A Genome-Wide Profiling of Glioma Patients with an IDH1 Mutation Using the Catalogue of Somatic Mutations in Cancer Database
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. COSMIC Database
2.2. Tissue Distributions of IDH1/2 Mutations
2.3. Occurrence of Mutations in Glioma with an IDH1 Mutation
2.4. CRAVAT Analysis
2.5. Gene Expression Levels in Glioma with an IDH1 Mutation
3. Results and Discussion
3.1. Tissue Distribution of IDH1/2 Mutations
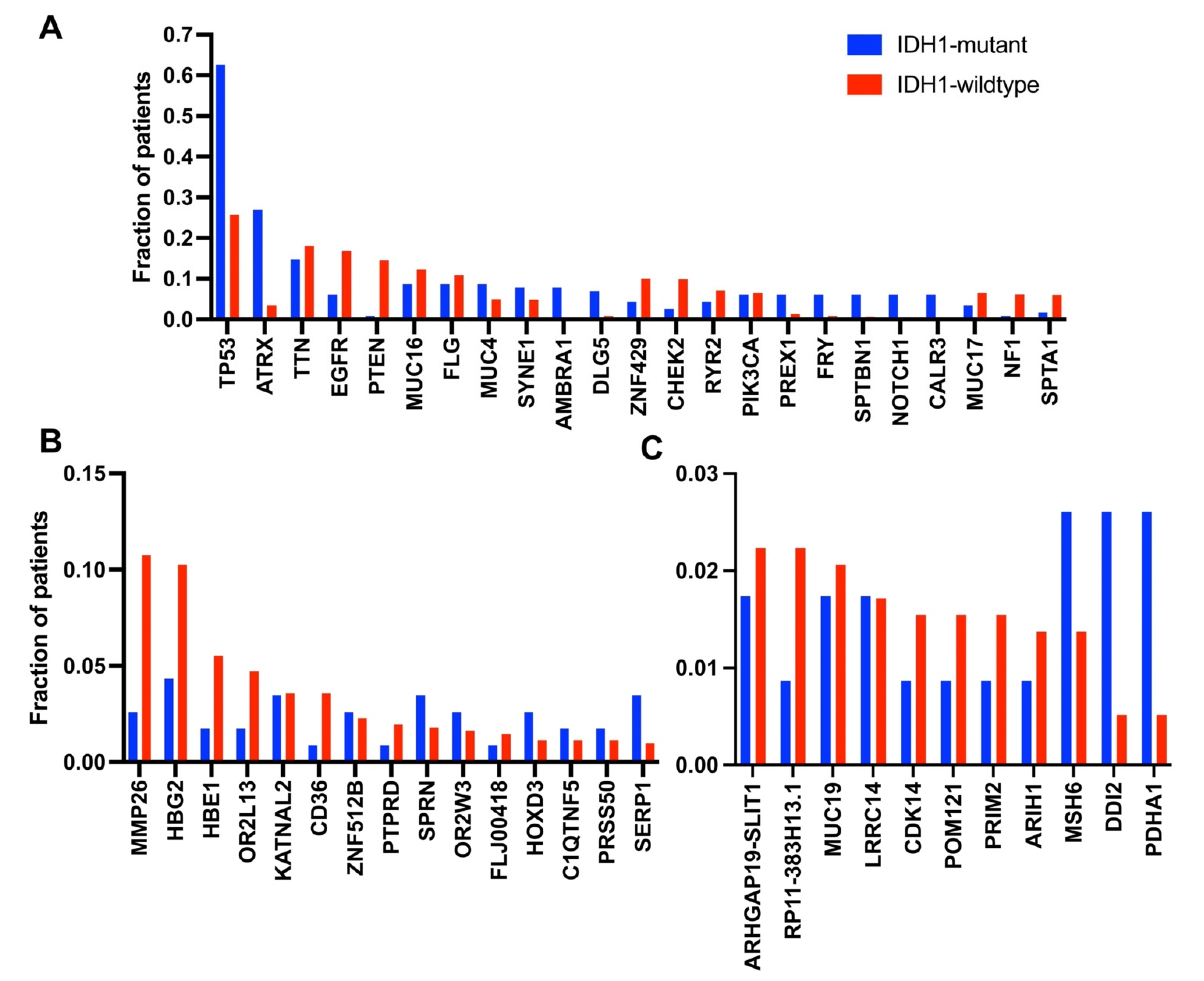
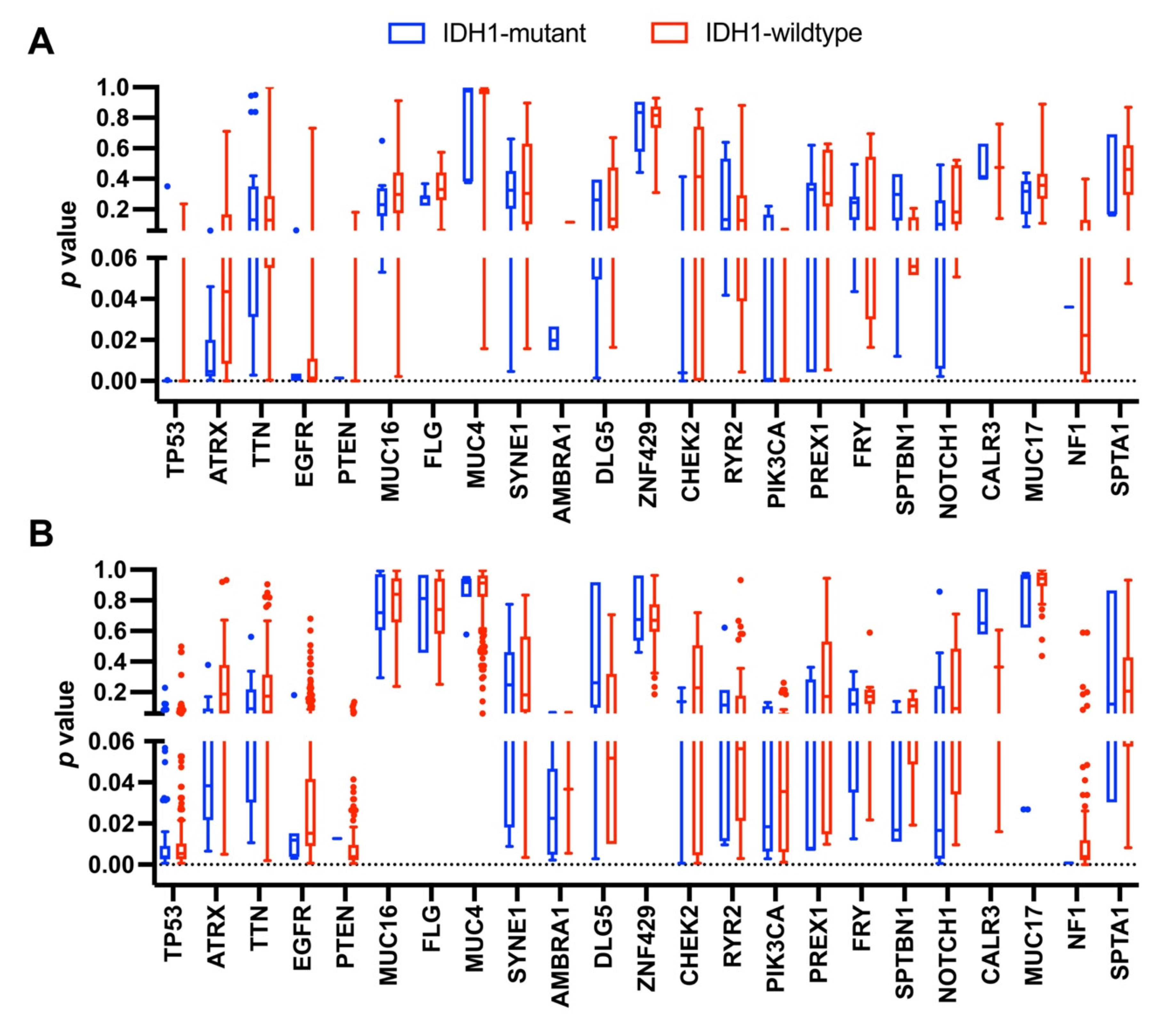
3.2. Occurrence of Mutations in Glioma with an IDH1 Mutation
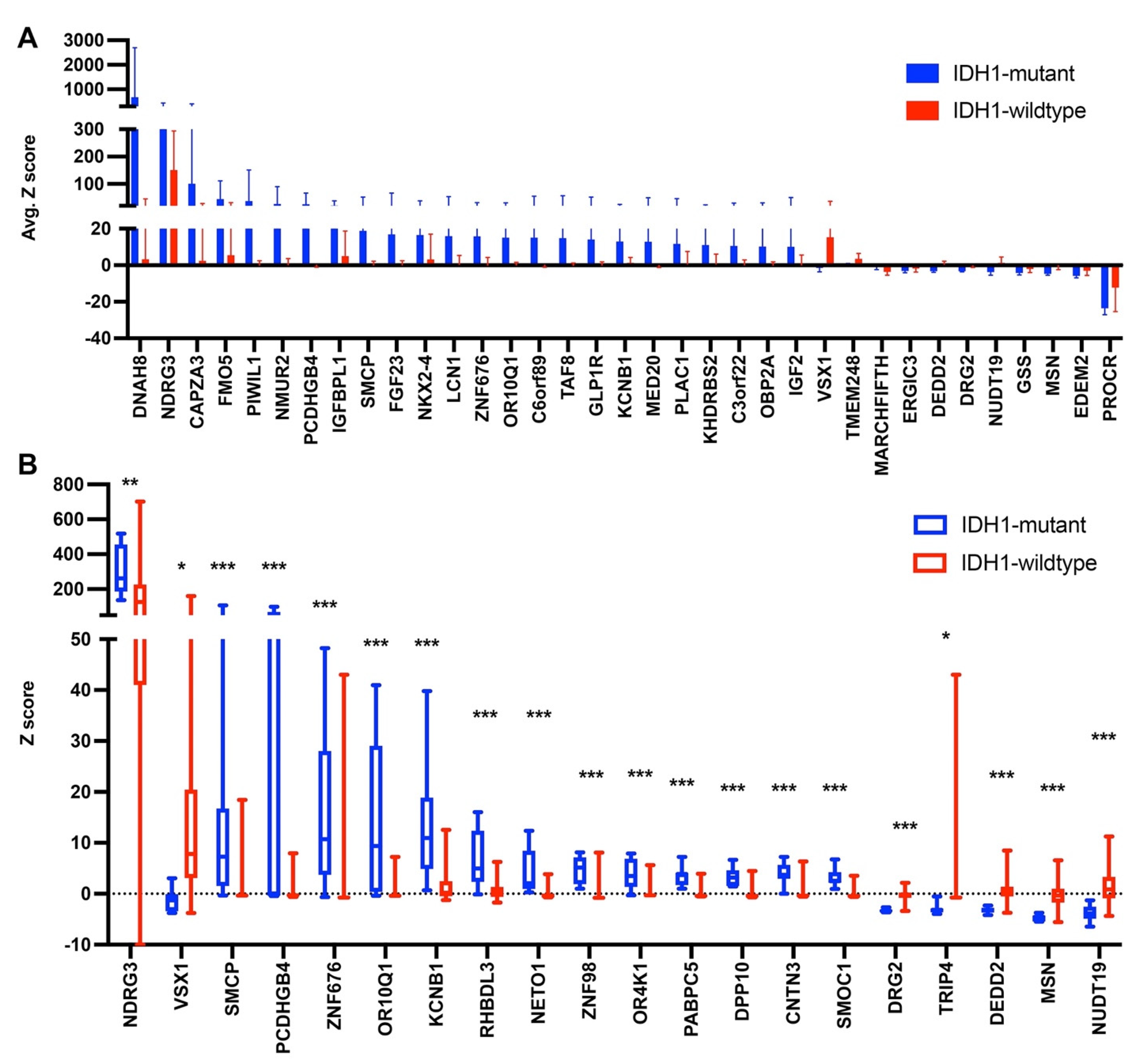
3.3. Gene Expression Levels in Glioma with an IDH1 Mutation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huse, J.T.; Holland, E.C. Targeting Brain Cancer: Advances in the Molecular Pathology of Malignant Glioma and Medulloblastoma. Nat. Rev. Cancer 2010, 10, 319–331. [Google Scholar] [CrossRef]
- Wen, P.Y.; Kesari, S. Malignant Gliomas in Adults. N. Engl. J. Med. 2008, 359, 492–507. [Google Scholar] [CrossRef]
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary Brain Tumours in Adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature 2008, 455, 1061–1068. [CrossRef]
- Brennan, C.W.; Verhaak, R.G.W.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Dang, L.; Jin, S.; Su, S.M. IDH Mutations in Glioma and Acute Myeloid Leukemia. Trends Mol. Med. 2010, 16, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.E. Friend or Foe-IDH1 Mutations in Glioma 10 Years On. Carcinogenesis 2019, 40, 1299–1307. [Google Scholar] [CrossRef]
- Tran, A.N.; Lai, A.; Li, S.; Pope, W.B.; Teixeira, S.; Harris, R.J.; Woodworth, D.C.; Nghiemphu, P.L.; Cloughesy, T.F.; Ellingson, B.M. Increased Sensitivity to Radiochemotherapy in IDH1 Mutant Glioblastoma as Demonstrated by Serial Quantitative MR Volumetry. Neuro Oncol. 2014, 16, 414–420. [Google Scholar] [CrossRef]
- Sanson, M.; Marie, Y.; Paris, S.; Idbaih, A.; Laffaire, J.; Ducray, F.; El Hallani, S.; Boisselier, B.; Mokhtari, K.; Hoang-Xuan, K.; et al. Isocitrate Dehydrogenase 1 Codon 132 Mutation Is an Important Prognostic Biomarker in Gliomas. J. Clin. Oncol. 2009, 27, 4150–4154. [Google Scholar] [CrossRef]
- Bleeker, F.E.; Atai, N.A.; Lamba, S.; Jonker, A.; Rijkeboer, D.; Bosch, K.S.; Tigchelaar, W.; Troost, D.; Vandertop, W.P.; Bardelli, A.; et al. The Prognostic IDH1(R132) Mutation Is Associated with Reduced NADP+-Dependent IDH Activity in Glioblastoma. Acta Neuropathol. 2010, 119, 487–494. [Google Scholar] [CrossRef]
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational Landscape and Clonal Architecture in Grade II and III Gliomas. Nat. Genet. 2015, 47, 458–468. [Google Scholar] [CrossRef]
- Liu, Y.; Lang, F.; Chou, F.-J.; Zaghloul, K.A.; Yang, C. Isocitrate Dehydrogenase Mutations in Glioma: Genetics, Biochemistry, and Clinical Indications. Biomedicines 2020, 8, 294. [Google Scholar] [CrossRef]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and Molecular Epidemiology of Adult Diffuse Glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef]
- Richardson, T.E.; Kumar, A.; Xing, C.; Hatanpaa, K.J.; Walker, J.M. Overcoming the Odds: Toward a Molecular Profile of Long-Term Survival in Glioblastoma. J. Neuropathol. Exp. Neurol. 2020, 79, 1031–1037. [Google Scholar] [CrossRef]
- Lucas, C.-H.G.; Solomon, D.A.; Perry, A. A Review of Recently Described Genetic Alterations in Central Nervous System Tumors. Hum. Pathol 2020, 96, 56–66. [Google Scholar] [CrossRef]
- Yan, H.; Bigner, D.D.; Velculescu, V.; Parsons, D.W. Mutant Metabolic Enzymes Are at the Origin of Gliomas. Cancer Res. 2009, 69, 9157–9159. [Google Scholar] [CrossRef]
- Minniti, G.; Scaringi, C.; Arcella, A.; Lanzetta, G.; Di Stefano, D.; Scarpino, S.; Bozzao, A.; Pace, A.; Villani, V.; Salvati, M.; et al. IDH1 Mutation and MGMT Methylation Status Predict Survival in Patients with Anaplastic Astrocytoma Treated with Temozolomide-Based Chemoradiotherapy. J. Neurooncol. 2014, 118, 377–383. [Google Scholar] [CrossRef]
- Yang, P.; Zhang, W.; Wang, Y.; Peng, X.; Chen, B.; Qiu, X.; Li, G.; Li, S.; Wu, C.; Yao, K.; et al. IDH Mutation and MGMT Promoter Methylation in Glioblastoma: Results of a Prospective Registry. Oncotarget 2015, 6, 40896–40906. [Google Scholar] [CrossRef]
- Hartmann, C.; Meyer, J.; Balss, J.; Capper, D.; Mueller, W.; Christians, A.; Felsberg, J.; Wolter, M.; Mawrin, C.; Wick, W.; et al. Type and Frequency of IDH1 and IDH2 Mutations Are Related to Astrocytic and Oligodendroglial Differentiation and Age: A Study of 1,010 Diffuse Gliomas. Acta Neuropathol. 2009, 118, 469–474. [Google Scholar] [CrossRef]
- Zhao, S.; Lin, Y.; Xu, W.; Jiang, W.; Zha, Z.; Wang, P.; Yu, W.; Li, Z.; Gong, L.; Peng, Y.; et al. Glioma-Derived Mutations in IDH1 Dominantly Inhibit IDH1 Catalytic Activity and Induce HIF-1alpha. Science 2009, 324, 261–265. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-Associated IDH1 Mutations Produce 2-Hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Chin, R.M.; Vergnes, L.; Hwang, H.; Deng, G.; Xing, Y.; Pai, M.Y.; Li, S.; Ta, L.; Fazlollahi, F.; et al. 2-Hydroxyglutarate Inhibits ATP Synthase and MTOR Signaling. Cell Metab. 2015, 22, 508–515. [Google Scholar] [CrossRef]
- Sulkowski, P.L.; Corso, C.D.; Robinson, N.D.; Scanlon, S.E.; Purshouse, K.R.; Bai, H.; Liu, Y.; Sundaram, R.K.; Hegan, D.C.; Fons, N.R.; et al. 2-Hydroxyglutarate Produced by Neomorphic IDH Mutations Suppresses Homologous Recombination and Induces PARP Inhibitor Sensitivity. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Chowdhury, R.; Yeoh, K.K.; Tian, Y.-M.; Hillringhaus, L.; Bagg, E.A.; Rose, N.R.; Leung, I.K.H.; Li, X.S.; Woon, E.C.Y.; Yang, M.; et al. The Oncometabolite 2-Hydroxyglutarate Inhibits Histone Lysine Demethylases. EMBO Rep. 2011, 12, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Waitkus, M.S.; Diplas, B.H.; Yan, H. Biological Role and Therapeutic Potential of IDH Mutations in Cancer. Cancer Cell 2018, 34, 186–195. [Google Scholar] [CrossRef]
- Carbonneau, M.; Gagné, L.M.; Lalonde, M.-E.; Germain, M.-A.; Motorina, A.; Guiot, M.-C.; Secco, B.; Vincent, E.E.; Tumber, A.; Hulea, L.; et al. The Oncometabolite 2-Hydroxyglutarate Activates the MTOR Signalling Pathway. Nat. Commun. 2016, 7, 12700. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, Y.; Chen, J.; Qiu, J.; Huang, K.; Wu, M.; Xia, C. IDH1 R132H Mutation Enhances Cell Migration by Activating AKT-MTOR Signaling Pathway, but Sensitizes Cells to 5-FU Treatment as NADPH and GSH Are Reduced. PLoS ONE 2017, 12, e0169038. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, Y.; Li, A.; Celiku, O.; Han, S.; Qian, M.; Yang, C. MTORC2/Rac1 Pathway Predisposes Cancer Aggressiveness in IDH1-Mutated Glioma. Cancers 2020, 12, 787. [Google Scholar] [CrossRef]
- Núñez, F.J.; Mendez, F.M.; Kadiyala, P.; Alghamri, M.S.; Savelieff, M.G.; Garcia-Fabiani, M.B.; Haase, S.; Koschmann, C.; Calinescu, A.-A.; Kamran, N.; et al. IDH1-R132H Acts as a Tumor Suppressor in Glioma via Epigenetic up-Regulation of the DNA Damage Response. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH Mutation Impairs Histone Demethylation and Results in a Block to Cell Differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.M.; Lu, C.; Ward, P.S.; et al. IDH1 Mutation Is Sufficient to Establish the Glioma Hypermethylator Phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Nobusawa, S.; Kleihues, P.; Ohgaki, H. IDH1 Mutations Are Early Events in the Development of Astrocytomas and Oligodendrogliomas. Am. J. Pathol. 2009, 174, 1149–1153. [Google Scholar] [CrossRef]
- Inoue, S.; Li, W.Y.; Tseng, A.; Beerman, I.; Elia, A.J.; Bendall, S.C.; Lemonnier, F.; Kron, K.J.; Cescon, D.W.; Hao, Z.; et al. Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer Cell 2016, 30, 337–348. [Google Scholar] [CrossRef]
- Ohba, S.; Mukherjee, J.; Johannessen, T.-C.; Mancini, A.; Chow, T.T.; Wood, M.; Jones, L.; Mazor, T.; Marshall, R.E.; Viswanath, P.; et al. Mutant IDH1 Expression Drives TERT Promoter Reactivation as Part of the Cellular Transformation Process. Cancer Res. 2016, 76, 6680–6689. [Google Scholar] [CrossRef]
- Turcan, S.; Makarov, V.; Taranda, J.; Wang, Y.; Fabius, A.W.M.; Wu, W.; Zheng, Y.; El-Amine, N.; Haddock, S.; Nanjangud, G.; et al. Mutant-IDH1-Dependent Chromatin State Reprogramming, Reversibility, and Persistence. Nat. Genet. 2018, 50, 62–72. [Google Scholar] [CrossRef]
- Leca, J.; Fortin, J.; Mak, T.W. Illuminating the Cross-Talk between Tumor Metabolism and Immunity in IDH-Mutated Cancers. Curt. Opin. Biotechnol. 2021, 68, 181–185. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH Mutation in Glioma: Molecular Mechanisms and Potential Therapeutic Targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, M.; Yan, J.; Youn, Y.; Drucker, K.L.; Kollmeyer, T.M.; McKinney, A.M.; Zazubovich, V.; Zhang, Y.; Costello, J.F.; Eckel-Passow, J.; et al. Functional Analysis of Low-Grade Glioma Genetic Variants Predicts Key Target Genes and Transcription Factors. Neuro Oncol. 2020. [Google Scholar] [CrossRef]
- Banan, R.; Stichel, D.; Bleck, A.; Hong, B.; Lehmann, U.; Suwala, A.; Reinhardt, A.; Schrimpf, D.; Buslei, R.; Stadelmann, C.; et al. Infratentorial IDH-Mutant Astrocytoma Is a Distinct Subtype. Acta Neuropathol. 2020, 140, 569–581. [Google Scholar] [CrossRef]
- Sun, C.; Xiao, L.; Zhao, Y.; Shi, J.; Yuan, Y.; Gu, Y.; Zhang, F.; Gao, X.; Yang, Y.; Yang, R.; et al. Wild-Type IDH1 and Mutant IDH1 Opposingly Regulate Podoplanin Expression in Glioma. Transl. Oncol. 2020, 13, 100758. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, A.; Casalini, B.; Chavez, L.; Hielscher, T.; Sill, M.; Ryzhova, M.; Sharma, T.; Schrimpf, D.; Stichel, D.; Capper, D.; et al. Integrated Molecular Characterization of IDH-Mutant Glioblastomas. Neuropathol. Appl. Neurobiol. 2019, 45, 108–118. [Google Scholar] [CrossRef]
- Wu, W.Y.-Y.; Johansson, G.; Wibom, C.; Brännström, T.; Malmström, A.; Henriksson, R.; Golovleva, I.; Bondy, M.L.; Andersson, U.; Dahlin, A.M.; et al. The Genetic Architecture of Gliomagenesis-Genetic Risk Variants Linked to Specific Molecular Subtypes. Cancers 2019, 11, 2001. [Google Scholar] [CrossRef]
- Atkins, I.; Kinnersley, B.; Ostrom, Q.T.; Labreche, K.; Il’yasova, D.; Armstrong, G.N.; Eckel-Passow, J.E.; Schoemaker, M.J.; Nöthen, M.M.; Barnholtz-Sloan, J.S.; et al. Transcriptome-Wide Association Study Identifies New Candidate Susceptibility Genes for Glioma. Cancer Res. 2019, 79, 2065–2071. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; He, D.; Yang, P.; He, J.; Zhang, Y. Genome-Wide Expression Profiling of Glioblastoma Using a Large Combined Cohort. Sci. Rep. 2018, 8, 15104. [Google Scholar] [CrossRef]
- Ohba, S.; Hirose, Y. Association between Mutant IDHs and Tumorigenesis in Gliomas. Med. Mol. Morphol. 2018, 51, 194–198. [Google Scholar] [CrossRef]
- Melin, B.S.; Barnholtz-Sloan, J.S.; Wrensch, M.R.; Johansen, C.; Il’yasova, D.; Kinnersley, B.; Ostrom, Q.T.; Labreche, K.; Chen, Y.; Armstrong, G.; et al. Genome-Wide Association Study of Glioma Subtypes Identifies Specific Differences in Genetic Susceptibility to Glioblastoma and Non-Glioblastoma Tumors. Nat. Genet. 2017, 49, 789–794. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, H.; Zhang, C.; Wang, Z.; Li, M.; Zhang, W.; Jiang, T. Methylation Associated Genes Contribute to the Favorable Prognosis of Gliomas with Isocitrate Dehydrogenase 1 Mutation. Am. J. Cancer Res. 2015, 5, 2745–2755. [Google Scholar]
- Rajaraman, P.; Melin, B.S.; Wang, Z.; McKean-Cowdin, R.; Michaud, D.S.; Wang, S.S.; Bondy, M.; Houlston, R.; Jenkins, R.B.; Wrensch, M.; et al. Genome-Wide Association Study of Glioma and Meta-Analysis. Hum. Genet. 2012, 131, 1877–1888. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic Cancer Genetics at High-Resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef]
- Bamford, S.; Dawson, E.; Forbes, S.; Clements, J.; Pettett, R.; Dogan, A.; Flanagan, A.; Teague, J.; Futreal, P.A.; Stratton, M.R.; et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) Database and Website. Br. J. Cancer 2004, 91, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.D.; Allaire, A.; Diamandis, P.; Bisaillon, M.; Scott, M.S.; Richer, M. A Machine Learning Analysis of a “Normal-like” IDH-WT Diffuse Glioma Transcriptomic Subgroup Associated with Prolonged Survival Reveals Novel Immune and Neurotransmitter-Related Actionable Targets. BMC Med. 2020, 18, 280. [Google Scholar] [CrossRef] [PubMed]
- Riobello, C.; López-Hernández, A.; Cabal, V.N.; García-Marín, R.; Suárez-Fernández, L.; Sánchez-Fernández, P.; Vivanco, B.; Blanco, V.; López, F.; Franchi, A.; et al. IDH2 Mutation Analysis in Undifferentiated and Poorly Differentiated Sinonasal Carcinomas for Diagnosis and Clinical Management. Am. J. Surg. Pathol. 2020, 44, 396–405. [Google Scholar] [CrossRef]
- Kang, M.R.; Kim, M.S.; Oh, J.E.; Kim, Y.R.; Song, S.Y.; Seo, S.I.; Lee, J.Y.; Yoo, N.J.; Lee, S.H. Mutational Analysis of IDH1 Codon 132 in Glioblastomas and Other Common Cancers. Int. J. Cancer 2009, 125, 353–355. [Google Scholar] [CrossRef]
- Stein, E.M. IDH2 Inhibition in AML: Finally Progress? Best Pract. Res. Clin. Haematol. 2015, 28, 112–115. [Google Scholar] [CrossRef]
- Fu, Y.; Huang, R.; Du, J.; Yang, R.; An, N.; Liang, A. Glioma-Derived Mutations in IDH: From Mechanism to Potential Therapy. Biochem. Biophys. Res. Commun. 2010, 397, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Largeaud, L.; Bérard, E.; Bertoli, S.; Dufrechou, S.; Prade, N.; Gadaud, N.; Tavitian, S.; Bories, P.; Luquet, I.; Sarry, A.; et al. Outcome of AML Patients with IDH2 Mutations in Real World before the Era of IDH2 Inhibitors. Leuk. Res. 2019, 81, 82–87. [Google Scholar] [CrossRef]
- Bullinger, L.; Döhner, K.; Döhner, H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J. Clin. Oncol. 2017, 35, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Dogan, S.; Frosina, D.; Geronimo, J.A.; Hernandez, E.; Mohanty, A.; Bale, T.; Hechtman, J.F.; Arcila, M.E.; Hameed, M.R.; Jungbluth, A.A. Molecular Epidemiology of IDH2 Hotspot Mutations in Cancer and Immunohistochemical Detection of R172K, R172G, and R172M Variants. Hum. Pathol. 2020, 106, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but Not the Same: The Causes and Consequences of Codon Bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Doherty, A.; McInerney, J.O. Translational Selection Frequently Overcomes Genetic Drift in Shaping Synonymous Codon Usage Patterns in Vertebrates. Mol. Biol. Evol. 2013, 30, 2263–2267. [Google Scholar] [CrossRef]
- Komar, A.A. The Yin and Yang of Codon Usage. Hum. Mol. Genet. 2016, 25, R77–R85. [Google Scholar] [CrossRef] [PubMed]
- Athey, J.; Alexaki, A.; Osipova, E.; Rostovtsev, A.; Santana-Quintero, L.V.; Katneni, U.; Simonyan, V.; Kimchi-Sarfaty, C. A New and Updated Resource for Codon Usage Tables. BMC Bioinform. 2017, 18, 391. [Google Scholar] [CrossRef]
- Lipman, D.J.; Wilbur, W.J. Interaction of Silent and Replacement Changes in Eukaryotic Coding Sequences. J. Mol. Evol. 1984, 21, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Sharp, P.M. Mammalian Gene Evolution: Nucleotide Sequence Divergence between Mouse and Rat. J. Mol. Evol. 1993, 37, 441–456. [Google Scholar] [CrossRef]
- Mouchiroud, D.; Gautier, C.; Bernardi, G. Frequencies of Synonymous Substitutions in Mammals Are Gene-Specific and Correlated with Frequencies of Nonsynonymous Substitutions. J. Mol. Evol. 1995, 40, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Comeron, J.M.; Kreitman, M. The Correlation between Synonymous and Nonsynonymous Substitutions in Drosophila: Mutation, Selection or Relaxed Constraints? Genetics 1998, 150, 767–775. [Google Scholar] [CrossRef]
- Creixell, P.; Schoof, E.M.; Tan, C.S.H.; Linding, R. Mutational Properties of Amino Acid Residues: Implications for Evolvability of Phosphorylatable Residues. Philos. Trans. R. Soc. Lond B Biol. Sci. 2012, 367, 2584–2593. [Google Scholar] [CrossRef]
- Anoosha, P.; Sakthivel, R.; Michael Gromiha, M. Exploring Preferred Amino Acid Mutations in Cancer Genes: Applications to Identify Potential Drug Targets. Biochim. Biophys. Acta 2016, 1862, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Brat, D.J.; Verhaak, R.G.W.; Aldape, K.D.; Yung, W.K.A.; Salama, S.R.; Cooper, L.A.D.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Ohba, S.; Kuwahara, K.; Yamada, S.; Abe, M.; Hirose, Y. Correlation between IDH, ATRX, and TERT Promoter Mutations in Glioma. Brain Tumor Pathol. 2020, 37, 33–40. [Google Scholar] [CrossRef]
- Gülten, G.; Yalçın, N.; Baltalarlı, B.; Doğu, G.; Acar, F.; Doğruel, Y. The Importance of IDH1, ATRX and WT-1 Mutations in Glioblastoma. Pol. J. Pathol. 2020, 71, 127–137. [Google Scholar] [CrossRef]
- Jiao, Y.; Killela, P.J.; Reitman, Z.J.; Rasheed, A.B.; Heaphy, C.M.; de Wilde, R.F.; Rodriguez, F.J.; Rosemberg, S.; Oba-Shinjo, S.M.; Nagahashi Marie, S.K.; et al. Frequent ATRX, CIC, FUBP1 and IDH1 Mutations Refine the Classification of Malignant Gliomas. Oncotarget 2012, 3, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, H.; Liu, Z.; Zhang, J. Oligodendroglial Tumours: Subventricular Zone Involvement and Seizure History Are Associated with CIC Mutation Status. BMC Neurol. 2019, 19, 134. [Google Scholar] [CrossRef] [PubMed]
- Eleveld, T.F.; Schild, L.; Koster, J.; Zwijnenburg, D.A.; Alles, L.K.; Ebus, M.E.; Volckmann, R.; Tijtgat, G.A.; van Sluis, P.; Versteeg, R.; et al. RAS-MAPK Pathway-Driven Tumor Progression Is Associated with Loss of CIC and Other Genomic Aberrations in Neuroblastoma. Cancer Res. 2018, 78, 6297–6307. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Yu, L.; Wei, W.; Lin, X.; Hou, X.; Tian, Y. ShRNA-Mediated AMBRA1 Knockdown Reduces the Cisplatin-Induced Autophagy and Sensitizes Ovarian Cancer Cells to Cisplatin. J. Toxicol. Sci. 2016, 41, 45–53. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Z.; Guo, J.; Wang, L.; Liu, X. Ambra1 Induces Autophagy and Desensitizes Human Prostate Cancer Cells to Cisplatin. Biosci. Rep. 2019, 39, BSR20170770. [Google Scholar] [CrossRef]
- Schoenherr, C.; Byron, A.; Griffith, B.; Loftus, A.; Wills, J.C.; Munro, A.F.; von Kriegsheim, A.; Frame, M.C. The Autophagy Protein Ambra1 Regulates Gene Expression by Supporting Novel Transcriptional Complexes. J. Biol. Chem. 2020, 295, 12045–12057. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Ren, Y.; Liu, P. DLG5 in Cell Polarity Maintenance and Cancer Development. Int. J. Biol. Sci. 2014, 10, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Y.; Shen, B.; Qin, Y.; Qiu, J. Methylation-Mediated Silencing of Dlg5 Facilitates Bladder Cancer Metastasis. Exp. Cell Res. 2015, 331, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Barrows, D.; He, J.Z.; Parsons, R. PREX1 Protein Function Is Negatively Regulated Downstream of Receptor Tyrosine Kinase Activation by P21-Activated Kinases (PAKs). J. Biol Chem. 2016, 291, 20042–20054. [Google Scholar] [CrossRef]
- Gont, A.; Daneshmand, M.; Woulfe, J.; Lavictoire, S.J.; Lorimer, I.A.J. PREX1 Integrates G Protein-Coupled Receptor and Phosphoinositide 3-Kinase Signaling to Promote Glioblastoma Invasion. Oncotarget 2017, 8, 8559–8573. [Google Scholar] [CrossRef][Green Version]
- Jiang, X.; Gillen, S.; Esposito, I.; Giese, N.A.; Michalski, C.W.; Friess, H.; Kleeff, J. Reduced Expression of the Membrane Skeleton Protein Beta1-Spectrin (SPTBN1) Is Associated with Worsened Prognosis in Pancreatic Cancer. Histol. Histopathol. 2010, 25, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zeng, J.; Chen, S.; Li, J.; Wu, H.; Dong, X.; Lei, Y.; Zhi, X.; Yao, L. SPTBN1 Suppresses the Progression of Epithelial Ovarian Cancer via SOCS3-Mediated Blockade of the JAK/STAT3 Signaling Pathway. Aging 2020, 12, 10896–10911. [Google Scholar] [CrossRef]
- Yi, L.; Zhou, X.; Li, T.; Liu, P.; Hai, L.; Tong, L.; Ma, H.; Tao, Z.; Xie, Y.; Zhang, C.; et al. Notch1 Signaling Pathway Promotes Invasion, Self-Renewal and Growth of Glioma Initiating Cells via Modulating Chemokine System CXCL12/CXCR4. J. Exp. Clin. Cancer Res. 2019, 38, 339. [Google Scholar] [CrossRef]
- Liao, Y.; Ma, Z.; Zhang, Y.; Li, D.; Lv, D.; Chen, Z.; Li, P.; Ai-Dherasi, A.; Zheng, F.; Tian, J.; et al. Targeted Deep Sequencing from Multiple Sources Demonstrates Increased NOTCH1 Alterations in Lung Cancer Patient Plasma. Cancer Med. 2019, 8, 5673–5686. [Google Scholar] [CrossRef]
- Peng, X.; Zhou, J.; Li, B.; Zhang, T.; Zuo, Y.; Gu, X. Notch1 and PI3K/Akt Signaling Blockers DAPT and LY294002 Coordinately Inhibit Metastasis of Gastric Cancer through Mutual Enhancement. Cancer Chemother. Pharmacol. 2020, 85, 309–320. [Google Scholar] [CrossRef]
- Aref, S.; El Agdar, M.; Salama, O.; Zeid, T.A.; Sabry, M. Significance of NOTCH1 Mutations Détections in T-Acute Lymphoblastic Leukemia Patients. Cancer Biomark 2020, 27, 157–162. [Google Scholar] [CrossRef]
- Larose, H.; Prokoph, N.; Matthews, J.D.; Schlederer, M.; Högler, S.; Alsulami, A.F.; Ducray, S.P.; Nuglozeh, E.; Fazaludeen, F.M.S.; Elmouna, A.; et al. Whole Exome Sequencing Reveals NOTCH1 Mutations in Anaplastic Large Cell Lymphoma and Points to Notch Both as a Key Pathway and a Potential Therapeutic Target. Haematologica 2021, 106, 1693–1704. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-M.; Lee, S.-H.; Lim, J.; Yoo, J.; Hwang, D.-Y. The Epidermal Growth Factor Receptor Variant Type III Mutation Frequently Found in Gliomas Induces Astrogenesis in Human Cerebral Organoids. Cell Prolif. 2021, 54, e12965. [Google Scholar] [CrossRef] [PubMed]
- Zannini, L.; Delia, D.; Buscemi, G. CHK2 Kinase in the DNA Damage Response and Beyond. J. Mol. Cell Biol. 2014, 6, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Higashiguchi, M.; Nagatomo, I.; Kijima, T.; Morimura, O.; Miyake, K.; Minami, T.; Koyama, S.; Hirata, H.; Iwahori, K.; Takimoto, T.; et al. Clarifying the Biological Significance of the CHK2 K373E Somatic Mutation Discovered in The Cancer Genome Atlas Database. FEBS Lett. 2016, 590, 4275–4286. [Google Scholar] [CrossRef]
- Sutcliffe, E.G.; Stettner, A.R.; Miller, S.A.; Solomon, S.R.; Marshall, M.L.; Roberts, M.E.; Susswein, L.R.; Arvai, K.J.; Klein, R.T.; Murphy, P.D.; et al. Differences in Cancer Prevalence among CHEK2 Carriers Identified via Multi-Gene Panel Testing. Cancer Genetics 2020, 246–247, 12–17. [Google Scholar] [CrossRef]
- Ansari, N.; Shahrabi, S.; Khosravi, A.; Shirzad, R.; Rezaeean, H. Prognostic Significance of CHEK2 Mutation in Progression of Breast Cancer. Lab. Med. 2019, 50, e36–e41. [Google Scholar] [CrossRef]
- Thibodeau, M.L.; Reisle, C.; Zhao, E.; Martin, L.A.; Alwelaie, Y.; Mungall, K.L.; Ch’ng, C.; Thomas, R.; Ng, T.; Yip, S.; et al. Genomic Profiling of Pelvic Genital Type Leiomyosarcoma in a Woman with a Germline CHEK2:C.1100delC Mutation and a Concomitant Diagnosis of Metastatic Invasive Ductal Breast Carcinoma. Cold Spring Harb. Mol. Case Stud. 2017, 3. [Google Scholar] [CrossRef]
- Deng, Y.; Li, W.; Li, Y.; Yang, H.; Xu, H.; Liang, S.; Zhang, L.; Li, Y. Expression of Matrix Metalloproteinase-26 Promotes Human Glioma U251 Cell Invasion in Vitro and in Vivo. Oncol. Rep. 2010, 23, 69–78. [Google Scholar]
- Zhang, Y.; Zhao, H.; Wang, Y.; Lin, Y.; Tan, Y.; Fang, X.; Zheng, L. Non-Small Cell Lung Cancer Invasion and Metastasis Promoted by MMP-26. Mol. Med. Rep. 2011, 4, 1201–1209. [Google Scholar] [CrossRef]
- Gutschalk, C.M.; Yanamandra, A.K.; Linde, N.; Meides, A.; Depner, S.; Mueller, M.M. GM-CSF Enhances Tumor Invasion by Elevated MMP-2, -9, and -26 Expression. Cancer Med. 2013, 2, 117–129. [Google Scholar] [CrossRef]
- Xu, X.; Ma, J.; Li, C.; Zhao, W.; Xu, Y. Regulation of Chondrosarcoma Invasion by MMP26. Tumour Biol. 2015, 36, 365–369. [Google Scholar] [CrossRef]
- Atschekzei, F.; Hennenlotter, J.; Jänisch, S.; Großhennig, A.; Tränkenschuh, W.; Waalkes, S.; Peters, I.; Dörk, T.; Merseburger, A.S.; Stenzl, A.; et al. SFRP1 CpG Island Methylation Locus Is Associated with Renal Cell Cancer Susceptibility and Disease Recurrence. Epigenetics 2012, 7, 447–457. [Google Scholar] [CrossRef]
- Ma, Q.; Wu, X.; Wu, J.; Liang, Z.; Liu, T. SERP1 Is a Novel Marker of Poor Prognosis in Pancreatic Ductal Adenocarcinoma Patients via Anti-Apoptosis and Regulating SRPRB/NF-ΚB Axis. Int. J. Oncol. 2017, 51, 1104–1114. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-Specific Expression by Genome-Wide Integration of Transcriptomics and Antibody-Based Proteomics. Mol. Cell Proteomics 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Yagi, T.; Takeichi, M. Cadherin Superfamily Genes: Functions, Genomic Organization, and Neurologic Diversity. Genes Dev. 2000, 14, 1169–1180. [Google Scholar] [PubMed]
- El Hajj, N.; Dittrich, M.; Haaf, T. Epigenetic Dysregulation of Protocadherins in Human Disease. Semin. Cell Dev. Biol. 2017, 69, 172–182. [Google Scholar] [CrossRef]
- Vega-Benedetti, A.F.; Loi, E.; Moi, L.; Blois, S.; Fadda, A.; Antonelli, M.; Arcella, A.; Badiali, M.; Giangaspero, F.; Morra, I.; et al. Clustered Protocadherins Methylation Alterations in Cancer. Clin. Epigenetics 2019, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Douville, C.; Carter, H.; Kim, R.; Niknafs, N.; Diekhans, M.; Stenson, P.D.; Cooper, D.N.; Ryan, M.; Karchin, R. CRAVAT: Cancer-Related Analysis of Variants Toolkit. Bioinformatics 2013, 29, 647–648. [Google Scholar] [CrossRef]
- Wang, W.; Li, Y.; Li, Y.; Hong, A.; Wang, J.; Lin, B.; Li, R. NDRG3 Is an Androgen Regulated and Prostate Enriched Gene That Promotes in Vitro and in Vivo Prostate Cancer Cell Growth. Int. J. Cancer 2009, 124, 521–530. [Google Scholar] [CrossRef]
- Luo, X.; Hou, N.; Chen, X.; Xu, Z.; Xu, J.; Wang, L.; Yang, S.; Liu, S.; Xu, L.; Chen, Y.; et al. High Expression of NDRG3 Associates with Unfavorable Overall Survival in Non-Small Cell Lung Cancer. Cancer Biomark 2018, 21, 461–469. [Google Scholar] [CrossRef]
- Jing, J.-S.; Li, H.; Wang, S.-C.; Ma, J.-M.; Yu, L.-Q.; Zhou, H. NDRG3 Overexpression Is Associated with a Poor Prognosis in Patients with Hepatocellular Carcinoma. Biosci. Rep. 2018, 38, BSR20180907. [Google Scholar] [CrossRef]
- Lee, G.Y.; Shin, S.-H.; Shin, H.-W.; Chun, Y.-S.; Park, J.-W. NDRG3 Lowers the Metastatic Potential in Prostate Cancer as a Feedback Controller of Hypoxia-Inducible Factors. Exp. Mol. Med. 2018, 50, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shetty, R.; Nuijts, R.M.M.A.; Nanaiah, S.G.; Anandula, V.R.; Ghosh, A.; Jayadev, C.; Pahuja, N.; Kumaramanickavel, G.; Nallathambi, J. Two Novel Missense Substitutions in the VSX1 Gene: Clinical and Genetic Analysis of Families with Keratoconus from India. BMC Med. Genet. 2015, 16, 33. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Wang, W.; Liu, Y.-W.; Li, M.-Y.; Liang, T.-Y.; Li, J.-Y.; Hu, H.-M.; Lu, Y.; Yao, C.; Ye, Y.-Y.; et al. Role of KCNB1 in the Prognosis of Gliomas and Autophagy Modulation. Sci. Rep. 2017, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Mangino, M.; Hwang, S.-J.; Spector, T.D.; Hunt, S.C.; Kimura, M.; Fitzpatrick, A.L.; Christiansen, L.; Petersen, I.; Elbers, C.C.; Harris, T.; et al. Genome-Wide Meta-Analysis Points to CTC1 and ZNF676 as Genes Regulating Telomere Homeostasis in Humans. Hum. Mol. Genet. 2012, 21, 5385–5394. [Google Scholar] [CrossRef]
- Levy, D.; Neuhausen, S.L.; Hunt, S.C.; Kimura, M.; Hwang, S.-J.; Chen, W.; Bis, J.C.; Fitzpatrick, A.L.; Smith, E.; Johnson, A.D.; et al. Genome-Wide Association Identifies OBFC1 as a Locus Involved in Human Leukocyte Telomere Biology. Proc. Natl. Acad. Sci. USA 2010, 107, 9293–9298. [Google Scholar] [CrossRef]
- Malnic, B.; Godfrey, P.A.; Buck, L.B. The Human Olfactory Receptor Gene Family. Proc. Natl. Acad. Sci. USA 2004, 101, 2584–2589. [Google Scholar] [CrossRef] [PubMed]
- Barbirou, M.; Sghaier, I.; Bedoui, S.; Ben Abderrazek, R.; Kraiem, H.; Farah, A.; Hassiki, R.; Mokrani, A.; Mezlini, A.; Almawi, W.Y.; et al. KCNB1 Gene Polymorphisms and Related Indel as Predictor Biomarkers of Treatment Response for Colorectal Cancer—Toward a Personalized Medicine. Tumour Biol. 2020, 42. [Google Scholar] [CrossRef]
- Barbirou, M.; Woldu, H.G.; Sghaier, I.; Bedoui, S.A.; Mokrani, A.; Aami, R.; Mezlini, A.; Yacoubi-Loueslati, B.; Tonellato, P.J.; Bouhaouala-Zahar, B. Western Influenced Lifestyle and Kv2.1 Association as Predicted Biomarkers for Tunisian Colorectal Cancer. BMC Cancer 2020, 20, 1086. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Kim, S.-I.; Ko, M.S.; Kim, H.J.; Heo, J.C.; Lee, H.J.; Lee, H.S.; Han, I.S.; Kwack, K.; Park, J.W. Overexpression of DRG2 Increases G2/M Phase Cells and Decreases Sensitivity to Nocodazole-Induced Apoptosis. J. Biochem. 2004, 135, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.J.; Yoo, S.S.; Choi, J.E.; Kang, H.-G.; Do, S.K.; Lee, J.H.; Lee, W.K.; Lee, J.; Lee, S.Y.; Cha, S.I.; et al. Functional Intronic Variant of SLC5A10 Affects DRG2 Expression and Survival Outcomes of Early-Stage Non-Small-Cell Lung Cancer. Cancer Sci. 2018, 109, 3902–3909. [Google Scholar] [CrossRef]
- Yoon, N.A.; Jung, S.J.; Choi, S.H.; Ryu, J.H.; Mani, M.; Lee, U.H.; Vo, M.-T.; Jeon, D.Y.; Chung, S.W.; Ju Lee, B.; et al. DRG2 Supports the Growth of Primary Tumors and Metastases of Melanoma by Enhancing VEGF-A Expression. FEBS J. 2020, 287, 2070–2086. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Schickling, O.; Stegh, A.H.; Oshima, R.G.; Dinsdale, D.; Cohen, G.M.; Peter, M.E. DEDD Regulates Degradation of Intermediate Filaments during Apoptosis. J. Cell Biol. 2002, 158, 1051–1066. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, B.C.; Lee, J.C.; Alappat, E.C.; Peter, M.E. The Death Effector Domain Protein Family. Oncogene 2003, 22, 8634–8644. [Google Scholar] [CrossRef][Green Version]
- Alcivar, A.; Hu, S.; Tang, J.; Yang, X. DEDD and DEDD2 Associate with Caspase-8/10 and Signal Cell Death. Oncogene 2003, 22, 291–297. [Google Scholar] [CrossRef]
- Wu, M.; Liu, D.-Y.; Yuan, X.-R.; Liu, Q.; Jiang, X.-J.; Yuan, D.; Huang, J.; Li, X.-J.; Yang, Z.-Q. The Expression of Moesin in Astrocytoma: Correlation with Pathologic Grade and Poor Clinical Outcome. Med. Oncol. 2013, 30, 372. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lu, X.; Zhao, S.; Pang, M.; Wu, X.; Wu, H.; Hoffman, R.M.; Yang, Z.; Zhang, Y. Moesin Expression Is Associated with Glioblastoma Cell Proliferation and Invasion. Anticancer Res. 2017, 37, 2211–2218. [Google Scholar] [CrossRef]
- Barros, F.B.A.; Assao, A.; Garcia, N.G.; Nonogaki, S.; Carvalho, A.L.; Soares, F.A.; Kowalski, L.P.; Oliveira, D.T. Moesin Expression by Tumor Cells Is an Unfavorable Prognostic Biomarker for Oral Cancer. BMC Cancer 2018, 18, 53. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, X.; Wang, J.; Yang, Z.; Hoffman, R.M.; Wu, X. Moesin Up-Regulation Is Associated with Enhanced Tumor Progression Imaged Non-Invasively in an Orthotopic Mouse Model of Human Glioblastoma. Anticancer Res. 2018, 38, 3267–3272. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pappula, A.L.; Rasheed, S.; Mirzaei, G.; Petreaca, R.C.; Bouley, R.A. A Genome-Wide Profiling of Glioma Patients with an IDH1 Mutation Using the Catalogue of Somatic Mutations in Cancer Database. Cancers 2021, 13, 4299. https://doi.org/10.3390/cancers13174299
Pappula AL, Rasheed S, Mirzaei G, Petreaca RC, Bouley RA. A Genome-Wide Profiling of Glioma Patients with an IDH1 Mutation Using the Catalogue of Somatic Mutations in Cancer Database. Cancers. 2021; 13(17):4299. https://doi.org/10.3390/cancers13174299
Chicago/Turabian StylePappula, Amrit L., Shayaan Rasheed, Golrokh Mirzaei, Ruben C. Petreaca, and Renee A. Bouley. 2021. "A Genome-Wide Profiling of Glioma Patients with an IDH1 Mutation Using the Catalogue of Somatic Mutations in Cancer Database" Cancers 13, no. 17: 4299. https://doi.org/10.3390/cancers13174299
APA StylePappula, A. L., Rasheed, S., Mirzaei, G., Petreaca, R. C., & Bouley, R. A. (2021). A Genome-Wide Profiling of Glioma Patients with an IDH1 Mutation Using the Catalogue of Somatic Mutations in Cancer Database. Cancers, 13(17), 4299. https://doi.org/10.3390/cancers13174299





