Low Doses of Silver Nanoparticles Selectively Induce Lipid Peroxidation and Proteotoxic Stress in Mesenchymal Subtypes of Triple-Negative Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Silver Nanoparticles
2.2. Cell Culture
2.3. MTT Assay
2.4. Clonogenic Growth Assay
2.5. IncuCyte Proliferation Assay
2.6. mRNA Expression
2.7. Inductively Coupled Plasma Mass Spectrometry (ICP-MS)
2.8. Lipid Peroxidation Assays
2.9. Western Blots
2.10. Protein Aggregation Assays
2.11. Protein Oxidation Assays
2.12. ImageJ Analysis
2.13. Statistical Analysis
3. Results
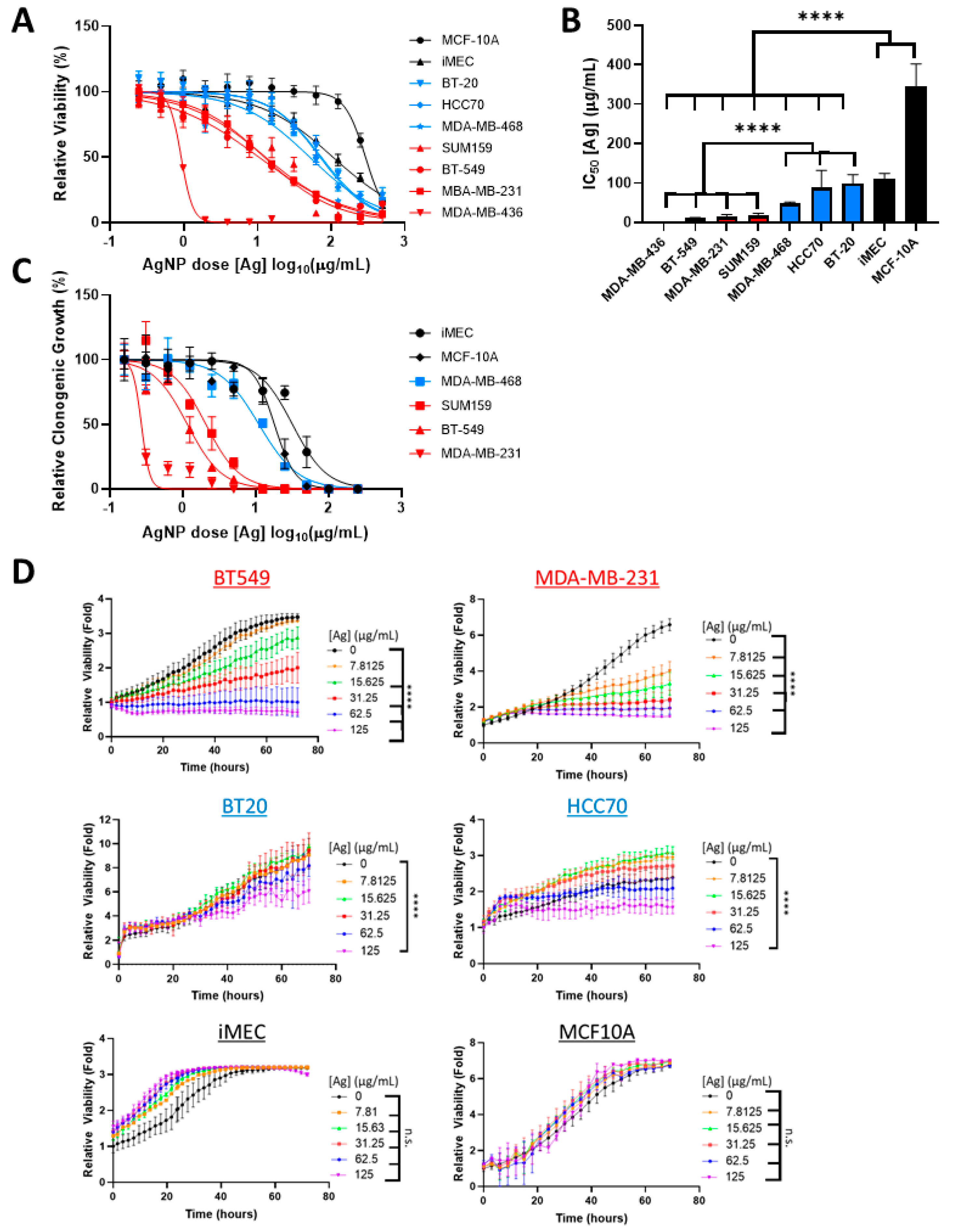
3.1. AgNPs Are Lethal to CLBCs at Doses That Have Little Effect on BLBCs and No Effect on Viability of Non-Malignant Breast Epithelial Cells
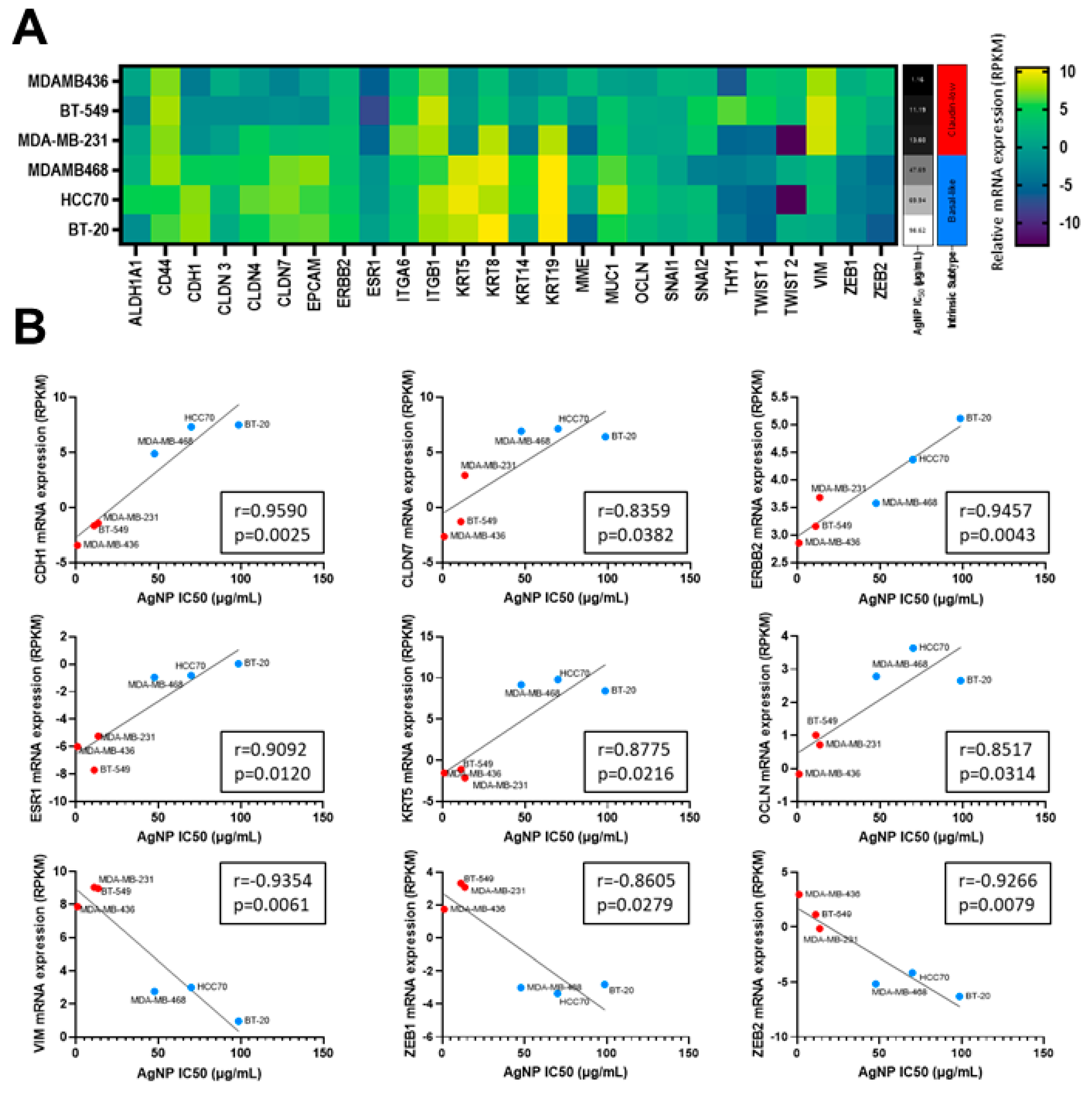
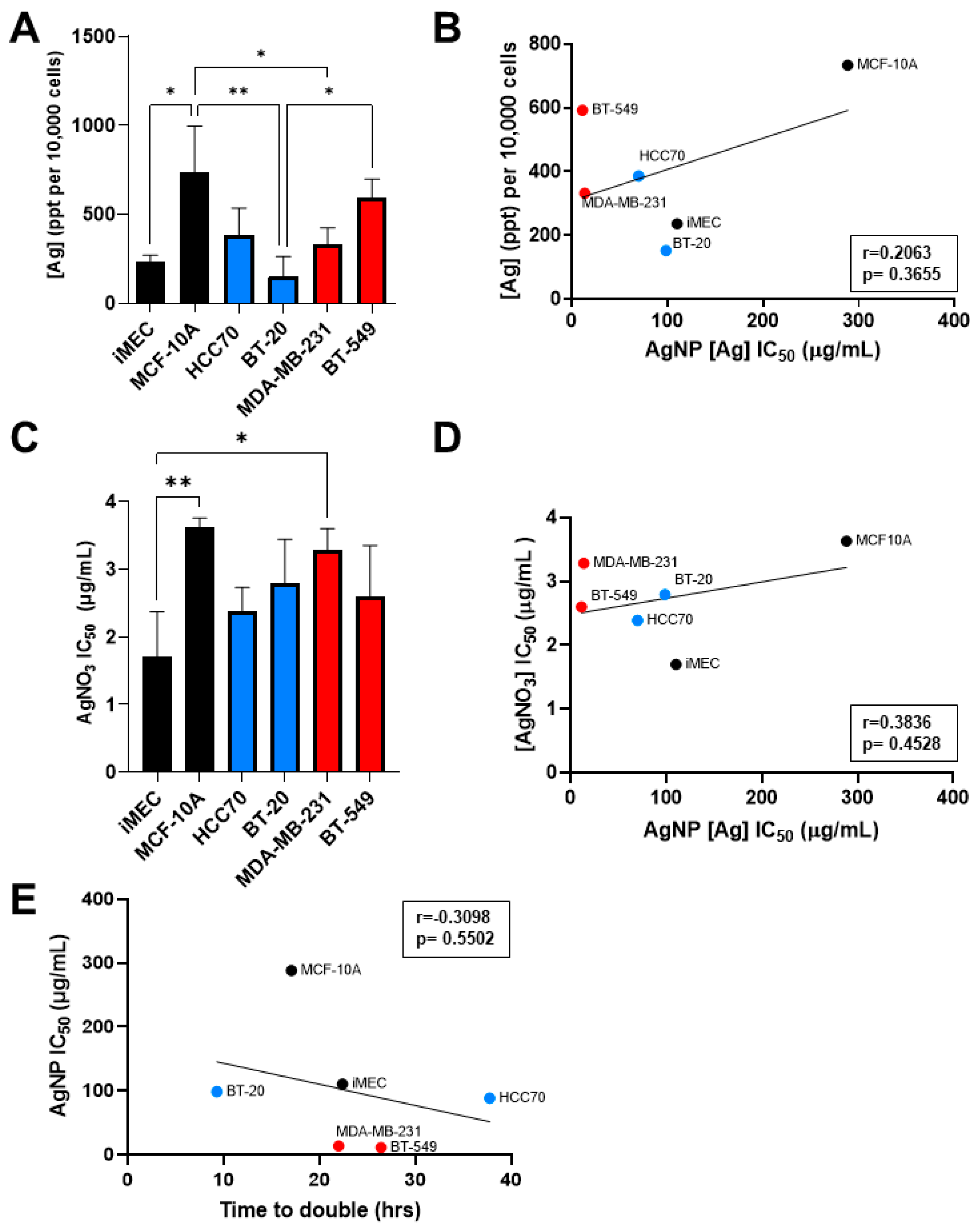
3.2. Sensitivity to AgNP Exposure Does Not Correlate with Mass of AgNPs Taken Up, Inherent Sensitivity to Silver Cations, or Proliferation Rate
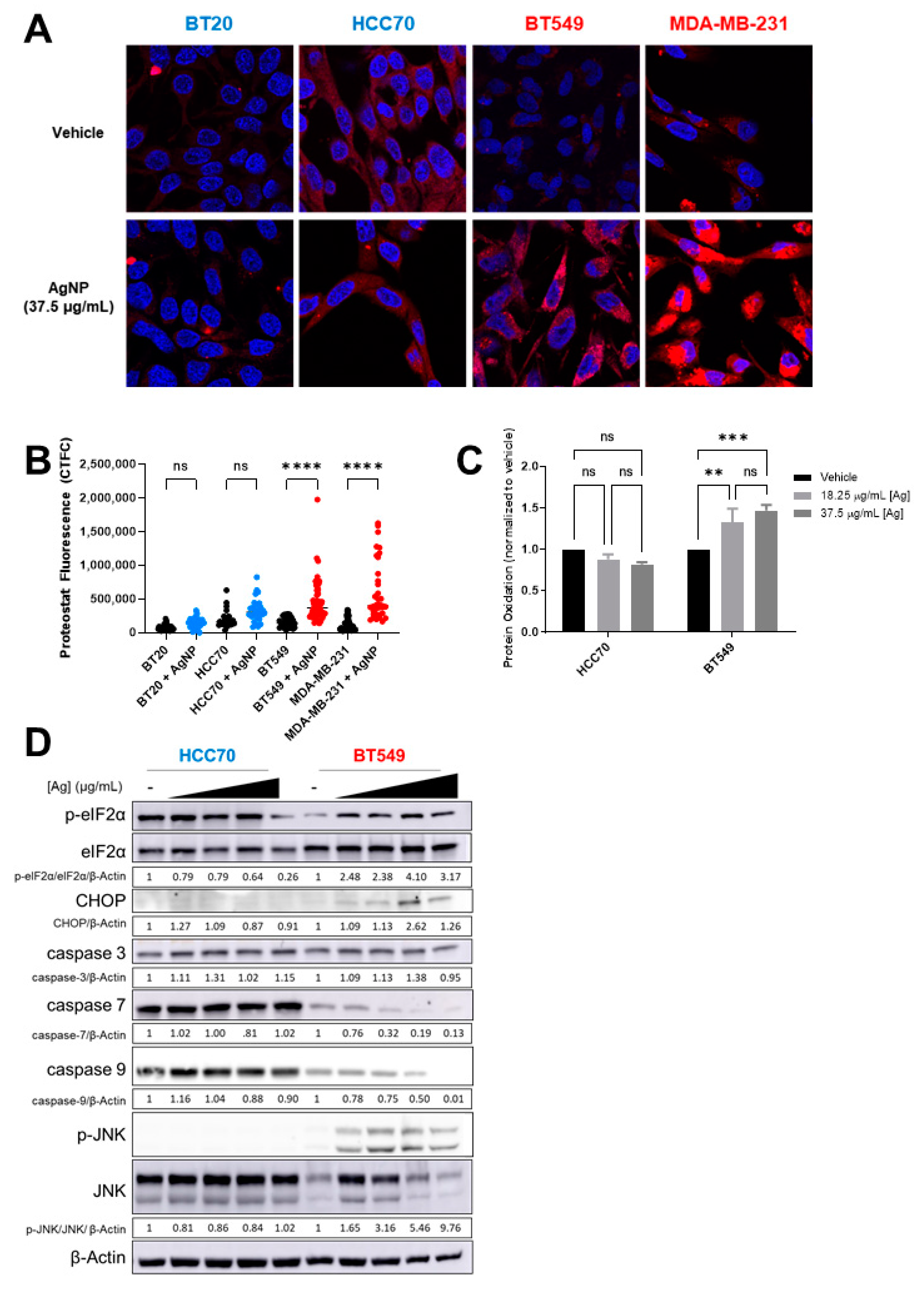
3.3. AgNPs Induce Lipid Peroxidation and 4-HNE-Histidine Adduct Formation in CLBC but Not in BLBC
3.4. AgNPs Induce Protein Aggregation, Protein Oxidation, and Proteotoxic Stress Responses in CLBC but in Not BLBC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yagata, H.; Kajiura, Y.; Yamauchi, H. Current strategy for triple-negative breast cancer: Appropriate combination of surgery, radiation, and chemotherapy. Breast Cancer 2011, 18, 165–173. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010, 12, R68. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Gerhard, R.; Ricardo, S.; Albergaria, A.; Gomes, M.; Silva, A.R.; Logullo, A.F.; Cameselle-Teijeiro, J.F.; Paredes, J.; Schmitt, F. Immunohistochemical features of claudin-low intrinsic subtype in metaplastic breast carcinomas. Breast 2012, 21, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Dias, K.; Dvorkin-Gheva, A.; Hallett, R.M.; Wu, Y.; Hassell, J.; Pond, G.R.; Levine, M.; Whelan, T.; Bane, A.L. Claudin-Low breast cancer; clinical & pathological characteristics. PLoS ONE 2017, 12, e0168669. [Google Scholar]
- Hennessy, B.T.; Gonzalez-Angulo, A.-M.; Stemke-Hale, K.; Gilcrease, M.Z.; Krishnamurthy, S.; Lee, J.-S.; Fridlyand, J.; A Sahin, A.; Agarwal, R.; Joy, C.; et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009, 69, 4116–4124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabatier, R.; Finetti, P.; Guille, A.; Adélaïde, J.; Chaffanet, M.; Viens, P.; Birnbaum, D.; Bertucci, F. Claudin-Low breast cancers: Clinical, pathological, molecular and prognostic characterization. Mol. Cancer 2014, 13, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perou, C.; Parker, J.S.; Prat, A.; Ellis, M.J.; Bernard, P.S. Clinical implementation of the intrinsic subtypes of breast cancer. Lancet Oncol. 2010, 11, 718–719. [Google Scholar] [CrossRef]
- Lu, W.; Kang, Y. Epithelial-Mesenchymal plasticity in Cancer progression and metastasis. Dev. Cell 2019, 49, 61–374. [Google Scholar] [CrossRef] [PubMed]
- Gooding, A.J.; Schiemann, W.P. Epithelial-Mesenchymal Transition programs and cancer stem cell phenotypes: Mediators of Breast cancer therapy resistance. Mol. Cancer Res. 2020, 18, 1257–1270. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.-X.; Jin, D.X.; Sokol, E.S.; Reinhardt, F.; Miller, D.H.; Gupta, P.B. Cancer-Specific PERK signaling drives invasion and metastasis through CREB3L1. Nat. Commun. 2017, 8, 1079. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.X.; Sokol, E.S.; Del Vecchio, C.A.; Sanduja, S.; Claessen, J.H.; Proia, T.A.; Jin, D.X.; Reinhardt, F.; Ploegh, H.L.; Wang, Q.; et al. Epithelial-to-mesenchymal transition activates PERK-eIF2α and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 2014, 4, 702–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; You, J.H.; Kim, M.-S.; Roh, J.-L. Epigenetic reprogramming of epithelial-mesenchymal transition promotes ferroptosis of head and neck cancer. Redox Biol. 2020, 37, 101697. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, V.; Ryan, M.; Dhruv, H.D.; Gill, S.; Eichhoff, O.M.; Seashore-Ludlow, B.; Kaffenberger, S.D.; Eaton, J.; Shimada, K.; Aguirre, A.J.; et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nat. Cell Biol. 2017, 547, 453–457. [Google Scholar] [CrossRef]
- Doll, S.; Proneth, B.; Tyurina, Y.; Panzilius, E.; Kobayashi, S.; Ingold, I.; Irmler, M.; Beckers, M.I.J.; Aichler, M.; Walch, M.A.A.; et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017, 13, 91–98. [Google Scholar] [CrossRef]
- Giudetti, A.M.; De Domenico, S.; Ragusa, A.; Lunetti, P.; Gaballo, A.; Franck, J.; Simeone, P.; Nicolardi, G.; De Nuccio, F.; Santino, A.; et al. A specific lipid metabolic profile is associated with the epithelial mesenchymal transition program. Biochimica Biophysica Acta Mol. Cell Biol. Lipids 2019, 1864, 344–357. [Google Scholar] [CrossRef]
- Li, Z.; Chen, L.; Chen, C.; Zhou, Y.; Hu, D.; Yang, J.; Chen, Y.; Zhuo, W.; Mao, M.; Zhang, X.; et al. Targeting ferroptosis in breast cancer. Biomark. Res. 2020, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Saha, M.; Ghosh, S.S. Nanoparticle mediated alteration of EMT dynamics: An approach to modulate cancer therapeutics. Mater. Adv. 2020, 1, 2614–2630. [Google Scholar] [CrossRef]
- Arvizo, R.R.; Saha, S.; Wang, E.; Robertson, J.; Bhattacharya, R.; Mukherjee, P. Inhibition of tumor growth and metastasis by a self-therapeutic nanoparticle. Proc. Natl. Acad. Sci. USA 2013, 110, 6700–6705. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Chen, C.; Qian, P.; Lu, X.; Sun, B.; Zhang, X.; Wang, L.; Gao, X.; Li, H.; Chen, Z.; et al. Gd-metallofullerenol nanomaterial as non-toxic breast cancer stem cell-specific inhibitor. Nat. Commun. 2015, 6, 5988. [Google Scholar] [CrossRef] [PubMed]
- Gowda, S.S.; Rajasowmiya, S.; Vadivel, V.; Devi, S.B.; Jerald, A.C.; Marimuthu, S.; Devipriya, N. Gallic acid-coated sliver nanoparticle alters the expression of radiation-induced epithelial-mesenchymal transition in non-small lung cancer cells. Toxicol. Vitro 2018, 52, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Swanner, J.; Mims, J.; Carroll, D.L.; Akman, S.A.; Furdui, C.M.; Torti, S.V.; Singh, R.N. Differential cytotoxic and radiosensitizing effects of silver nanoparticles on triple-negative breast cancer and non-triple-negative breast cells. Int. J. Nanomed. 2015, 10, 3937–3953. [Google Scholar]
- Swanner, J.; Fahrenholtz, C.D.; Tenvooren, I.; Bernish, B.W.; Sears, J.J.; Hooker, A.; Furdui, C.M.; Alli, E.; Li, W.; Donati, G.L.; et al. Silver nanoparticles selectively treat triple-negative breast cancer cells without affecting non-malignant breast epithelial cells in vitro and in vivo. FASEB BioAdv. 2019, 1, 639–660. [Google Scholar] [CrossRef]
- Simard, J.-C.; Durocher, I.; Girard, D. Silver nanoparticles induce irremediable endoplasmic reticulum stress leading to unfolded protein response dependent apoptosis in breast cancer cells. Apoptosis 2016, 21, 1279–1290. [Google Scholar] [CrossRef]
- Sears, J.; Swanner, J.; Fahrenholtz, C.D.; Snyder, C.; Rohde, M.; Levi-Polyachenko, N.; Singh, R. Combined photothermal and ionizing radiation sensitization of triple-negative breast cancer using triangular silver nanoparticles. Int. J. Nanomed. 2021, 16, 851–865. [Google Scholar] [CrossRef]
- Lewis, J.E.; Singh, N.; Holmila, R.J.; Sumer, B.D.; Williams, N.S.; Furdui, C.M.; Kemp, M.L.; Boothman, D.A. Targeting NAD+ metabolism to enhance radiation therapy responses. Semin. Radiat. Oncol. 2019, 29, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Simard, J.-C.; Vallieres, F.; de Liz, R.; Lavastre, V.; Girard, D. Silver nanoparticles Induce degradation of the endoplasmic reticulum stress sensor activating transcription factor-6 leading to activation of the NLRP-3 inflammasome. J. Biol. Chem. 2015, 290, 5926–5939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmila, R.; Wu, H.; Lee, J.; Tsang, A.W.; Singh, R.; Furdui, C.M. Integrated redox proteomic analysis highlights new mechanisms of sensitivity to silver nanoparticles. Mol. Cell. Proteom. 2021, 20, 100073. [Google Scholar] [CrossRef]
- Saleeb, N.; Robinson, B.; Cavanagh, J.; Ross, J.; Munir, K.; Gooneratne, R. Antioxidant enzyme activity and lipid peroxidation in Aporrectodea caliginosa earthworms Exposed to silver nanoparticles and silver nitrate in spiked soil. Environ. Toxicol. Chem. 2020, 39, 1257–1266. [Google Scholar] [CrossRef]
- Xiang, Q.-Q.; Wang, D.; Zhang, J.-L.; Ding, C.-Z.; Luo, X.; Tao, J.; Ling, J.; Shea, D.; Chen, L.-Q. Effect of silver nanoparticles on gill membranes of common carp: Modification of fatty acid profile, lipid peroxidation and membrane fluidity. Environ. Pollut. 2020, 256, 113504. [Google Scholar] [CrossRef] [PubMed]
- Paciorek, P.; Żuberek, M.; Grzelak, A. Products of lipid peroxidation as a factor in the toxic effect of silver nanoparticles. Matererials 2020, 13, 2460. [Google Scholar] [CrossRef] [PubMed]
- Fahrenholtz, C.D.; Swanner, J.; Ramirez-Perez, M.; Singh, R.N. Heterogeneous Responses of ovarian cancer cells to silver nanoparticles as a single agent and in combination with cisplatin. J. Nanomater. 2017, 2017, 5107485. [Google Scholar] [CrossRef]
- Guo, D.; Zhu, L.; Huang, Z.; Zhou, H.; Ge, Y.; Ma, W.; Wu, J.; Zhang, X.; Zhou, X.; Zhang, Y.; et al. Anti-leukemia activity of PVP-coated silver nanoparticles via generation of reactive oxygen species and release of silver ions. Biomaterials 2013, 34, 7884–7894. [Google Scholar] [CrossRef] [PubMed]
- Fougner, C.; Bergholtz, H.; Norum, J.H.; Sørlie, T. Re-Definition of claudin-low as a breast cancer phenotype. Nat. Commun. 2020, 11, 1787. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, K.; Saito, Y.; Sakiyama, J.; Ohuchi, Y.; Oseto, F.; Noguchi, N. A novel fluorescent probe with high sensitivity and selective detection of lipid hydroperoxides in cells. R. Soc. Chem. Adv. 2012, 2, 7894–7900. [Google Scholar] [CrossRef]
- Hackett, M.; E Smith, S.; Caine, S.; Nichol, H.; George, G.; Pickering, I.; Paterson, P.G. Novel bio-spectroscopic imaging reveals disturbed protein homeostasis and thiol redox with protein aggregation prior to hippocampal CA1 pyramidal neuron death induced by global brain ischemia in the rat. Free. Radic. Biol. Med. 2015, 89, 806–818. [Google Scholar] [CrossRef] [Green Version]
- Castro, J.P.; Jung, T.; Grune, T.; Siems, W. 4-Hydroxynonenal (HNE) modified proteins in metabolic diseases. Free. Radic. Biol. Med. 2017, 111, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.; Carroll, K.S. Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 847–875. [Google Scholar] [CrossRef] [Green Version]
- Holmila, R.J.; Vance, S.A.; Chen, X.; Wu, H.; Shukla, K.; Bharadwaj, M.S.; Mims, J.; Wary, Z.; Marrs, G.; Singh, R.; et al. Mitochondria-targeted probes for imaging protein sulfenylation. Sci. Rep. 2018, 8, 6635. [Google Scholar] [CrossRef] [Green Version]
- McConkey, D.J. The integrated stress response and proteotoxicity in cancer therapy. Biochem. Biophys. Res. Commun. 2017, 482, 450–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darling, N.J.; Cook, S.J. The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 2150–2163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnedos, M.; Vicier, C.; Loi, S.; Lefebvre, C.; Michiels, S.; Bonnefoi, H.; Andre, F. Precision medicine for metastatic breast cancer—Limitations and solutions. Nat. Rev. Clin. Oncol. 2015, 12, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-C. Triple-Negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Navarro, E.; Piccapietra, F.; Wagner, B.; Marconi, F.; Kaegi, R.; Odzak, N.; Sigg, L.; Behra, R. Toxicity of silver nanoparticles to Chlamydomonas reinhardtii. Environ. Sci. Technol. 2008, 42, 8959–8964. [Google Scholar] [CrossRef] [PubMed]
- Beer, C.; Foldbjerg, R.; Hayashi, Y.; Sutherland, D.; Autrup, H. Toxicity of silver nanoparticles—Nanoparticle or silver ion? Toxicol. Lett. 2012, 208, 286–292. [Google Scholar] [CrossRef]
- Kawata, K.; Osawa, M.; Okabe, S. In vitro toxicity of silver nanoparticles at noncytotoxic doses to HepG2 human hepatoma cells. Environ. Sci. Technol. 2009, 43, 6046–6051. [Google Scholar] [CrossRef]
- Smith, J.N.; Thomas, D.G.; Jolley, H.; Kodali, V.K.; Littke, M.H.; Munusamy, P.; Baer, D.R.; Gaffrey, M.J.; Thrall, B.D.; Teeguarden, J.G. All that is silver is not toxic: Silver ion and particle kinetics reveals the role of silver ion aging and dosimetry on the toxicity of silver nanoparticles. Part. Fibre Toxicol. 2018, 15, 47. [Google Scholar] [CrossRef]
- Casciano, J.C.; Perry, C.; Cohen-Nowak, A.J.; Miller, K.D.; Voorde, J.V.; Zhang, Q.; Chalmers, S.; Sandison, M.E.; Liu, Q.; Hedley, A.; et al. MYC regulates fatty acid metabolism through a multigenic program in claudin-low triple negative breast cancer. Br. J. Cancer 2020, 122, 868–884. [Google Scholar] [CrossRef]
- Angeli, J.P.F.; Krysko, D.V.; Conrad, M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer 2019, 19, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Ding, W.; Ji, X.; Ao, X.; Liu, Y.; Yu, W.; Wang, J. Molecular mechanisms of ferroptosis and its role in cancer therapy. J. Cell. Mol. Med. 2019, 23, 4900–4912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Ding, C.-K.; Wu, J.; Sjol, J.; Wardell, S.; Spasojevic, I.; George, D.; McDonnell, D.P.; Hsu, D.S.; Chang, J.T.; et al. Cystine addiction of triple-negative breast cancer associated with EMT augmented death signaling. Oncogene 2017, 36, 4235–4242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timmerman, L.A.; Holton, T.; Yuneva, M.; Louie, R.J.; Padro, M.; Daemen, A.; Hu, M.; Chan, D.A.; Ethier, S.P.; van ’t Veer, L.J.; et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 2013, 24, 450–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [Green Version]
- Gout, P.W.; Buckley, A.R.; Simms, C.R.; Bruchovsky, N. Sulfasalazine, a potent suppressor of lymphoma growth by inhibition of the xc− cystine transporter: A new action for an old drug. Leukemia 2001, 15, 1633–1640. [Google Scholar] [CrossRef] [Green Version]
- Ooko, E.; Saeed, M.E.; Kadioglu, O.; Sarvi, S.; Colak, M.; Elmasaoudi, K.; Janah, R.; Greten, J.; Efferth, T. Artemisinin derivatives induce iron-dependent cell death (ferroptosis) in tumor cells. Phytomedicine 2015, 22, 1045–1054. [Google Scholar] [CrossRef]
- Kumari, K.; Keshari, S.; Sengupta, D.; Sabat, S.C.; Mishra, S.K. Transcriptome analysis of genes associated with breast cancer cell motility in response to Artemisinin treatment. BMC Cancer 2017, 17, 858. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Yang, C.; Jian, L.; Guo, S.; Chen, R.; Li, K.; Qu, F.; Tao, K.; Fu, Y.; Luo, F.; et al. Sulfasalazine-induced ferroptosis in breast cancer cells is reduced by the inhibitory effect of estrogen receptor on the transferrin receptor. Oncol. Rep. 2019, 42, 826–838. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, H.; Daniels, J.D.; Zandkarimi, F.; Liu, H.; Brown, L.M.; Uchida, K.; O’Connor, O.A.; Stockwell, B.R. Imidazole Ketone Erastin Induces Ferroptosis and Slows Tumor Growth in a Mouse Lymphoma Model. Cell Chem. Biol. 2019, 26, 623–633.e9. [Google Scholar] [CrossRef]
- Hetz, C.; Papa, F.R. The Unfolded protein response and cell fate control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Kasetti, R.B.; Maddineni, P.; Millar, J.C.; Clark, A.F.; Zode, G.S. Increased synthesis and deposition of extracellular matrix proteins leads to endoplasmic reticulum stress in the trabecular meshwork. Sci. Rep. 2017, 7, 14591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, J. The proteasome: A suitable antineoplastic target. Nat. Rev. Cancer 2004, 4, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Wathieu, H.; Issa, N.T.; Fernandez, A.; Mohandoss, M.; Tiek, D.M.; Franke, J.L.; Byers, S.W.; Riggins, R.B.; Dakshanamurthy, S. Differential prioritization of therapies to subtypes of triple negative breast cancer using a systems medicine method. Oncotarget 2017, 8, 92926–92942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matossian, M.D.; Burks, H.E.; Elliott, S.; Hoang, V.T.; Bowles, A.C.; Sabol, R.A.; Wahba, B.; Anbalagan, M.; Rowan, B.; Abazeed, M.E.; et al. Drug resistance profiling of a new triple negative breast cancer patient-derived xenograft model. BMC Cancer 2019, 19, 205. [Google Scholar] [CrossRef] [PubMed]
- Tate, C.R.; Rhodes, L.V.; Segar, H.C.; Driver, J.L.; Pounder, F.N.; Burow, M.E.; Collins-Burow, B.M. Targeting triple-negative breast cancer cells with the histone deacetylase inhibitor panobinostat. Breast Cancer Res. 2012, 14, R79. [Google Scholar] [CrossRef] [Green Version]
- Terranova-Barberio, M.; Thomas, S.; Ali, N.; Pawlowska, N.; Park, J.; Krings, G.; Rosenblum, M.D.; Budillon, A.; Munster, P.N. HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget 2017, 8, 114156–114172. [Google Scholar] [CrossRef] [Green Version]
- Pongas, G.; Fojo, T. BEZ235: When promising science meets clinical reality. Oncolologist 2016, 21, 1033–1034. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Song, L.; Hu, X.; Liu, C.; Shi, J.; Wang, H.; Zhan, L.; Song, H. Inhibition of epithelial-mesenchymal transition and tissue regeneration by waterborne titanium dioxide nanoparticles. ACS Appl. Mater. Interfaces 2018, 10, 3449–3458. [Google Scholar] [CrossRef]
- Huang, K.-J.; Wei, Y.-H.; Chiu, Y.-C.; Wu, S.-R.; Shieh, D.-B. Assessment of zero-valent iron-based nanotherapeutics for ferroptosis induction and resensitization strategy in cancer cells. Biomater. Sci. 2019, 7, 1311–1322. [Google Scholar] [CrossRef]
- Wen, J.; Chen, H.; Ren, Z.; Zhang, P.; Chen, J.; Jiang, S. Ultrasmall iron oxide nanoparticles induced ferroptosis via Beclin1/ATG5-dependent autophagy pathway. Nano Converg. 2021, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.K.; Kumar, A.; Pandey, A.K.; Singh, S.S.; Dhawan, A. Titanium dioxide nanoparticles induce oxidative stress-mediated apoptosis in human keratinocyte cells. J. Biomed. Nanotechnol. 2011, 7, 100–101. [Google Scholar] [CrossRef]
- Kim, S.E.; Zhang, L.; Ma, K.; Riegman, M.; Chen, F.; Ingold, I.; Conrad, M.; Turker, M.Z.; Gao, M.; Jiang, X.; et al. Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat. Nanotechnol. 2016, 11, 977–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, G.; Gliga, A.; Hedberg, J.; Serra, A.; Greco, D.; Odnevall Wallinder, I.; Fadeel, B. Cobalt nanoparticles trigger ferrop-tosis-like cell death (oxytosis) in neuronal cells: Potential implications for neurodegenerative disease. FASEB J. 2020, 34, 5262–5281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Li, F.; Qiao, R.; Hu, X.; Liao, H.; Chen, L.; Wu, J.; Wu, H.; Zhao, M.; Liu, J.; et al. Arginine-Rich manganese silicate nanobubbles as a ferroptosis-inducing agent for tumor-targeted theranostics. ACS Nano 2018, 12, 12380–12392. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [Green Version]
- Chavez, K.J.; Garimella, S.V.; Lipkowitz, S. Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010, 32, 35–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Cell Line | Media Formulation |
|---|---|
| BT-20 | EMEM supplemented with penicillin (250 units/mL), streptomycin (250 μg/mL), 2 mM L-glutamine, and 10% fetal bovine serum |
| BT-549 | RPMI supplemented with penicillin (250 units/mL), streptomycin (250 μg/mL), and 10% fetal bovine serum |
| HCC70 | RPMI supplemented with penicillin (250 units/mL), streptomycin (250 μg/mL), and 10% fetal bovine serum |
| iMEC | DMEM/F12 supplemented with 10 µg/mL insulin, 20 ng/mL human epidermal growth factor (hEGF), and 0.5 μg/mL hydrocortisone, and 10% fetal bovine serum |
| MCF-10A | DMEM/F12 supplemented with penicillin (250 units/mL), streptomycin (250 μg/mL), 2 mM L-glutamine, 10 μg/mL insulin, 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, and 100 ng/mL cholera toxin, and 5% heat-inactivated horse serum |
| MDA- MB-231 | DMEM/F12 supplemented with penicillin (250 units/mL), streptomycin (250 μg/mL), 2 mM L-glutamine, and 10% fetal bovine serum |
| MDA- MB-436 | DMEM supplemented with penicillin (250 units/mL), streptomycin (250 μg/mL), 2 mM L-glutamine, and 10% fetal bovine serum |
| MDA- MB-468 | DMEM supplemented with penicillin (250 units/mL), streptomycin (250 μg/mL), 2 mM L-glutamine, and 10% fetal bovine serum |
| SUM-159 | HAM’s F12 supplemented with penicillin (250 units/mL), streptomycin (250 μg/mL), 2mM L-glutamine, 5 μg/mL insulin, 1 μg/mL hydrocortisone, 10 μM HEPES, and 5% fetal bovine serum |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Snyder, C.M.; Rohde, M.M.; Fahrenholtz, C.D.; Swanner, J.; Sloop, J.; Donati, G.L.; Furdui, C.M.; Singh, R. Low Doses of Silver Nanoparticles Selectively Induce Lipid Peroxidation and Proteotoxic Stress in Mesenchymal Subtypes of Triple-Negative Breast Cancer. Cancers 2021, 13, 4217. https://doi.org/10.3390/cancers13164217
Snyder CM, Rohde MM, Fahrenholtz CD, Swanner J, Sloop J, Donati GL, Furdui CM, Singh R. Low Doses of Silver Nanoparticles Selectively Induce Lipid Peroxidation and Proteotoxic Stress in Mesenchymal Subtypes of Triple-Negative Breast Cancer. Cancers. 2021; 13(16):4217. https://doi.org/10.3390/cancers13164217
Chicago/Turabian StyleSnyder, Christina M., Monica M. Rohde, Cale D. Fahrenholtz, Jessica Swanner, John Sloop, George L. Donati, Cristina M. Furdui, and Ravi Singh. 2021. "Low Doses of Silver Nanoparticles Selectively Induce Lipid Peroxidation and Proteotoxic Stress in Mesenchymal Subtypes of Triple-Negative Breast Cancer" Cancers 13, no. 16: 4217. https://doi.org/10.3390/cancers13164217
APA StyleSnyder, C. M., Rohde, M. M., Fahrenholtz, C. D., Swanner, J., Sloop, J., Donati, G. L., Furdui, C. M., & Singh, R. (2021). Low Doses of Silver Nanoparticles Selectively Induce Lipid Peroxidation and Proteotoxic Stress in Mesenchymal Subtypes of Triple-Negative Breast Cancer. Cancers, 13(16), 4217. https://doi.org/10.3390/cancers13164217








