Non-Invasive Ultrasonic Description of Tumor Evolution
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experiment
2.2. Tumor Volume
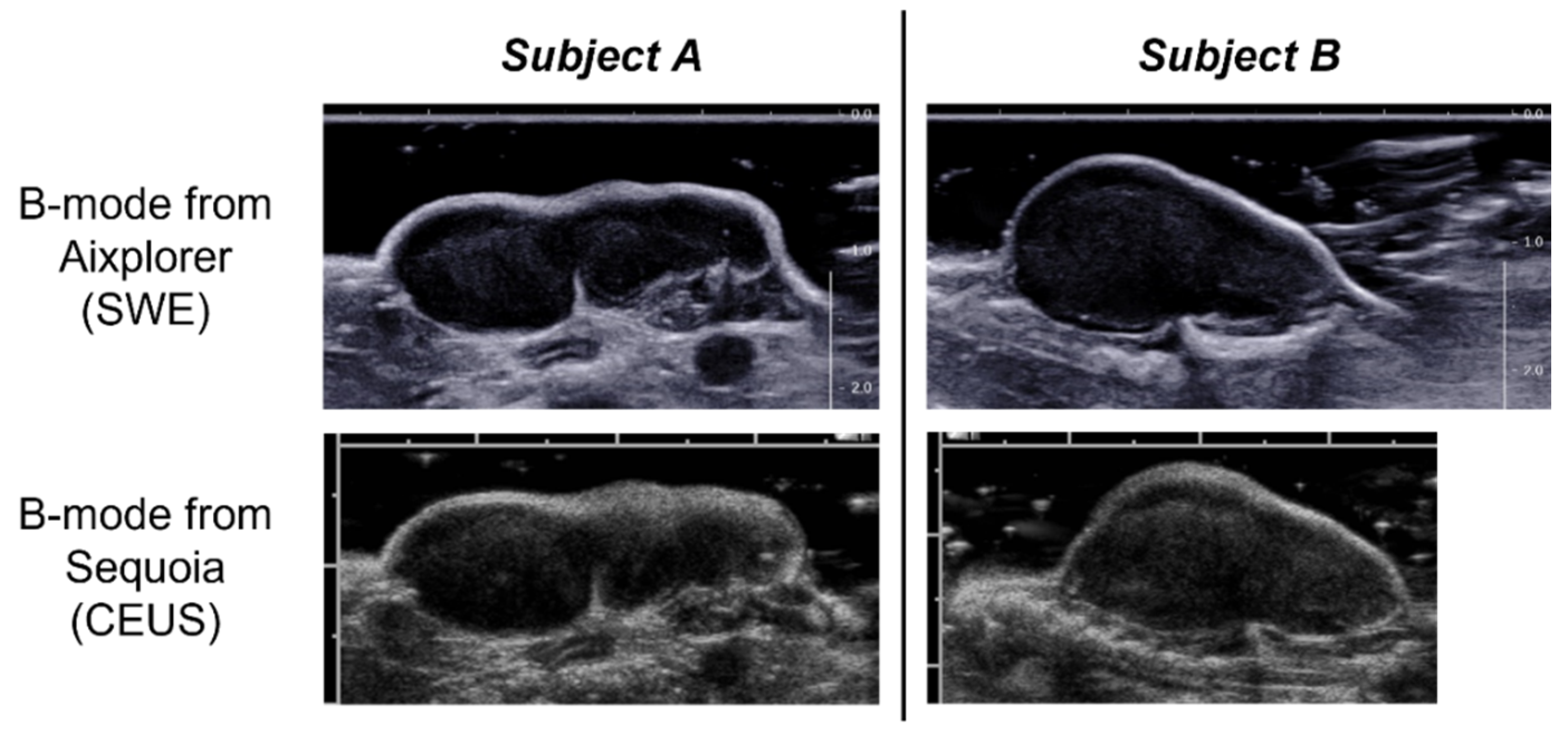
2.3. SWE
2.4. CEUS
2.5. Immunohistology
2.6. Statistical Tests
3. Results
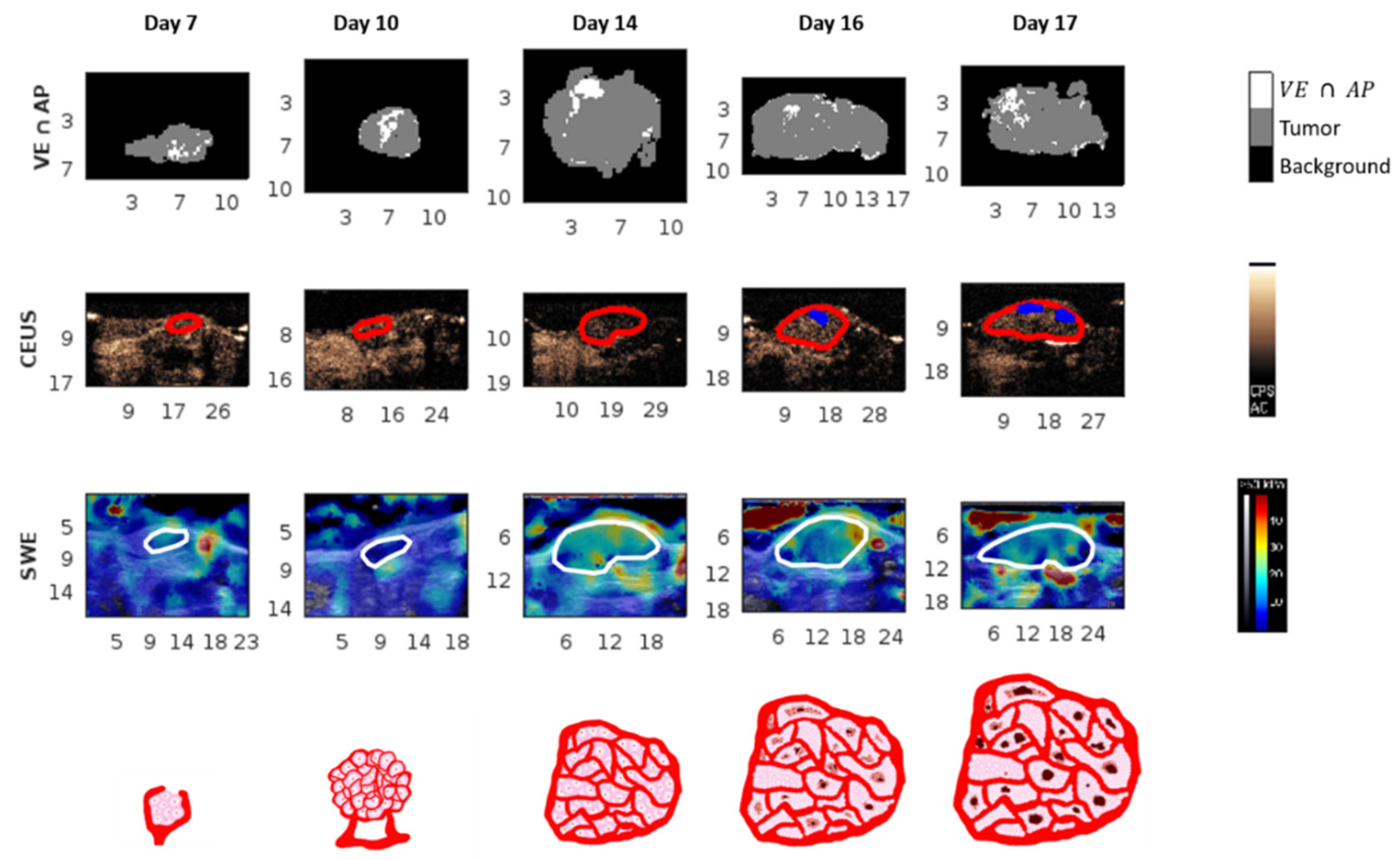
3.1. Evolution over Time
3.1.1. Tumor Volume and Immunohistological Parameters
3.1.2. Ultrasound Parameters
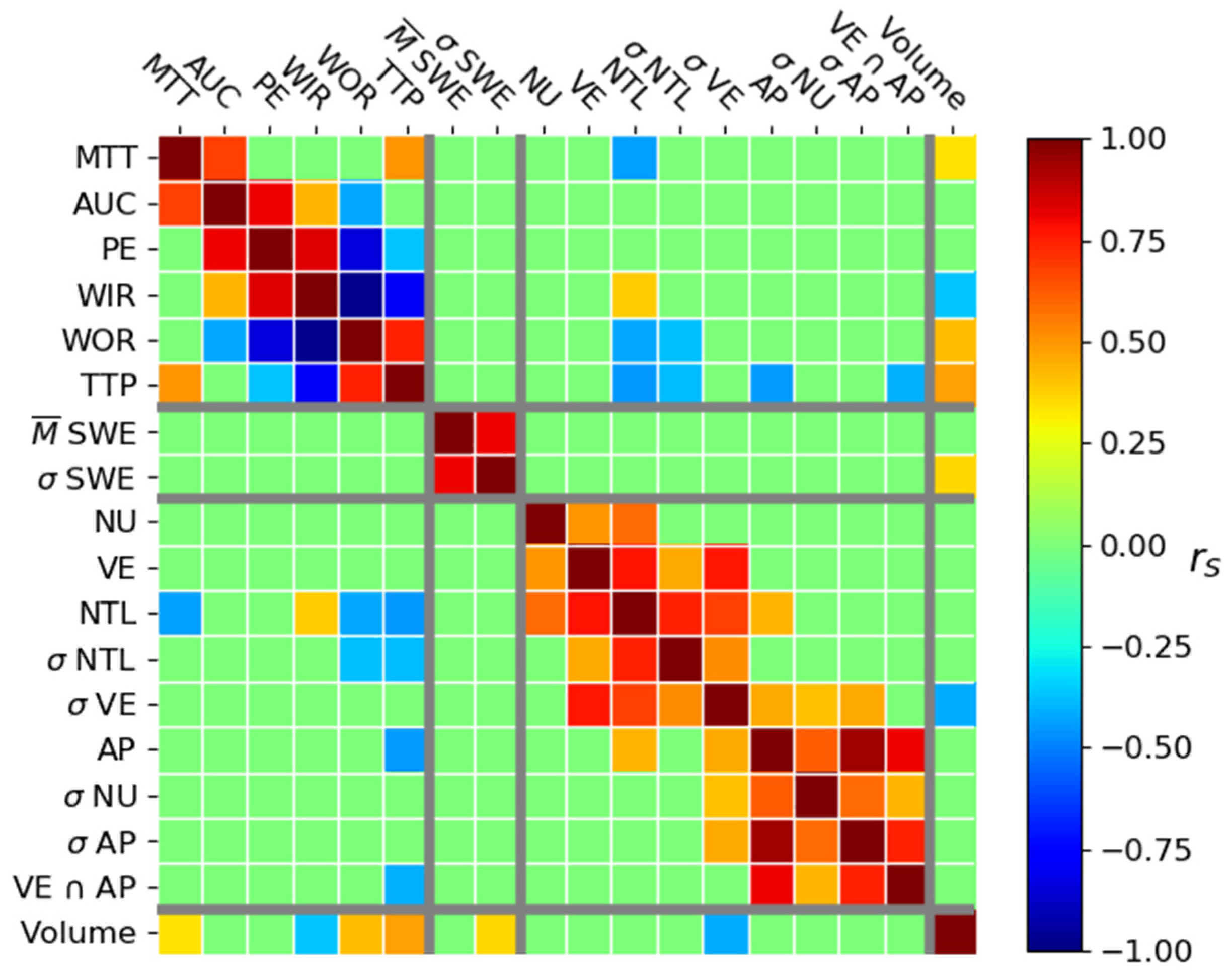
3.2. Correlation
3.2.1. Correlation between Tumor Volume and Histological Parameters
3.2.2. Correlation between US Parameters
3.2.3. Correlation between Volume and US Parameters
3.2.4. Correlation between Immunohistological and US Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Folkman, J.; Merler, E.; Abernathy, C.; Williams, G. Isolation of a tumor factor responsible for angiogenesis. J. Exp. Med. 1971, 133, 275–288. [Google Scholar] [CrossRef]
- Carmeliet, P.; Jain, R.K. Angiogenesis in cancer and other diseases. Nature 2000, 407, 249–257. [Google Scholar] [CrossRef]
- Rajendran, J.G.; Krohn, K.A. Imaging hypoxia and angiogenesis in tumors. Radiol. Clin. N. Am. 2005, 43, 169–187. [Google Scholar] [CrossRef]
- Minchinton, A.I.; Tannock, I.F. Drug penetration in solid tumours. Nat. Rev. Cancer 2006, 6, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Egeblad, M.; Ewald, A.J.; Askautrud, H.A.; Truitt, M.L.; Welm, B.E.; Bainbridge, E.; Peeters, G.; Krummel, M.F.; Werb, Z. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis. Model. Mech. 2008, 1, 155–167, discussion 165. [Google Scholar] [CrossRef]
- Egeblad, M.; Nakasone, E.S.; Werb, Z. Tumors as organs: Complex tissues that interface with the entire organism. Dev. Cell 2010, 18, 884–901. [Google Scholar] [CrossRef]
- Egeblad, M.; Rasch, M.G.; Weaver, V.M. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 2010, 22, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Erler, J.T.; Weaver, V.M. Three-dimensional context regulation of metastasis. Clin. Exp. Metastasis 2009, 26, 35–49. [Google Scholar] [CrossRef]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Eliceiri, K.W.; Campbell, J.M.; Inman, D.R.; White, J.G.; Keely, P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Therasse, P.; Arbuck, S.G.; Eisenhauer, E.A.; Wanders, J.; Kaplan, R.S.; Rubinstein, L.; Verweij, J.; Van Glabbeke, M.; van Oosterom, A.T.; Christian, M.C.; et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 2000, 92, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Brindle, K. New approaches for imaging tumour responses to treatment. Nat. Rev. Cancer 2008, 8, 94–107. [Google Scholar] [CrossRef]
- Huang, D.; Ding, Y.; Li, Y.; Luo, W.-M.; Zhang, Z.-F.; Snider, J.; Vandenbeldt, K.; Qian, C.-N.; Teh, B.T. Sunitinib acts primarily on tumor endothelium rather than tumor cells to inhibit the growth of renal cell carcinoma. Cancer Res. 2010, 70, 1053–1062. [Google Scholar] [CrossRef]
- Bogaerts, J.; Ford, R.; Sargent, D.; Schwartz, L.H.; Rubinstein, L.; Lacombe, D.; Eisenhauer, E.; Verweij, J.; Therasse, P.; RECIST Working Party. Individual patient data analysis to assess modifications to the RECIST criteria. Eur. J. Cancer 2009, 45, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Charnsangavej, C.; Faria, S.C.; Macapinlac, H.A.; Burgess, M.A.; Patel, S.R.; Chen, L.L.; Podoloff, D.A.; Benjamin, R.S. Correlation of computed tomography and positron emission tomography in patients with metastatic gastrointestinal stromal tumor treated at a single institution with imatinib mesylate: Proposal of new computed tomography response criteria. J. Clin. Oncol. 2007, 25, 1753–1759. [Google Scholar] [CrossRef]
- Lamuraglia, M.; Raslan, S.; Elaidi, R.; Oudard, S.; Escudier, B.; Slimane, K.; Penna, R.R.; Wagner, M.; Lucidarme, O. mTOR-inhibitor treatment of metastatic renal cell carcinoma: Contribution of Choi and modified Choi criteria assessed in 2D or 3D to evaluate tumor response. Eur. Radiol. 2016, 26, 278–285. [Google Scholar] [CrossRef]
- Seymour, L.; Bogaerts, J.; Perrone, A.; Ford, R.; Schwartz, L.H.; Mandrekar, S.; Lin, N.U.; Litière, S.; Dancey, J.; Chen, A.; et al. iRECIST: Guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017, 18, e143–e152. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Chamming’s, F.; Le-Frère-Belda, M.-A.; Latorre-Ossa, H.; Fitoussi, V.; Redheuil, A.; Assayag, F.; Pidial, L.; Gennisson, J.-L.; Tanter, M.; Cuénod, C.-A.; et al. Supersonic Shear Wave Elastography of Response to Anti-cancer Therapy in a Xenograft Tumor Model. Ultrasound Med. Biol. 2016, 42, 924–930. [Google Scholar] [CrossRef]
- Seguin, J.; Mignet, N.; Latorre Ossa, H.; Tanter, M.; Gennisson, J.-L. Evaluation of Antivascular Combretastatin A4 P Efficacy Using Supersonic Shear Imaging Technique of Ectopic Colon Carcinoma CT26. Ultrasound Med. Biol. 2017, 43, 2352–2361. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Cheng, W.; Li, Z.-Y.; Ying, L.; Wang, Q.-C.; Wu, T.; Tian, J.-W. Early Evaluation of Relative Changes in Tumor Stiffness by Shear Wave Elastography Predicts the Response to Neoadjuvant Chemotherapy in Patients With Breast Cancer. J. Ultrasound Med. 2016, 35, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Lassau, N.; Bonastre, J.; Kind, M.; Vilgrain, V.; Lacroix, J.; Cuinet, M.; Taieb, S.; Aziza, R.; Sarran, A.; Labbe-Devilliers, C.; et al. Validation of dynamic contrast-enhanced ultrasound in predicting outcomes of antiangiogenic therapy for solid tumors: The French multicenter support for innovative and expensive techniques study. Investig. Radiol. 2014, 49, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Lamuraglia, M.; Barrois, G.; Le Guillou-Buffello, D.; Santin, M.; Kerbol, A.; Comperat, E.; Coron, A.; Lucidarme, O.; Bridal, S.L. Monitoring Dual VEGF Inhibition in Human Pancreatic Tumor Xenografts With Dynamic Contrast-Enhanced Ultrasound. Technol. Cancer Res. Treat. 2020, 19, 1533033819886896. [Google Scholar] [CrossRef] [PubMed]
- Shiina, T.; Nightingale, K.R.; Palmeri, M.L.; Hall, T.J.; Bamber, J.C.; Barr, R.G.; Castera, L.; Choi, B.I.; Chou, Y.-H.; Cosgrove, D.; et al. WFUMB guidelines and recommendations for clinical use of ultrasound elastography: Part 1: Basic principles and terminology. Ultrasound Med. Biol. 2015, 41, 1126–1147. [Google Scholar] [CrossRef]
- Chamming’s, F.; Latorre-Ossa, H.; Le Frère-Belda, M.A.; Fitoussi, V.; Quibel, T.; Assayag, F.; Marangoni, E.; Autret, G.; Balvay, D.; Pidial, L.; et al. Shear wave elastography of tumour growth in a human breast cancer model with pathological correlation. Eur. Radiol. 2013, 23, 2079–2086. [Google Scholar] [CrossRef]
- Pochon, S.; Tardy, I.; Bussat, P.; Bettinger, T.; Brochot, J.; von Wronski, M.; Passantino, L.; Schneider, M. BR55: A lipopeptide-based VEGFR2-targeted ultrasound contrast agent for molecular imaging of angiogenesis. Investig. Radiol. 2010, 45, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M. SonoVue, a new ultrasound contrast agent. Eur. Radiol. 1999, 9 (Suppl. 3), S347–S348. [Google Scholar] [CrossRef]
- Seguin, J.; Doan, B.-T.; Latorre Ossa, H.; Jugé, L.; Gennisson, J.-L.; Tanter, M.; Scherman, D.; Chabot, G.G.; Mignet, N. Evaluation of Nonradiative Clinical Imaging Techniques for the Longitudinal Assessment of Tumour Growth in Murine CT26 Colon Carcinoma. Int. J. Mol. Imaging 2013, 2013, 983534. [Google Scholar] [CrossRef]
- Payen, T.; Dizeux, A.; Baldini, C.; Le Guillou-Buffello, D.; Lamuraglia, M.; Comperat, E.; Lucidarme, O.; Bridal, S.L. VEGFR2-Targeted Contrast-Enhanced Ultrasound to Distinguish between Two Anti-Angiogenic Treatments. Ultrasound Med. Biol. 2015, 41, 2202–2211. [Google Scholar] [CrossRef]
- Turco, S.; El Kaffas, A.; Zhou, J.; Lutz, A.M.; Wijkstra, H.; Willmann, J.K.; Mischi, M. Pharmacokinetic Modeling of Targeted Ultrasound Contrast Agents for Quantitative Assessment of Anti-Angiogenic Therapy: A Longitudinal Case-Control Study in Colon Cancer. Mol. Imaging Biol. 2019, 21, 633–643. [Google Scholar] [CrossRef]
- Jugé, L.; Doan, B.-T.; Seguin, J.; Albuquerque, M.; Larrat, B.; Mignet, N.; Chabot, G.G.; Scherman, D.; Paradis, V.; Vilgrain, V.; et al. Colon tumor growth and antivascular treatment in mice: Complementary assessment with MR elastography and diffusion-weighted MR imaging. Radiology 2012, 264, 436–444. [Google Scholar] [CrossRef]
- Cheung, A.M.Y.; Brown, A.S.; Hastie, L.A.; Cucevic, V.; Roy, M.; Lacefield, J.C.; Fenster, A.; Foster, F.S. Three-dimensional ultrasound biomicroscopy for xenograft growth analysis. Ultrasound Med. Biol. 2005, 31, 865–870. [Google Scholar] [CrossRef]
- Dizeux, A.; Payen, T.; Barrois, G.; Le Guillou Buffello, D.; Bridal, S.L. Reproducibility of Contrast-Enhanced Ultrasound in Mice with Controlled Injection. Mol. Imaging Biol. 2016, 18, 651–658. [Google Scholar] [CrossRef]
- Payen, T.; Coron, A.; Lamuraglia, M.; Le Guillou-Buffello, D.; Gaud, E.; Arditi, M.; Lucidarme, O.; Bridal, S.L. Echo-power estimation from log-compressed video data in dynamic contrast-enhanced ultrasound imaging. Ultrasound Med. Biol. 2013, 39, 1826–1837. [Google Scholar] [CrossRef][Green Version]
- Dietrich, C.F.; Averkiou, M.A.; Correas, J.-M.; Lassau, N.; Leen, E.; Piscaglia, F. An EFSUMB introduction into Dynamic Contrast-Enhanced Ultrasound (DCE-US) for quantification of tumour perfusion. Ultraschall Med. 2012, 33, 344–351. [Google Scholar] [CrossRef]
- Hudson, J.M.; Williams, R.; Tremblay-Darveau, C.; Sheeran, P.S.; Milot, L.; Bjarnason, G.A.; Burns, P.N. Dynamic contrast enhanced ultrasound for therapy monitoring. Eur. J. Radiol. 2015, 84, 1650–1657. [Google Scholar] [CrossRef]
- Alaarg, A.; Pérez-Medina, C.; Metselaar, J.M.; Nahrendorf, M.; Fayad, Z.A.; Storm, G.; Mulder, W.J.M. Applying nanomedicine in maladaptive inflammation and angiogenesis. Adv. Drug Deliv. Rev. 2017, 119, 143–158. [Google Scholar] [CrossRef]
- McDonald, D.M.; Choyke, P.L. Imaging of angiogenesis: From microscope to clinic. Nat. Med. 2003, 9, 713–725. [Google Scholar] [CrossRef]
- Opacic, T.; Dencks, S.; Theek, B.; Piepenbrock, M.; Ackermann, D.; Rix, A.; Lammers, T.; Stickeler, E.; Delorme, S.; Schmitz, G.; et al. Motion model ultrasound localization microscopy for preclinical and clinical multiparametric tumor characterization. Nat. Commun. 2018, 9, 1527. [Google Scholar] [CrossRef]
- O’Connor, J.P.B.; Rose, C.J.; Waterton, J.C.; Carano, R.A.D.; Parker, G.J.M.; Jackson, A. Imaging intratumor heterogeneity: Role in therapy response, resistance, and clinical outcome. Clin. Cancer Res. 2015, 21, 249–257. [Google Scholar] [CrossRef]
- Wildeboer, R.R.; van Sloun, R.J.G.; Postema, A.W.; Mannaerts, C.K.; Gayet, M.; Beerlage, H.P.; Wijkstra, H.; Mischi, M. Accurate validation of ultrasound imaging of prostate cancer: A review of challenges in registration of imaging and histopathology. J. Ultrasound 2018, 21, 197–207. [Google Scholar] [CrossRef]
- Wildeboer, R.R.; Van Sloun, R.J.G.; Schalk, S.G.; Mannaerts, C.K.; Van Der Linden, J.C.; Huang, P.; Wijkstra, H.; Mischi, M. Convective-Dispersion Modeling in 3D Contrast-Ultrasound Imaging for the Localization of Prostate Cancer. IEEE Trans. Med. Imaging 2018, 37, 2593–2602. [Google Scholar] [CrossRef]
- Mamou, J.; Oelze, M.L.; O’Brien, W.D.; Zachary, J.F. Identifying ultrasonic scattering sites from three-dimensional impedance maps. J. Acoust. Soc. Am. 2005, 117, 413–423. [Google Scholar] [CrossRef]
- Streeter, J.E.; Gessner, R.C.; Tsuruta, J.; Feingold, S.; Dayton, P.A. Assessment of molecular imaging of angiogenesis with three-dimensional ultrasonography. Mol. Imaging 2011, 10, 460–468. [Google Scholar] [CrossRef]
- Heng, J.; Heng, H.H. Genome chaos: Creating new genomic information essential for cancer macroevolution. Semin Cancer Biol. 2020. S1044-579X(20)30224-8. [Google Scholar] [CrossRef]
- Henry, H. Genome Chaos: Rethinking Genetics, Evolution, and Molecular Medicine; Chapter 8—The Rationale and Challenges of Molecular Medicine; Academic Press: Cambridge, MA, USA, 2019; pp. 427–479. [Google Scholar] [CrossRef]

| Parameters | D7 | D10 | D14 | D16 | D17 |
|---|---|---|---|---|---|
| Volume | 28.3 (9.7) | 64.4 (22.7) | 554.4 (263.4) | 862.6 (369.3) | 978.1 (510.3) |
| NU | 29.98 (−) | 55.42 (7.77) | 58.15 (13.36) | 46.17 (0.99) | 48.04 (3.83) |
| σ NU | 19.71 (−) | 18.57 (1.19) | 17.55 (3.64) | 20.15 (3.94) | 17.79 (1.47) |
| NTL | 861.18 (−) | 597.38 (143.19) | 636.09 (233.60) | 282.78 (61.15) | 321.02 (76.50) |
| σ NTL | 2.88 × 103 (−) | 760.49 (148.61) | 637.93 (289.35) | 527.22 (129.09) | 361.31 (105.64) |
| AP | 2.29 (−) | 2.36 (0.74) | 1.00 (0.56) | 0.80 (0.92) | 1.63 (0.84) |
| σ AP | 3.63 (−) | 5.28 (2.26) | 2.70 (0.56) | 2.92 (1.95) | 4.78 (2.06) |
| VE | 27.32 (−) | 38.49 (21.98) | 37.48 (5.02) | 20.18 (15.26) | 32.93 (7.02) |
| σ VE | 19.50 (−) | 18.21 (0.91) | 16.58 (2.72) | 13.14 (4.57) | 15.39 (2.50) |
| VE ∩ AP | 11.24 (−) | 12.97 (4.65) | 4.36 (3.13) | 4.05 (2.49) | 8.25 (3.11) |
| SWE | 10.8 (3.3) | 12.9 (4.5) | 16.2 (3.9) | 17.4 (2.9) | 16.6 (2.2) |
| σ SWE | 3.0 (1.1) | 3.6 (0.4) | 5.4 (1.3) | 6.5 (1.7) | 5.8 (0.6) |
| AUC | 3.21 (0.84) | 5.56 (2.32) | 6.48 (3.35) | 6.33 (1.29) | 4.66 (2.52) |
| PE | 7.3 × 10−2 (2.7 × 10−2) | 0.12 (4.1 × 10−2) | 9.2 × 10−2 (2.0 × 10−2) | 9.6 × 10−2 (2.3 × 10−2) | 6.0 × 10−2 (2.4 × 10−2) |
| TTP | 6.23 (1.59) | 7.63 (1.33) | 14.77 (6.34) | 15.37 (5.78) | 17.19 (2.67) |
| MTT | 68.53 (22.75) | 64.58 (6.65) | 68.90 (16.70) | 76.22 (15.99) | 67.10 (14.45) |
| WIR | 2.8 × 10−2 (1.2 × 10−2) | 4.0 × 10−2 (1.7 × 10−2) | 1.6 × 10−2 (6 × 10−3) | 1.5 × 10−2 (5 × 10−3) | 7 × 10−3 (1 × 10−3) |
| WOR | −2 × 10−3 (2 × 10−3) | −3 × 10−3 (6 × 10−4) | −2 × 10−3 (1 × 10−4) | −2 × 10−3 (3 × 10−4) | −9 × 10−4 (2 × 10−4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Griffon, J.; Buffello, D.; Giron, A.; Bridal, S.L.; Lamuraglia, M. Non-Invasive Ultrasonic Description of Tumor Evolution. Cancers 2021, 13, 4560. https://doi.org/10.3390/cancers13184560
Griffon J, Buffello D, Giron A, Bridal SL, Lamuraglia M. Non-Invasive Ultrasonic Description of Tumor Evolution. Cancers. 2021; 13(18):4560. https://doi.org/10.3390/cancers13184560
Chicago/Turabian StyleGriffon, Jerome, Delphine Buffello, Alain Giron, S. Lori Bridal, and Michele Lamuraglia. 2021. "Non-Invasive Ultrasonic Description of Tumor Evolution" Cancers 13, no. 18: 4560. https://doi.org/10.3390/cancers13184560
APA StyleGriffon, J., Buffello, D., Giron, A., Bridal, S. L., & Lamuraglia, M. (2021). Non-Invasive Ultrasonic Description of Tumor Evolution. Cancers, 13(18), 4560. https://doi.org/10.3390/cancers13184560






