number of mutations in a sample exceeds 1000 [21].
MSI: as a measure of MSI, we took the MSIsensor scores which were derived from standard tumor-normal paired sequence data from Niu et al. [22]. MSI cases were defined as those with an MSIsensor score > 4 [22] according to the TCGA PanCanAtlas [21].
Genetic principal components of TCGA cohort: given that the ancestral genetic background may influence genomic alterations, we obtain the pre-calculated principal components (PCs) from the WashU genetic ancestry analysis of the TCGA PanCanAtlas project [12,23]. The PCs were calculated on 298,004 variants with MAF > 0.15 and low missingness, and PC1 and PC2 accounted for 51.6% and 29.2% of the variations across the first 20 PCs [23] and were included as covariates in the regression analysis.
2.3. Association Analyses of Germline and Somatic Mutations Using Multivariate Regression Models
We use a linear regression model to estimate the influence of germline or somatic alterations on the HRD with the “glm” function of the “base” package of the R-project [24]. Since the HRD may be affected by age and genetic background, the analysis was controlled for covariates, including patients’ age at diagnosis and population substructure (first two PCs) [23]. The model is:
Only genes with predisposing variants harbored at least 4 individuals within the cancer cohort were included in the regression analysis. We perform this analysis within each cancer type.
2.4. Association Analyses of Gene-Level CNV Using Multivariate Regression Models
We used a linear regression model to estimate the influence of gene-level CNV events on the HRD with the “glm” function of the “base” package of the R-project. Given that the copy number variation may be affected by age, gender, arm-level CNV, the analysis was controlled for covariates, including patients’ age at diagnosis, gender, and arm-level CNV events, as well as TMB and the first two PCs [15,23,25]. The model is:
We performed this analysis within each cancer type. For breast cancer and prostate cancer, we excluded gender from the covariates.
2.5. Independent Contribution of Germline and Somatic Mutations
We use a Partial Least Squares Path Modeling (PLS-PM) analysis to investigate the contribution of germline and somatic mutations (Figure S3). The PLS-PM is a multivariate data analysis method which introduces latent variables for analyzing systems of relationships between multiple variables [26]. We include two latent variables (germline and somatic) in our analysis. The PLS-PM algorithm includes measurement model and structural model. The measurement model represents the relationships between the individual genes and the latent variables which generates coefficient (β) denoting the contribution of mutant genes to latent variable. The structural model represents the relationships between the latent variables which also provides coefficient and p value measuring the relative contribution of latent variables to HRD. In our analysis, we identified that germline and somatic mutations of 30 DNA damage repair genes were associated with HRD. We only use the 9512 overlapped samples with germline, somatic, and signature data available in this analysis. We perform PLS-PM analysis with the R packages “plspm” [27]. We compared the HRD score in cases with germline, somatic, both germline and somatic HR mutations, and the wildtype cases using the two-sided Mann–Whitney test.
2.6. Co-Occurrence and Mutual Exclusivity of Germline and Somatic Mutations
We estimate the interaction between germline predisposing variants and somatic driver mutations by using Fisher’s exact test. We assign 0 if wild type and 1 if mutated to each sample in the TCGA for each gene at both germline and somatic levels. We perform this analysis across all samples and within each cancer types separately. We compared the HRD score in cases with germline BRCA1, somatic TP53, both germline BRCA1 and somatic TP53 mutations, and the wildtype cases using a two-sided Mann–Whitney test.
2.7. Adjustment for Multiple Comparisons
p values were adjusted by the Benjamini–Hochberg procedure for false discovery rate (FDR) were computed globally across all analyses by genes and cancer types. All the analyses in this study were performed using scripts written with the R programming language. The significant effects of germline variants, somatic mutations, and somatic copy number variations were defined as FDR < 0.05.
3. Results
3.1. Germline and Somatic Mutations Associated with HRD in 32 Cancer Types
We first examined if genomic alterations in 30 genes involved with HR are associated with HRD phenotype captured by the HRD score of Knijnenburg et al. [13]. The HR genes included 21 genes in the homologous recombination pathway and nine DNA damage sensor-related genes (ATM, ATR, ATRIP, CHEK1, CHEK2, MDC1, RNMT, TOPBP1, TREX1) (Table S2) [28,29,30]. We found that microsatellite instability (MSI) was correlated with HRD score in several cancer types (Figure S2). To mitigate potential noise caused by large variations of genome instability in some cancers, we excluded hypermutators (n = 344) and microsatellite instability (n = 226) tumors and limited subsequent analyses to the remaining 9512 cases, comprising 32 cancer types (Figure S1, Table S1). We found that 3.5% of the 9512 cases carried germline pathogenic variants (from now on referred to as germline variants) and 4.5% harbored somatic driver mutations in HR genes. The percentages of cases carried germline or somatic HR mutations varied across cancer types: ovarian cancer (OV) showed the highest frequencies of germline variants (17.8%) and somatic mutations (8.1%), followed by pancreatic cancer (PAAD), breast cancer (BRCA), stomach cancer (STAD), and urothelial bladder carcinoma (BLCA) (Figure 1). These results illustrate the distinct germline and somatic landscapes of HR mutations across cancer types.
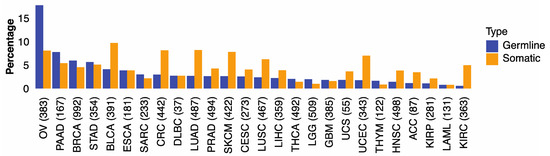
Figure 1.
The percentage of germline and somatic Homologous Recombination mutations in 32 TCGA cancer types. The proportion of TCGA cases within each cancer type carrying germline variants (blue) or somatic mutations (orange) in 30 HR genes.
We next sought to identify the germline and somatic alterations that are associated with the tumors’ HRD phenotype by using an HRD score, which was previously calculated as a weighted sum of Loss of Heterozygosity (LOH), Telomeric Allelic Imbalance (TAI), and Large-scale State Transitions (LST) events in tumor of the TCGA PanCanAtlas project across 32 cancer types [13].
Using a multivariate regression model, corrected for age and genetic ancestry represented by principal components, we found 12 positive correlations between the HRD score and germline variants in five (i.e., BRCA1, BRCA2, PABL2, ATM, ATR) genes across cancer types (Table 1). Both, BRCA1 and BRCA2 variants were associated with higher HRD scores in BRCA (FDR < 1.3 × 10−42) and OV (FDR < 5.0 × 10−12), BRCA2 (but not BRCA1) variants were also associated with higher HRD of PAAD (FDR = 1.3 × 10−17) and stomach cancer (STAD) (FDR = 6.7 × 10−17) (Table 1). We also found that germline PALB2 variants are associated with higher HRD in STAD (FDR = 1.2 × 10−41). Germline ATR and ATM were associated with higher HRD in BRCA (FDR < 8.5 × 10−8), and germline ATM variants in prostate cancer (PRAD, FDR = 9.0 × 10−6), lung adenocarcinoma (LUAD, FDR = 5.8 × 10−5), and STAD (FDR = 0.007) (Table 1). While BRCA1/2 germline variants in BRCA, OV, PAAD and prostate cancer are well known to be associated with HRD phenotype, our results identify a much broader range of cancers with different germline alterations in HR-related genes that are significantly associated with HRD (Table 1 and Table S3).

Table 1.
The significant associations (FDR < 0.05) of germline variants and somatic mutations with HRD phenotype.
We conducted a similar multivariate regression analysis for somatic mutations, and identified 24 significant associations in five (i.e., BRCA1, BRCA2, ATM, ATR, CHEK2) genes and higher HRD (FDR < 0.05) across cancer types. Somatic mutations of BRCA1 and BRCA2 were significantly associated with HRD in BRCA, OV, and BLCA, and lung squamous cell carcinoma (LUSC) (FDR < 0.002, Table 1). Somatic BRCA2 mutations were also associated with HRD in STAD, PRAD, and colon rectum adenocarcinoma (CRC) (FDR < 0.01). Somatic mutations of ATM and ATR were associated with higher HRD in BLCA, LUAD, CRC, PRAD, head–neck squamous cell carcinoma (HNSC), BRCA, and kidney renal clear cell carcinoma (KIRC) (FDR < 0.008, Table 1). Somatic mutations of CHEK2 significantly increased HRD in uterine corpus endometrial carcinoma (UCEC, FDR < 3.3×10−7, Table 1). Of note, we also identified negative associations between HRD scores and somatic HR mutations in cancer types including UCEC, LUAD, skin cutaneous melanoma (SKCM), and LUSC (FDR < 0.03, Table 1). Overall, these results highlight that both germline and somatic mutations can be associated with high HRD and the importance of different HR genes to maintain genomic stability can vary from cancer type to cancer type.
3.2. Relative Contributions of Germline and Somatic Mutations on HRD
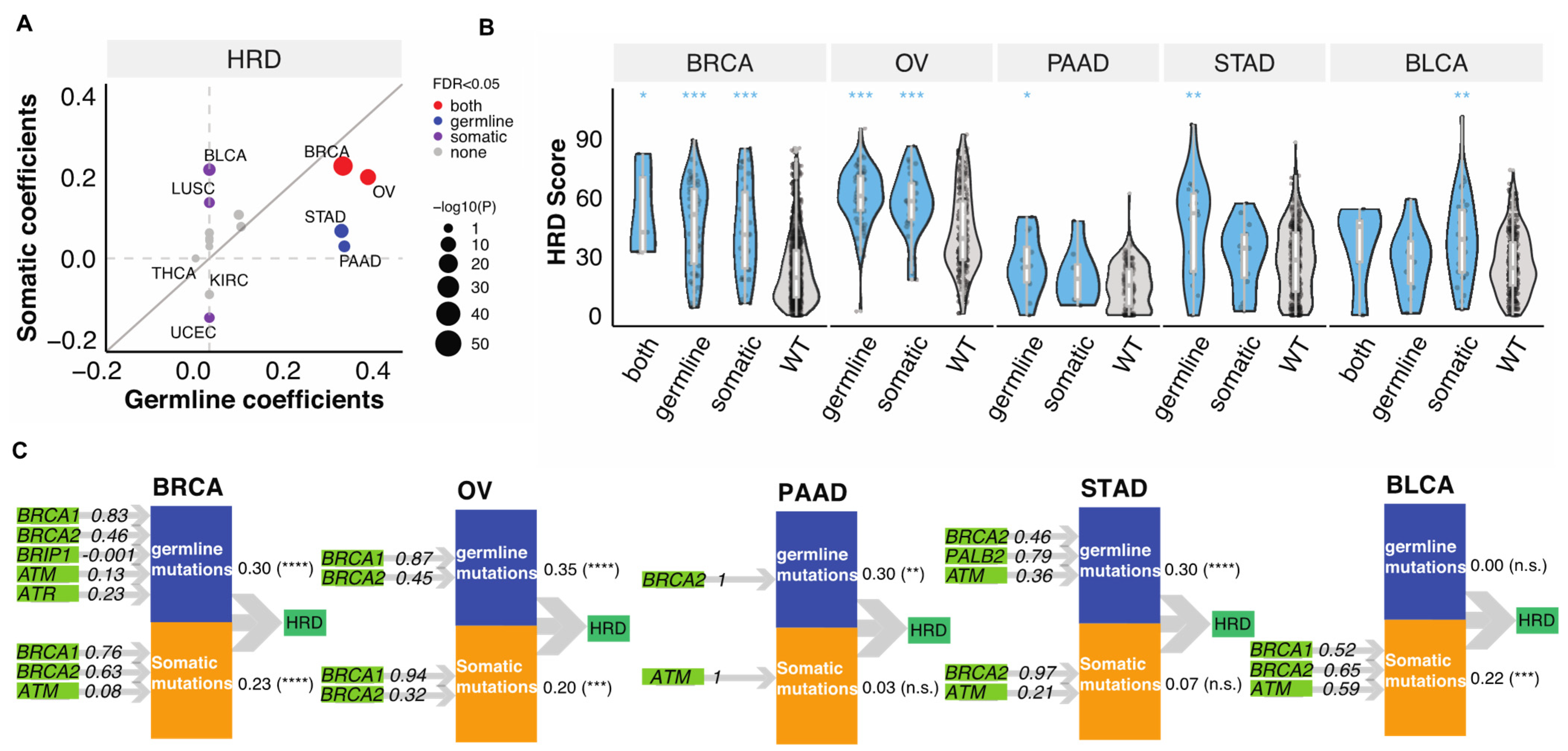
To delineate the relative contributions of germline variants vs. somatic mutations to the tumor HRD phenotype, we applied Partial Least Squares Path Modeling (PLS-PM) [26]. In this analysis, we only considered genes that were affected in at least four TCGA cases within a cancer type and therefore only seven genes (BRCA1/2, ATM, ATR, PALB2, CHEK2, BRIP1) were included. This may neglect the contribution of other HR genes with lower mutation frequencies, but we have limited power to identify these in the current data set. Both germline and somatic mutations significantly contributed to the HRD of BRCA (FDR < 1.5 × 10−12) and OV (FDR < 2.4 × 10−4, (Figure 2A)). In STAD and PAAD, mostly germline variants contributed to high HRD (FDR < 7.2 × 10−4), whereas in BLCA and LUSC high HRD were “driven” mainly by somatic mutations (FDR < 0.01, (Figure 2A)). We show the distribution of HRD scores in these cancer types by mutation status on Figure 2B.
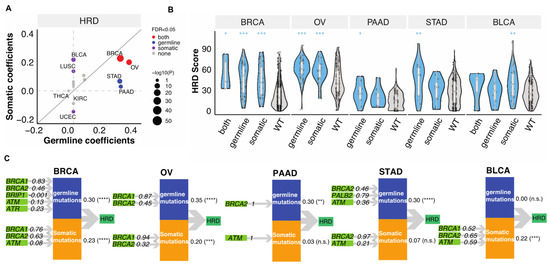
Figure 2.
The combined contributions of germline variants and somatic mutations to HRD. (A). The combined germline and somatic contributions represented by coefficients obtained from the PLS-PM analysis. Each dot represents a cancer type. The size of the circles represents −log10(p). Grey, purple, blue, and red represent none of, somatic, germline, or both FDR meet the criteria FDR < 0.05. The grey line indicates the diagonal with slope of one. (B). The distribution of HRD score in combined germline and somatic carriers in BRCA (n = 5 (both), 53 (germline), 38 (somatic), 850 (wildtype)), OV (n = 66 (germline), 31 (somatic), 272 (wildtype)), PAAD (n = 12 (germline), 8 (somatic), 131 (wildtype)), STAD (n = 18 (germline), 15 (somatic), 309 (wildtype)), BLCA (n = 5 (both), 10 (germline), 32 (somatic), 334 (wildtype)). p-value was calculated using a two-sided Mann–Whitney test and FDR corrected. “***”, “**”, “*” denote FDR less than 0.0001, 0.001, and 0.05. (C). PLS-PM models show the significant combined contribution of germline variants and somatic mutations with FDR < 0.05 of combined effect in selected cancer types, including BRCA, OV, PAAD, STAD, and BLCA. The numbers between the latent variables and HRD indicated combined correlation coefficients identified by the PM-PLS model, the value. “****”, “***”, “**” denote FDR less than 0.00001, 0.0001, 0.001. “n.s.” denotes “not significant” (FDR > 0.05).
The PLS-PM analysis also allows to quantify the relative contribution of individual gene’s to predicting higher HRD when combining all the contributors together. Both germline variants and somatic mutations of BRCA1/2 (β > 0.32) dominantly contributed to HRD in BRCA and OV, while only germline BRCA2 contributed to HRD in PAAD (Figure 2C). Germline PALB2 (β = 0.79) and BRCA2 (β = 0.46) and ATM (β = 0.36) variants contributed to HRD of STAD, with non-significant effects from somatic BRCA2 (β = 0.97) and ATM (β = 0.21) (Figure 2C). On the other hand, somatic BRCA1 (β = 0.52), BRCA2 (β = 0.65) and ATM (β = 0.59) contributed to HRD in BLCA (Figure 2C). These results highlight that somatic and germline alterations in specific HR genes contribute differently to HRD in different cancer types.
3.3. Germline-Somatic Interactions Shaping HRD
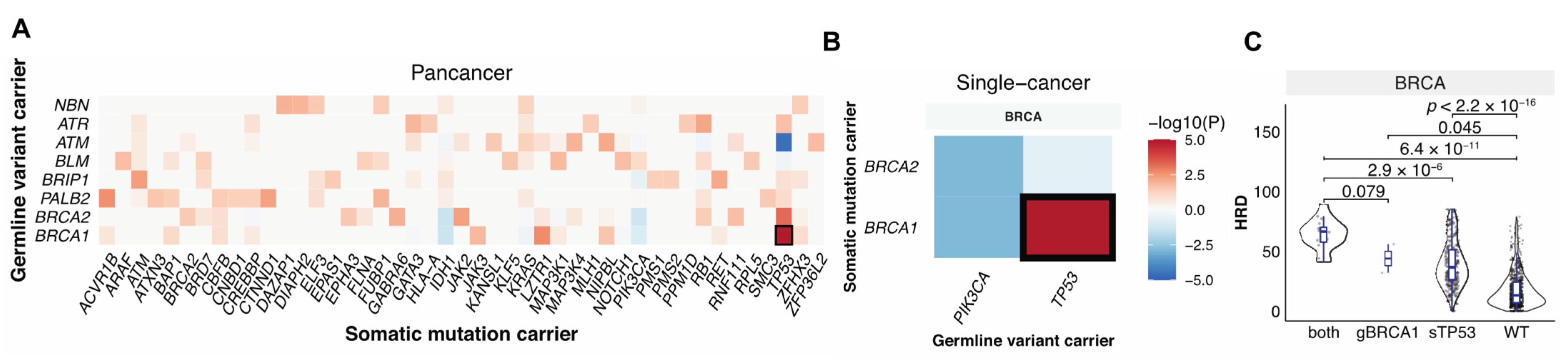
Germline variants and somatic mutations can complement each other in the malignant transformation process [12,31,32], and together they can shape tumor phenotypes including HRD. To identify germline–somatic mutation pairs that may cooperate to affect HRD, we first identified the significant co-occurring germline–somatic mutation pairs considering germline variants in the 30 HR genes vs. the PanCanAtlas-defined somatic driver genes (n = 299) across all TCGA cancer cases and also within each cancer types using the Fisher’s exact test. Germline BRCA1 mutations significantly co-occurred with somatic TP53 mutations at both the pan-cancer level (FDR = 1.0 × 10−6, Figure 3A) and within breast cancer (FDR = 6.2 × 10−4, Figure 3B). We also detected trends of other germline-somatic co-occurrences, including mutually exclusive alterations between germline BRCA1/2 and somatic PIK3CA (FDR = 0.2, p = 0.001), we found no other germline–somatic pairs that reached significance. In BRCA, samples that carried both germline BRCA1 and somatic TP53 mutations had higher HRD scores compared to cases with only germline BRCA1 alteration (p = 0.08), or with somatic TP53 mutation only (p = 2.9 × 10−6), or cases that were wild type for both (p = 6.4 × 10−11) (Figure 3C). These results demonstrate that while germline and somatic mutations may induce independent effects, tumors carrying both pathogenic BRCA1 germline variants and TP53 driver mutations show markedly increased HRD.
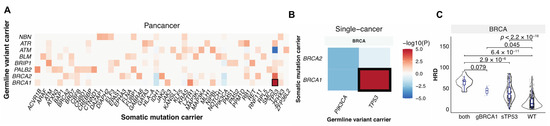
Figure 3.
The interaction between germline pathogenic variants and somatic driver mutations. (A). Co-occurring germline variants and somatic mutations across all cancers. (B). Co-occurring germline variants and somatic mutations within each cancer type. The x-axis shows germline-affected genes, and the y-axis shows somatic-affected genes. Color denotes the Fisher exact test −log10(p value) showing the significance of the germline and somatic interaction. Red and blue indicate co-occurrence and mutual-exclusivity, respectively. (C). The distribution of HRD score in only germline (g) BRCA1 (n = 2), only somatic (s) TP53 (n = 306), and both carriers (n = 16). WT indicates samples without either of the mutations (n = 622).
3.4. Somatic Copy Number Variations Associated with HRD
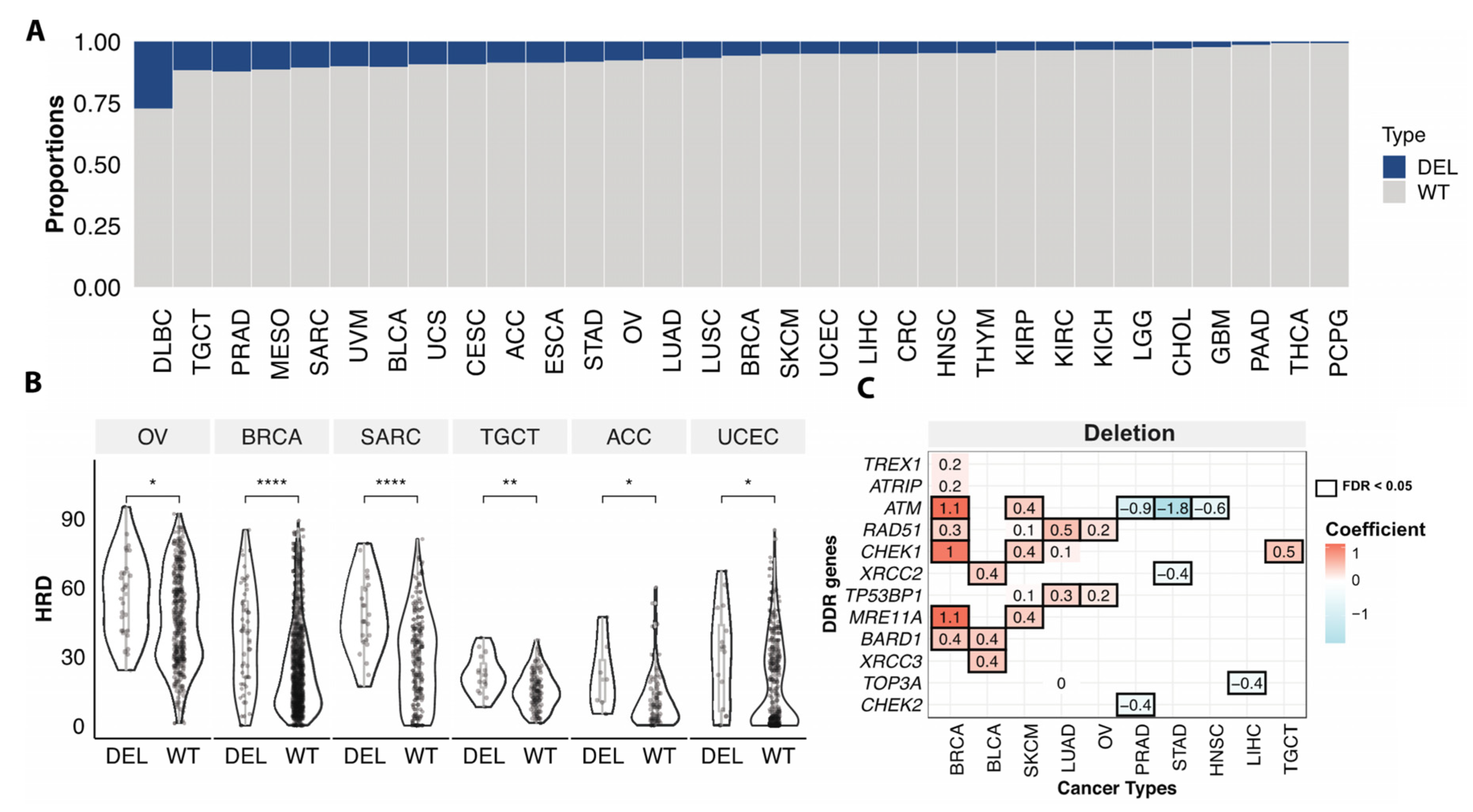
Somatic copy number deletion of genes involved in HR may also compromise HR. Given that the calculation of HRD scores included copy number variation of BRCA1/2 genes, we assessed the associations between HRD and copy number deletions focusing only on the rest of 28 HR genes. Overall, 7.0% of the non-hypermutator/microsatellite stable cases had somatic copy number deletions of genes involved in HR, excluding BRCA1 and BRCA2. Across cancer types, diffuse large B-cell lymphoma (DLBC) had the highest frequency of deletions (24.3%) in the HR genes, followed by PRAD (11.5%), and testicular germ cell tumors (TGCT) (11.7%) (Figure 4A). About 5.6% of BRCA and 7.6% of OV cases were affected by HR gene deletions (excluding BRCA1/2) (Figure 4A).
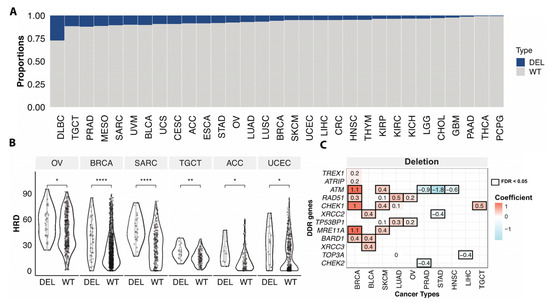
Figure 4.
Somatic copy number deletions of HR genes associated with HRD. (A). The proportions of samples with somatic copy number deletion of homologous recombination and damage sensor genes in each cancer type. Blue (loss) indicates samples with somatic copy number deletions in any of 28 HR genes (excluding BRCA1/2), grey (WT) denotes non-mutated cases. (B). The HRD score between cases with somatic copy number deletion (DEL) of homologous recombination and damage sensor genes and wildtype (WT) cases excluding BRCA1/2 copy number variation affected cases in OV (n = 28 (DEL), 332 (WT)), BRCA (n = 53 (DEL), 860 (WT)), SARC (n = 23 (DEL), 193 (WT)), TGCT (n = 15 (DEL), 113 (WT)), ACC (n = 7 (DEL), 74 (WT)), UCEC (n = 16 (DEL), 303 (WT)). p-value was calculated using a two-sided Mann–Whitney test and FDR corrected. “****”, “**”, “*” denote FDR less than 0.00001, 0.001, and 0.05. (C). The heatmap shows the significant (FDR < 0.05, black box) association of somatic copy number deletion of individual genes with the HRD of TCGA cancer types. Both value and color in each cell represent the coefficient obtained from the multivariate logistic regression model.
Cases with at least one somatic copy-number deletion in any of the HR genes (excluding BRCA1/2) showed significantly higher HRD scores compare to cases without deletions in OV, BRCA, Sarcoma (SARC), TGCT, adrenocortical carcinoma (ACC), low grade glioma (LGG), and UCEC (FDR < 0.05) (Figure 4B). Applying a multivariate regression model, we further compared the HRD scores between copy number deletion vs. wildtype cases at the gene level. Deletions of ATM, RAD51, MRE11A, CHEK1, and BARD1 were associated with higher HRD of BRCA. We also identified other deletions associated with higher HRD in multiple cancer types, such as XRCC2/3 and BARD1 deletion in BLCA, TP53BP1, and RAD51 deletion in OV and LUAD, and CHEK1 deletion in TCGT and SKCM, etc. (Figure 4C). These analyses show that not only mutations but copy number deletions in HR genes can also be associated with HRD and might confer sensitivity to PARPi, warranting further mechanistic and clinical investigations.
4. Discussion
This study comprehensively evaluated the associations between deleterious germline variants, somatic driver mutations, and somatic copy number deletions in 30 HR-related genes and the HRD phenotype of 32 cancer types. We demonstrated that aside from BRCA1/2 in well-known BRCA-associated cancer types (breast, ovarian, pancreatic, and prostate), several other HR gene mutations are associated with HRD in a broad range of cancers. Both germline variants and somatic mutations of HR genes can be associated with HRD phenotype. Further, their individual contributions (i.e., strength of association) and synergistic interactions between them varied by cancer type. We also demonstrated that somatic copy number deletions of HR genes are associated with HRD. These results expand the repertoire of genes and type of genomic abnormalities that may cause HRD. Further clinical studies could further validate whether these alterations could serve as potential biomarkers for treatments exploiting synthetic lethality to treat tumors with impaired HR in a broader range of cancer types.
Several PARPi have already been approved for clinical use. The US FDA approved olaparib for the treatment of germline BRCA-mutated breast, prostate, ovarian, and pancreatic cancer patients; talazoparib is also approved for germline BRCA mutated breast and ovarian cancers [33,34]. The NOVA study demonstrated that niraparib improved survival of somatic BRCA1/2 mutant or BRCA normal but HRD positive by the Myriad myChoice CDx assay ovarian cancer [4]. The TBCRC-048 trial, demonstrated the efficacy of Olaparib in somatic BRCA mutant and germline PALB2 mutant metastatic breast cancers [7]. Consistent with these clinical observations, our results demonstrate that either germline or somatic mutations in BRCA1/2 genes can be associated with HRD phenotype of breast and ovarian cancer. In addition to the known associations, our data also suggests that there could be additional subsets of patients who might benefit from HR targeted therapies. For example, germline PALB2 variants have recently been associated with familial and sporadic stomach cancer [35]. We found that germline PALB2 variants were positively correlated with HRD in stomach cancers. Deleterious germline variants in ATR and ATM were also associated with HRD phenotype in breast cancer, ATM in prostate, lung, and stomach cancers, and BRCA2 in stomach cancers.
Somatic mutations of BRCA1/2 were also significantly associated with HRD in breast and ovarian cancers and these associations are already clinically exploited. Notably, BRCA2 mutations were also associated with HRD in urothelial, squamous cell lung, and stomach carcinomas, where tumor response to PARPi has not yet been associated with HR mutations or HRD. We also identified previously unreported associations of somatic mutations of several other HR genes with HRD phenotype, suggesting new patient subsets that may benefit from PARPi. For example, somatic BRCA2 and ATM mutations were correlated with HRD in bladder cancer, lung cancer, and colon cancer. Moreover, ATR mutations were associated with high HRD scores in head and neck cancer and kidney cancer (Table 1). We also noted that germline and somatic mutations contribute differently to the HRD phenotype in different cancer types. Both germline and somatic mutations significantly contributed almost equally to HRD in breast cancer. In stomach and prostate cancers, HRD was predominantly influenced by germline variants, whereas in bladder cancer somatic mutations contributed to the majority of tumors with HRD phenotype.
The effects of copy number variants on HRD and PARPi response have rarely been investigated, and only a few studies have reported ATM, BRCA1, and BRCA2 deletions could affect sensitivity to DNA damaging chemotherapies [36,37,38]. In our analysis, we found that somatic copy number deletions in several HR genes, such as ATM, RAD51, and CHEK1 were associated with increased HRD phenotype in OV, BRCA, SARC, and TGCT. The results suggest that somatic copy number deletion of HR genes should also be evaluated as potential biomarkers for predicting HRD and PARPi sensitivity.
One limitation of the study is that the identified associations do not imply causality, and require further experimental validation. We also recognize that results from a clinically used HRD test is not available for any of the TCGA cases. Our HRD scores were derived from the SNP array and WES data that resemble the myChoice® CDx HRD assay according to the TCGA Pan-Cancer Atlas project. The HRD scores used in this study may limit the detection of HRD tumors. Further, observing an HRD phenotype may not guarantee PARPi sensitivity in tumors. Future studies using clinical cohorts with genomic and PARPi response data are needed to validate whether these tumors may be effectively treated by therapies exploiting synthetic lethality.
5. Conclusions
Our results showed that the HRD phenotype is associated with several genomic alterations in genes involved with homologous recombination across both BRCA-associated and non-BRCA-associated cancer types. These findings raise the possibility that many subpopulations of cancer patients might benefit from PARP inhibitors and other HR-targeted therapies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13184572/s1, Figure S1: CONSORT diagram, TCGA cases included in this study, Figure S2: The association between MSIsensor score and HRD score in all TCGA cancer cases, Figure S3: A schema of Partial Least Squares Path Modeling analysis, Table S1. TCGA samples, Table S2: 30 HR genes, Table S3: The associations of germline variants and mutations with HRD, the full list of Table 1.
Author Contributions
Conception and design, K.L.-H. and T.Q.; analysis and interpretation of data, T.Q., X.W., and T.J.; drafting the article or revising it critically for important intellectual content, T.Q. and K.L.-H.; writing—review and editing, supervision, K.L.-H. and L.P.; resources, K.L.-H., L.P., and L.D.; agreement to be accountable for all aspects of the work, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Breast Cancer Research Foundation (Investigator award BCRF 19-133) and the Susan G. Komen Foundation (grant number SAC160076) from Yale Cancer Center, and NIGMS R35GM138113 and seed fund from the Icahn School of Medicine at Mount Sinai.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The TCGA germline variants are available at https://gdc.cancer.gov/about-data/publications/PanCanAtlas-Germline-AWG (accessed on 11 August 2021). The TCGA somatic mutations are available at https://gdc.cancer.gov/about-data/publications/mc3-2017 (accessed on 11 August 2021). Data supporting the findings of this study are available in the Article, Supplementary Information, or from the authors upon reasonable request.
Acknowledgments
The authors wish to acknowledge the participating patients and family who generously contributed to the datasets.
Conflicts of Interest
L.P. has received consulting fees and honoraria from Pfizer, Astra Zeneca, Merck, Novartis, Bristol-Myers Squibb, Genentech, Eisai, Pieris, Immunomedics, Seattle Genetics, Clovis, Syndax, H3Bio, and Daiichi. No disclosures were reported by the other authors.
References
- Dottino, J.A.; Moss, H.A.; Lu, K.H.; Secord, A.A.; Havrilesky, L.J. U.S. Food and drug administration-approved poly (Adp-ribose) Polymerase inhibitor maintenance therapy for recurrent ovarian cancer: A cost-effectiveness analysis. Obstet. Gynecol. 2019, 133, 795–802. [Google Scholar] [CrossRef]
- Hoppe, M.M.; Sundar, R.; Tan, D.S.P.; Jeyasekharan, A.D. Biomarkers for homologous recombination deficiency in cancer. J. Natl. Cancer Inst. 2018, 110, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. New Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.R.; Monk, B.J.; Herrstedt, J.; Oza, A.; Mahner, S.; Redondo, A.; Fabbro, M.; Ledermann, J.A.; Lorusso, D.; Vergote, I.; et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. New Engl. J. Med. 2016, 375, 2154–2164. [Google Scholar] [CrossRef] [PubMed]
- FDA U. 2019 FDA Approves Niraparib For HRD-Positive Advanced Ovarian Cancer. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-niraparib-hrd-positive-advanced-ovarian-cancer#:~:text=The%20FDA%20also%20approved%20the,genomic%20instability%20for%20this%20indication (accessed on 23 October 2019).
- Kaufman, B.; Shapira-Frommer, R.; Schmutzler, R.K.; Audeh, M.W.; Friedlander, M.; Balmana, J.; Mitchell, G.; Fried, G.; Stemmer, S.M.; Hubertet, A.; et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 2015, 33, 244–250. [Google Scholar] [CrossRef]
- Tung, N.M.; Robson, M.E.; Ventz, S.; Santa-Maria, C.A.; Nanda, R.; Marcom, P.K.; Shah, P.D.; Ballinger, T.J.; Yang, E.S.; Vinayak, S.; et al. TBCRC 048: Phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J. Clin. Oncol. 2020, 38, 4274–4282. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.-J.; Xu, R.-H.; Chin, K.; Lee, K.-W.; Park, S.H.; Rha, S.Y.; Shen, L.; Qin, S.; Xu, N.; Im, S.-A.; et al. Olaparib in combination with paclitaxel in patients with advanced gastric cancer who have progressed following first-line therapy (GOLD): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 1637–1651. [Google Scholar] [CrossRef]
- Vikas, P.; Borcherding, N.; Chennamadhavuni, A.; Garje, R. Therapeutic potential of combining PARP inhibitor and immunotherapy in solid tumors. Front. Oncol. 2020, 10, 570. [Google Scholar] [CrossRef]
- Sztupinszki, Z.; Diossy, M.; Krzystanek, M.; Borcsok, J.; Pomerantz, M.M.; Tisza, V.; Spisak, S.; Rusz, O.; Csabai, I.; Freedmanet, M.L.; et al. Detection of molecular signatures of homologous recombination deficiency in prostate cancer with or without BRCA1/2 mutations. Clin. Cancer Res. 2020, 26, 2673–2680. [Google Scholar] [CrossRef]
- Rahman, N. Realizing the promise of cancer predisposition genes. Nature 2014, 505, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-L.; Mashl, R.J.; Wu, Y.; Ritter, D.I.; Wang, J.; Oh, C.; Paczkowska, M.; Reynolds, S.; Wyczalkowski, M.; Oak, N.; et al. Pathogenic germline variants in 10,389 adult cancers. Cell 2018, 173, 355–370.e14. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef] [PubMed]
- Mermel, C.H.; Schumacher, E.S.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018, 33, 676–689 e673. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.L.; Cibulskis, K.; Helman, E.; McKenna, A.; Shen, H.; Zack, T.; Laird, P.W.; Onofrio, R.C.; Winckler, W.; Weir, B.A.; et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012, 30, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Birkbak, N.J.; Wang, Z.C.; Kim, J.Y.; Eklund, A.C.; Li, Q.; Tian, R.; Bowman-Colin, C.; Li, Y.; Greene-Colozzi, A.; Iglehart, J.D.; et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2012, 2, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Popova, T.; Manie, E.; Rieunier, G.; Caux-Moncoutier, V.; Tirapo, C.; Dubois, T.; Delattre, O.; Sigal-Zafrani, B.; Bollet, M.; Longy, M.; et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012, 72, 5454–5462. [Google Scholar] [CrossRef] [PubMed]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018, 6, 271–281.e7. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018, 174, 1034–1035. [Google Scholar] [CrossRef]
- Ding, L.; Bailey, M.H.; Porta-Pardo, E.; Thorsson, V.; Colaprico, A.; Bertrand, D.; Gibbs, D.L.; Weerasinghe, A.; Huang, K.L.; Tokheim, C.; et al. Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell 2018, 173, 305–320.e310. [Google Scholar] [CrossRef] [PubMed]
- Carrot-Zhang, J.; Chambwe, N.; Damrauer, J.S.; Knijnenburg, T.A.; Robertson, A.G.; Yau, C.; Zhou, W.; Berger, A.C.; Huang, K.L.; Newberg, J.Y.; et al. Comprehensive Analysis of Genetic Ancestry and Its Molecular Correlates in Cancer. Cancer Cell 2020, 37, 639–654.e636. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2020. Available online: https://www.R-project.org/ (accessed on 11 August 2021).
- Yuan, Y.; Liu, L.; Chen, H.; Wang, Y.; Xu, Y.; Mao, H.; Li, J.; Mills, G.B.; Shu, Y.; Li, L.; et al. Comprehensive characterization of molecular differences in cancer between male and female patients. Cancer Cell 2016, 29, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Wold, H. Causal flows with latent variables: Partings of the ways in the light of NIPALS modelling. Eur. Eco. Rev. 1974, 5, 67–86. [Google Scholar] [CrossRef]
- Gaston Sanchez LTaGR. plspm: Tools for Partial Least Squares Path Modeling (PLS-PM). 2015. Available online: https://github.com/gastonstat/plspm (accessed on 11 August 2021).
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Rodrigues, D.N.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-repair defects and olaparib in metastatic prostate cancer. New Engl. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- McCabe, N.; Turner, N.C.; Lord, C.; Kluzek, K.; Białkowska, A.; Swift, S.; Giavara, S.; O’Connor, M.J.; Tutt, A.N.; Zdzienicka, M.Z.; et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) Polymerase inhibition. Cancer Res. 2006, 66, 8109–8115. [Google Scholar] [CrossRef]
- Moudry, P.; Watanabe, K.; Wolanin, K.M.; Bartkova, J.; Wassing, I.E.; Watanabe, S.; Strauss, R.; Pedersen, R.T.; Oestergaard, V.; Lisby, M.; et al. TOPBP1 regulates RAD51 phosphorylation and chromatin loading and determines PARP inhibitor sensitivity. J. Cell Biol. 2016, 212, 281–288. [Google Scholar] [CrossRef]
- Qing, T.; Mohsen, H.; Marczyk, M.; Ye, Y.; O’Meara, T.; Zhao, H.; Townsend, J.P.; Gerstein, M.; Hatzis, C.; Kluger, Y.; et al. Germline variant burden in cancer genes correlates with age at diagnosis and somatic mutation burden. Nat. Commun. 2020, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Carter, H.; Marty, R.; Hofree, M.; Gross, A.M.; Jensen, J.; Fisch, K.; Wu, X.; DeBoever, C.; Van Nostrand, E.; Song, Y.; et al. Interaction landscape of inherited polymorphisms with somatic events in cancer. Cancer Discov. 2017, 7, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Hoy, S.M. Talazoparib: First global approval. Drugs 2018, 78, 1939–1946. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Ison, G.; McKee, A.E.; Zhang, H.; Tang, S.; Gwise, T.; Sridhara, R.; Lee, E.; Tzou, A.; Philip, R.; et al. FDA approval summary: Olaparib monotherapy in patients with deleterious germline brca-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin. Cancer Res. 2015, 21, 4257–4261. [Google Scholar] [CrossRef] [PubMed]
- Fewings, E.; Larionov, A.; Redman, J.; Goldgraben, M.; Scarth, J.; Richardson, S.; Brewer, C.; Davidson, R.; Ellis, I.; Evans, D.G.; et al. Germline pathogenic variants in PALB2 and other cancer-predisposing genes in families with hereditary diffuse gastric cancer without CDH1 mutation: A whole-exome sequencing study. Lancet Gastroenterol. Hepatol. 2018, 3, 489–498. [Google Scholar] [CrossRef]
- Watkins, J.A.; Irshad, S.; Grigoriadis, A.; Tutt, A.N.J. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014, 16, 211. [Google Scholar] [CrossRef] [PubMed]
- Den Brok, W.D.; Schrader, K.A.; Sun, S.; Tinker, A.V.; Zhao, E.Y.; Aparicio, S.; Gelmon, K.A. Homologous recombination deficiency in breast cancer: A clinical review. JCO Precision Oncol. 2017, 1, 1–13. [Google Scholar] [CrossRef]
- Rafiei, S.; Fitzpatrick, K.; Liu, D.; Cai, M.-Y.; Elmarakeby, H.A.; Park, J.; Ricker, C.; Kochupurakkal, B.S.; Choudhury, A.D.; Hahn, W.C.; et al. ATM Loss Confers Greater Sensitivity to ATR Inhibition Than PARP Inhibition in Prostate Cancer. Cancer Res. 2020, 80, 2094–2100. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).