A Novel Orthotopic Liver Cancer Model for Creating a Human-like Tumor Microenvironment
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Establishment of Stable Luciferase-Expressing Huh7 Cells (Huh7-Luci)
2.3. Generation of the HCC Organoid
2.4. Generation of the Pancreatic Cancer Organoid
2.5. Animal
2.6. HCC Organoid Implantation
2.7. Pancreatic Cancer Organoid Implantation
2.8. Induction of Liver Fibrosis in Mice
2.9. Induction of Non-Alcoholic Fatty Liver Disease (NAFLD) in Mice
2.10. Oil Red O (ORO) Staining
2.11. Hematoxylin and Eosin (HE) Staining
2.12. Immunofluorescence Staining
2.13. Sirius Red Staining
2.14. Extraction of the Total Lipid Fraction from Liver
2.15. Human Albumin (ALB) Concentration in Culture Medium
2.16. Luciferase Assay In Vitro
2.17. In Vivo Bioluminescence Imaging (BLI)
2.18. Statistical Analysis
3. Results
3.1. Generation of HCC Organoid from Huh7-Luci Co-Cultured with EC plus MC Derived from Human iPSC
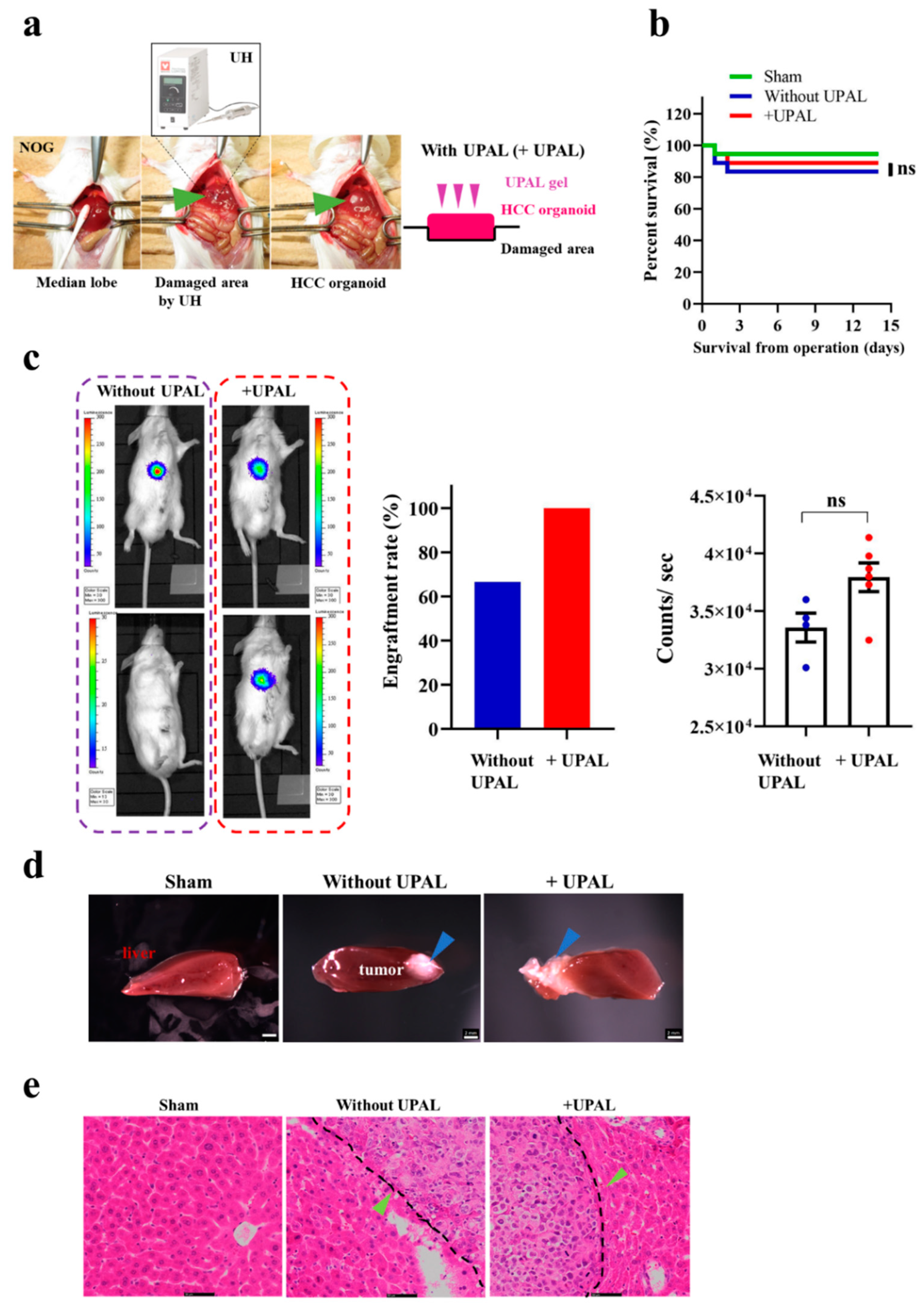
3.2. Implantation of HCC Organoid Onto the Mouse Liver Surface
3.3. Generation of Liver Cancer Mouse Model after Different Liver Cancer Organoid Implantation In Vivo
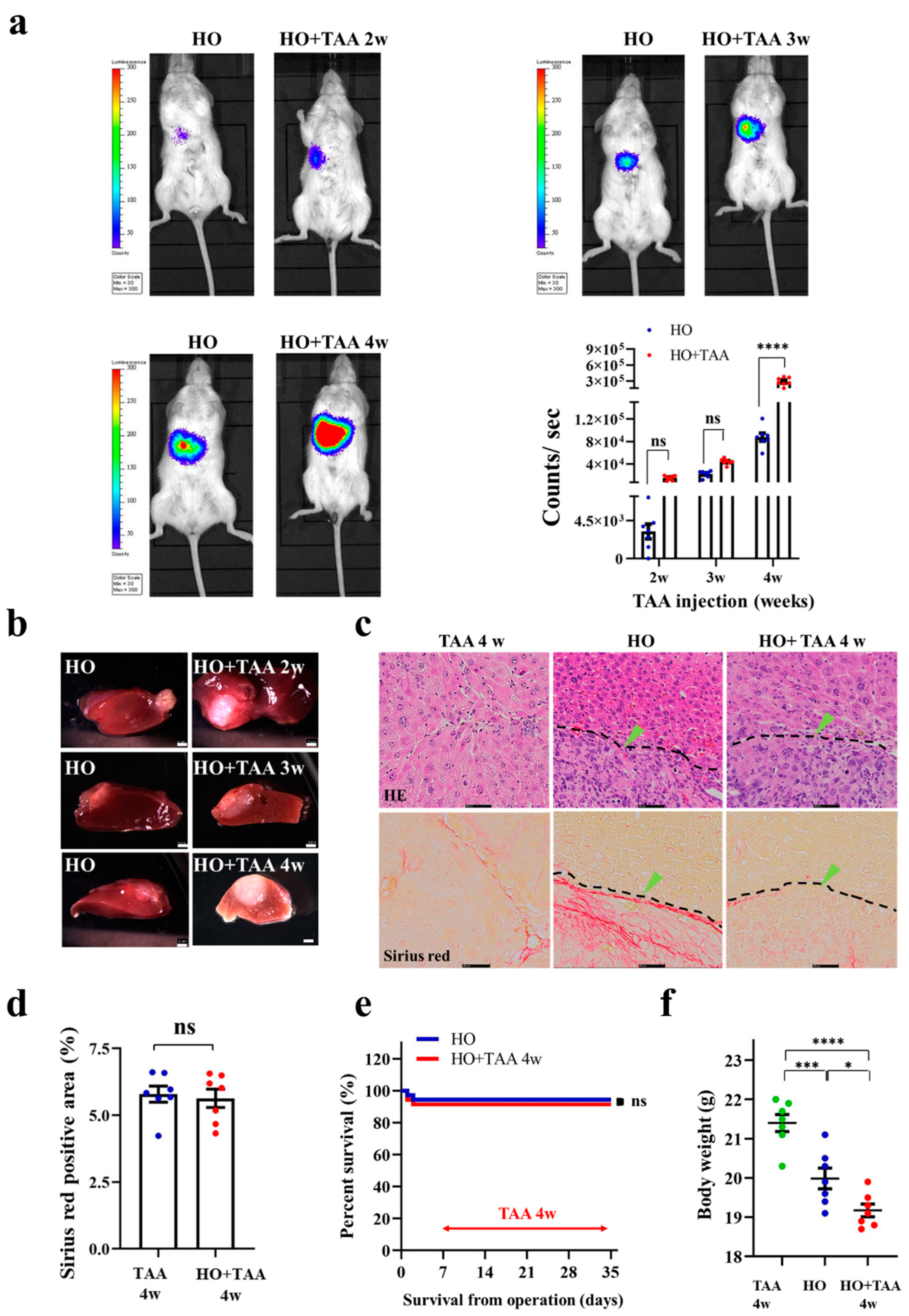
3.4. Implantation of HCC Organoid to the Liver with Fibrosis TME
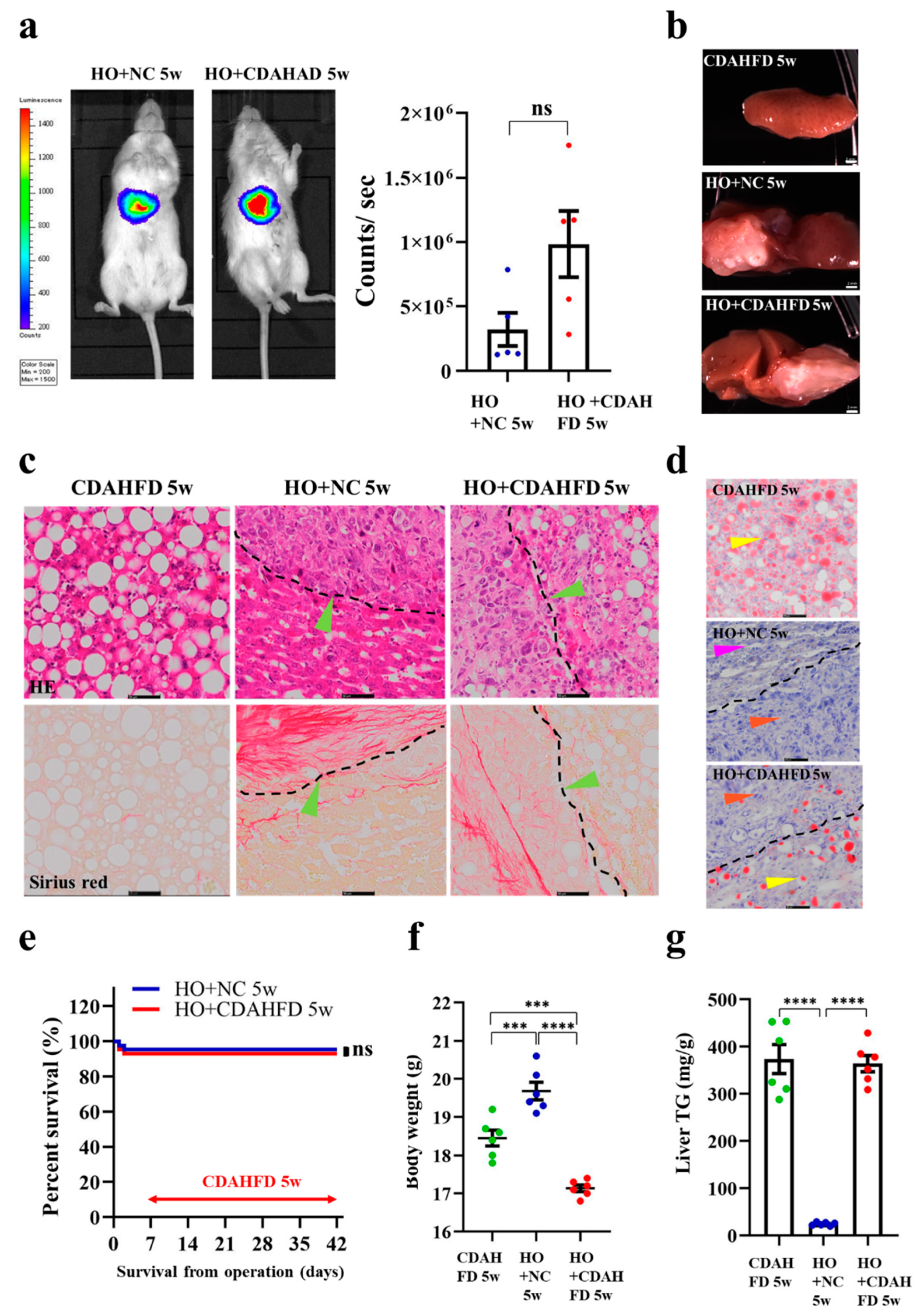
3.5. Implantation of HCC Organoid to the Liver with NAFLD TME
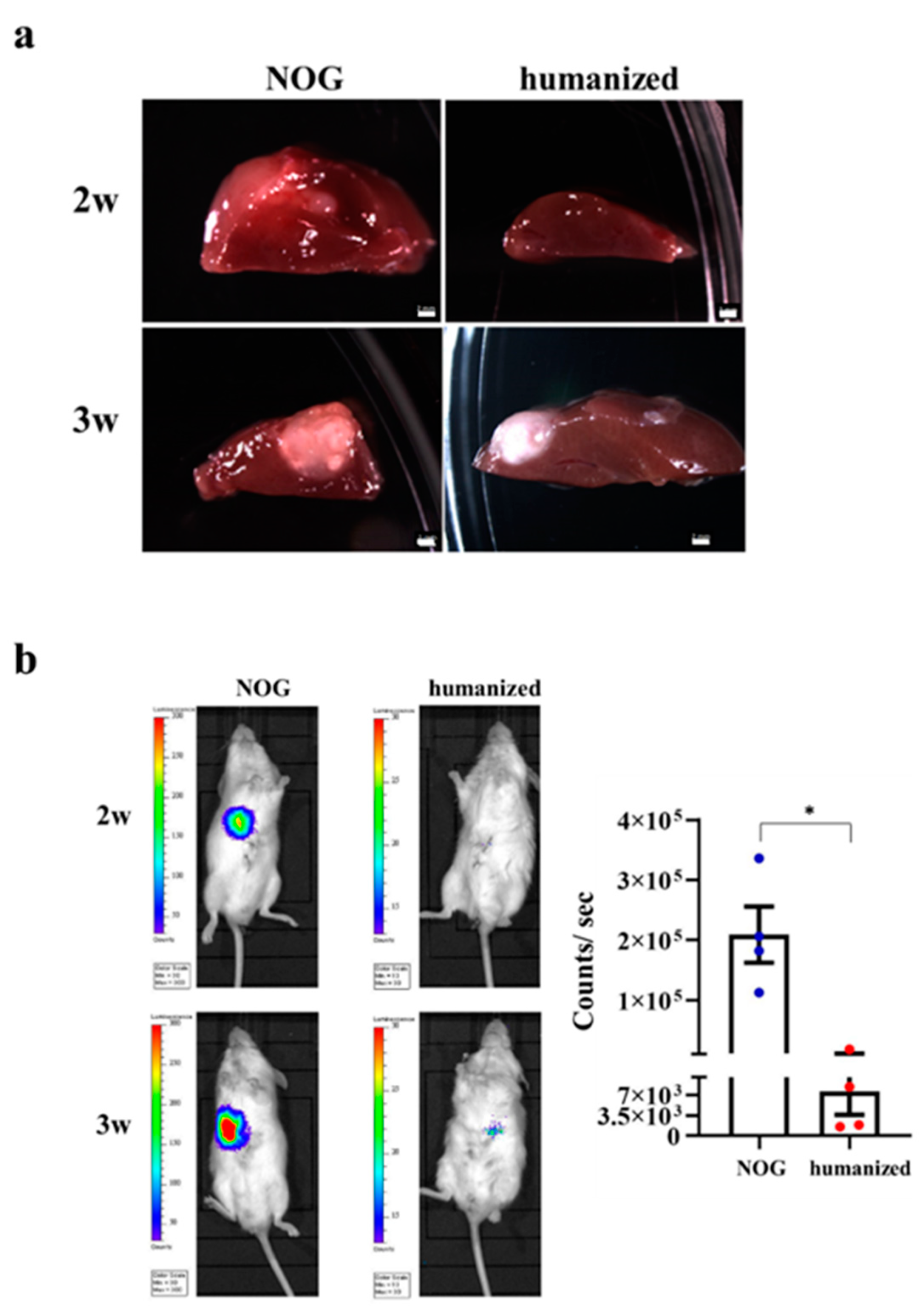
3.6. Implantation of HO in the Humanized Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Fitzmaurice, C.; Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level results from the global burden of disease study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar] [CrossRef]
- Wang, F.S.; Fan, J.G.; Zhang, Z.; Gao, B.; Wang, H.Y. The global burden of liver disease: The major impact of China. Hepatology 2014, 60, 2099–2108. [Google Scholar] [CrossRef]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Kim, W.R.; Coelho, R.; Mettler, T.A.; Benson, J.T.; Sanderson, S.O.; Therneau, T.M.; Kim, B.; Roberts, L.R. Cirrhosis is present in most patients with hepatitis B and hepatocellular carcinoma. Clin. Gastroenterol. Hepatol. 2011, 9, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.-F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.-L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Anson, M.; Crain-Denoyelle, A.M.; Baud, V.; Chereau, F.; Gougelet, A.; Terris, B.; Yamagoe, S.; Colnot, S.; Viguier, M.; Perret, C.; et al. Oncogenic β-catenin triggers an inflammatory response that determines the aggressiveness of hepatocellular carcinoma in mice. J. Clin. Investig. 2012, 122, 586–599. [Google Scholar] [CrossRef]
- Thorgeirsson, S.S.; Santoni-Rugiu, E. Transgenic mouse models in carcinogenesis: Interaction of c-myc with transforming growth factor a and hepatocyte growth factor in hepatocarcinogenesis. Br. J. Clin. Pharmacol. 1996, 42, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Coulouarn, C.; Factor, V.M.; Conner, E.A.; Thorgeirsson, S.S. Genomic modeling of tumor onset and progression in a mouse model of aggressive human liver cancer. Carcinogenesis 2011, 32, 1434–1440. [Google Scholar] [CrossRef]
- Uehara, T.; Pogribny, I.P.; Rusyn, I. The DEN and CCl4-induced mouse model of fibrosis and inflammation-associated hepatocellular carcinoma. Curr. Protoc. Pharmacol. 2014, 2014, 14.30.1–14.30.10. [Google Scholar] [CrossRef]
- Kung, A.L. Practices and pitfalls of mouse cancer models in drug discovery. Adv. Cancer Res. 2006, 96, 191–212. [Google Scholar] [CrossRef]
- Shiga, K.; Hara, M.; Nagasaki, T.; Sato, T.; Takahashi, H.; Takeyama, H. Cancer-Associated fibroblasts: Their characteristics and their roles in tumor growth. Cancers (Basel) 2015, 7, 2443–2458. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Bae, J.-S. Tumor-Associated macrophages and neutrophils in tumor microenvironment. Mediat. Inflamm. 2016, 2016, 6058147. [Google Scholar] [CrossRef] [PubMed]
- Baluk, P.; Hashizume, H.; McDonald, D.M. Cellular abnormalities of blood vessels as targets in cancer. Curr. Opin. Genet. Dev. 2005, 15, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; Yang, Y.; Hass, R. Interaction of MSC with tumor cells. Cell Commun. Signal. 2016, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Mbeunkui, F.; Johann, D.J.J. Cancer and the tumor microenvironment: A review of an essential relationship. Cancer Chemother. Pharmacol. 2009, 63, 571–582. [Google Scholar] [CrossRef]
- Nii, T.; Makino, K.; Tabata, Y. Three-Dimensional culture system of cancer cells combined with biomaterials for drug screening. Cancers (Basel) 2020, 12, 2754. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Y.; Xu, X.-X.; Guo, X.; Sun, G.-W.; Ma, X.-J. Mesenchymal stem cells enhance the metastasis of 3D-cultured hepatocellular carcinoma cells. BMC Cancer 2016, 16, 566. [Google Scholar] [CrossRef]
- Miyazaki, K.; Oyanagi, J.; Hoshino, D.; Togo, S.; Kumagai, H.; Miyagi, Y. Cancer cell migration on elongate protrusions of fibroblasts in collagen matrix. Sci. Rep. 2019, 9, 292. [Google Scholar] [CrossRef]
- Han, H.-W.; Hsu, S.-H. Chitosan-hyaluronan based 3D co-culture platform for studying the crosstalk of lung cancer cells and mesenchymal stem cells. Acta Biomater. 2016, 42, 157–167. [Google Scholar] [CrossRef]
- Sasser, A.K.; Mundy, B.L.; Smith, K.M.; Studebaker, A.W.; Axel, A.E.; Haidet, A.M.; Fernandez, S.A.; Hall, B.M. Human bone marrow stromal cells enhance breast cancer cell growth rates in a cell line-dependent manner when evaluated in 3D tumor environments. Cancer Lett. 2007, 254, 255–264. [Google Scholar] [CrossRef]
- Burdett, E.; Kasper, F.K.; Mikos, A.G.; Ludwig, J.A. Engineering tumors: A tissue engineering perspective in cancer biology. Tissue Eng. Part B Rev. 2010, 16, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Schwartz, M.; Mazzaferro, V. Resection and liver transplantation for hepatocellular carcinoma. Semin. Liver Dis. 2005, 25, 181–200. [Google Scholar] [CrossRef]
- Nii, T.; Kuwahara, T.; Makino, K.; Tabata, Y. A Co-Culture System of Three-Dimensional Tumor-Associated Macrophages and Three-Dimensional Cancer-Associated Fibroblasts Combined with Biomolecule Release for Cancer Cell Migration. Tissue Eng. Part A 2020, 26, 1272–1282. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef]
- Zechmann, C.M.; Woenne, E.C.; Brix, G.; Radzwill, N.; Ilg, M.; Bachert, P.; Peschke, P.; Kirsch, S.; Kauczor, H.-U.; Delorme, S.; et al. Impact of stroma on the growth, microcirculation, and metabolism of experimental prostate tumors. Neoplasia 2007, 9, 57–67. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Céspedes, M.V.; Espina, C.; García-Cabezas, M.A.; Trias, M.; Boluda, A.; Gómez del Pulgar, M.T.; Sancho, F.J.; Nistal, M.; Lacal, J.C.; Mangues, R. Orthotopic microinjection of human colon cancer cells in nude mice induces tumor foci in all clinically relevant metastatic sites. Am. J. Pathol. 2007, 170, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, A.; Suijkerbuijk, S.J.E.; Begthel, H.; Beerling, E.; Oost, K.C.; Snippert, H.J.; van Rheenen, J.; Drost, J. A surgical orthotopic organoid transplantation approach in mice to visualize an study colorectal cancer progression. Nat. Protoc. 2018, 13, 235–247. [Google Scholar] [CrossRef]
- Takebe, T.; Sekine, K.; Kimura, M.; Yoshizawa, E.; Ayano, S.; Koido, M.; Funayama, S.; Nakanishi, N.; Hisai, T.; Kobayashi, T.; et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 2017, 21, 2661–2670. [Google Scholar] [CrossRef] [PubMed]
- Takebe, T.; Sekine, K.; Enomura, M.; Koike, H.; Kimura, M.; Ogaeri, T.; Zhang, R.R.; Ueno, Y.; Zheng, Y.W.; Koike, N.; et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013, 499, 481–484. [Google Scholar] [CrossRef]
- Camp, J.G.; Sekine, K.; Gerber, T.; Loeffler-Wirth, H.; Binder, H.; Gac, M.; Kanton, S.; Kageyama, J.; Damm, G.; Seehofer, D.; et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017, 546, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.; Murata, S.; Oshiro, K.; Hatada, Y.; Taniguchi, H. Transplantation of fetal liver tissue coated by ultra-purified alginate gel over liver improves hepatic function in the cirrhosis rat model. Sci. Rep. 2020, 10, 4–6. [Google Scholar] [CrossRef]
- DePolo, N.J.; Reed, J.D.; Sheridan, P.L.; Townsend, K.; Sauter, S.L.; Jolly, D.J.; Dubensky, T.W.J. VSV-G pseudotyped lentiviral vector particles produced in human cells are inactivated by human serum. Mol. Ther. 2000, 2, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Konermann, S.; Gootenberg, J.S.; Abudayyeh, O.O.; Platt, R.J.; Brigham, M.D.; Sanjana, N.E.; Zhang, F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 2017, 12, 828–863. [Google Scholar] [CrossRef] [PubMed]
- Takebe, T.; Zhang, R.-R.; Koike, H.; Kimura, M.; Yoshizawa, E.; Enomura, M.; Koike, N.; Sekine, K.; Taniguchi, H. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat. Protoc. 2014, 9, 396–409. [Google Scholar] [CrossRef]
- Mori, A.; Murata, S.; Tashiro, N.; Tadokoro, T.; Okamoto, S.; Otsuka, R.; Wada, H.; Murata, T.; Takahashi, T.; Seino, K.-I.; et al. Establishment of human leukocyte antigen-mismatched immune responses after transplantation of human liver bud in humanized mouse models. Cells 2021, 10, 476. [Google Scholar] [CrossRef]
- Kahlert, C.; Kalluri, R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 2013, 91, 431–437. [Google Scholar] [CrossRef]
- Gupta, M.K.; Qin, R.Y. Mechanism and its regulation of tumor-induced angiogenesis. World J. Gastroenterol. 2003, 9, 1144–1155. [Google Scholar] [CrossRef]
- Tonini, T.; Rossi, F.; Claudio, P.P. Molecular basis of angiogenesis and cancer. Oncogene 2003, 22, 6549–6556. [Google Scholar] [CrossRef]
- Carmeliet, P. VEGF as a key mediator of angiogenesis in cancer. Oncology 2005, 69, 4–10. [Google Scholar] [CrossRef]
- He, L.; Tian, D.A.; Li, P.Y.; He, X.X. Mouse models of liver cancer: Progress and recommendations. Oncotarget 2015, 6, 23306–23322. [Google Scholar] [CrossRef]
- Williams, J.W.; Hodgson, W.J. Histologic evaluation of tissues sectioned by ultrasonically powered instruments (a preliminary report). Mt. Sinai J. Med. 1979, 46, 105–106. [Google Scholar]
- Takayama, T.; Makuuchi, M.; Kubota, K.; Harihara, Y.; Hui, A.M.; Sano, K.; Ijichi, M.; Hasegawa, K. Randomized comparison of ultrasonic vs clamp transection of the liver. Arch. Surg. 2001, 136, 922–928. [Google Scholar] [CrossRef]
- Lesurtel, M.; Selzner, M.; Petrowsky, H.; McCormack, L.; Clavien, P.A.; Bismuth, B.; Heberer, H. How should transection of the liver be performed? A prospective randomized study in 100 consecutive patients: Comparing four different transection strategies. Ann. Surg. 2005, 242, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Bodzin, A.S.; Leiby, B.E.; Ramirez, C.G.; Frank, A.M.; Doria, C. Liver resection using cavitron ultrasonic surgical aspirator (CUSA) versus harmonic scalpel: A retrospective cohort study. Int. J. Surg. 2014, 12, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Ura, K.; Yamada, K.; Tsujimoto, T.; Ukeba, D.; Iwasaki, N.; Sudo, H. Ultra-purified alginate gel implantation decreases inflammatory cytokine levels, prevents intervertebral disc degeneration, and reduces acute pain after discectomy. Sci. Rep. 2021, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.; Wang, T.C. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell 2009, 15, 79–80. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.; Karin, M. Autocrine IL-6 signaling: A key event in tumorigenesis? Cancer Cell 2008, 13, 7–9. [Google Scholar] [CrossRef]
- Mauer, J.; Denson, J.L.; Brüning, J.C. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 2015, 36, 92–101. [Google Scholar] [CrossRef]
- White, J.P.; Puppa, M.J.; Gao, S.; Sato, S.; Welle, S.L.; Carson, J.A. Muscle mTORC1 suppression by IL-6 during cancer cachexia: A role for AMPK. Am. J. Physiol. Endocrinol. Metab. 2013, 304, 1042–1052. [Google Scholar] [CrossRef]
- Lin, M.T.; Juan, C.Y.; Chang, K.J.; Chen, W.J.; Kuo, M.L. IL-6 inhibits apoptosis and retains oxidative DNA lesions in human gastric cancer AGS cells through up-regulation of anti-apoptotic gene mcl-1. Carcinogenesis 2001, 22, 1947–1953. [Google Scholar] [CrossRef]
- Jourdan, M.; Veyrune, J.L.; De Vos, J.; Redal, N.; Couderc, G.; Klein, B. A major role for Mcl-1 antiapoptotic protein in the IL-6-induced survival of human myeloma cells. Oncogene 2003, 22, 2950–2959. [Google Scholar] [CrossRef]
- Chang, C.H.; Hsiao, C.F.; Yeh, Y.M.; Chang, G.C.; Tsai, Y.H.; Chen, Y.M.; Huang, M.S.; Chen, H.L.; Li, Y.J.; Yang, P.C.; et al. Circulating interleukin-6 level is a prognostic marker for survival in advanced nonsmall cell lung cancer patients treated with chemotherapy. Int. J. Cancer 2013, 132, 1977–1985. [Google Scholar] [CrossRef]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Yeoh, G.C.T.; Ernst, M.; Rose-John, S.; Akhurst, B.; Payne, C.; Long, S.; Alexander, W.; Croker, B.; Grail, D.; Matthews, V.B. Opposing roles of gp130-mediated STAT-3 and ERK-1/2 signaling in liver progenitor cell migration and proliferation. Hepatology 2007, 45, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Wegiel, B.; Bjartell, A.; Culig, Z.; Persson, J.L. Interleukin-6 activates PI3K/Akt pathway and regulates cyclin A1 to promote prostate cancer cell survival. Int. J. Cancer 2008, 122, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.H.; Kuo, M.L.; Chen, C.A.; Chou, C.H.; Lai, K.B.; Lee, C.N.; Hsieh, C.Y. Interleukin-6 promotes cervical tumor growth by VEGF-dependent angiogenesis via a STAT3 pathway. Oncogene 2003, 22, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Lee, O.Y.; Shon, S.Y.; Nam, O.; Ryu, P.M.; Seo, M.W.; Lee, D.S. A mutual activation loop between breast cancer cells and myeloid-derived suppressor cells facilitates spontaneous metastasis through IL-6 trans-signaling in a murine model. Breast Cancer Res. 2013, 15, R79. [Google Scholar] [CrossRef]
- Zou, M.; Zhang, X.; Xu, C. IL6-induced metastasis modulators p-STAT3, MMP-2 and MMP-9 are targets of 3,3′-diindolylmethane in ovarian cancer cells. Cell. Oncol. 2016, 39, 47–57. [Google Scholar] [CrossRef]
- Katayama, Y.; Uchino, J.; Chihara, Y.; Tamiya, N.; Kaneko, Y.; Yamada, T.; Takayama, K. Tumor neovascularization and developments in therapeutics. Cancers (Basel) 2019, 11, 316. [Google Scholar] [CrossRef] [PubMed]
- Folkman, J.; Klagsbrun, M. Angiogenic factors. Science 1987, 235, 442–447. [Google Scholar] [CrossRef]
- Folkman, J. Angiogenesis: An organizing principle for drug discovery? Nat. Rev. Drug Discov. 2007, 6, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Rawal, P.; Siddiqui, H.; Hassan, M.; Choudhary, M.C.; Tripathi, D.M.; Nain, V.; Trehanpati, N.; Kaur, S. Endothelial cell-derived TGF-β promotes epithelial-mesenchymal transition via CD133 in HBx-infected hepatoma cells. Front. Oncol. 2019, 9, 308. [Google Scholar] [CrossRef]
- Fattovich, G.; Stroffolini, T.; Zagni, I.; Donato, F. Hepatocellular carcinoma in cirrhosis: Incidence and risk factors. Gastroenterology 2004, 127, 35–50. [Google Scholar] [CrossRef]
- O’Rourke, J.M.; Sagar, V.M.; Shah, T.; Shetty, S. Carcinogenesis on the background of liver fibrosis: Implications for the management of hepatocellular cancer. World J. Gastroenterol. 2018, 24, 4436–4447. [Google Scholar] [CrossRef] [PubMed]
- Baffy, G.; Brunt, E.M.; Caldwell, S.H. Hepatocellular carcinoma in non-alcoholic fatty liver disease: An emerging menace. J. Hepatol. 2012, 56, 1384–1391. [Google Scholar] [CrossRef]
- Kim, M.N.; Kim, S.U.; Kim, B.K.; Park, J.Y.; Kim, D.Y.; Ahn, S.H.; Song, K.J.; Park, Y.N.; Han, K.H. Increased risk of hepatocellular carcinoma in chronic hepatitis B patients with transient elastography-defined subclinical cirrhosis. Hepatology 2015, 61, 1851–1859. [Google Scholar] [CrossRef] [PubMed]
- Akima, T.; Tamano, M.; Hiraishi, H. Liver stiffness measured by transient elastography is a predictor of hepatocellular carcinoma development in viral hepatitis. Hepatol. Res. 2011, 41, 965–970. [Google Scholar] [CrossRef]
- Farazi, P.A.; DePinho, R.A. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat. Rev. Cancer 2006, 6, 674–687. [Google Scholar] [CrossRef]
- Maeda, S.; Kamata, H.; Luo, J.L.; Leffert, H.; Karin, M. IKKβ couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 2005, 121, 977–990. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Z.; Ye, Y.; Xie, L.; Li, W. Oxidative stress and liver cancer: Etiology and therapeutic targets. Oxid. Med. Cell. Longev. 2016, 2016. [Google Scholar] [CrossRef]
- De La Rochere, P.; Guil-Luna, S.; Decaudin, D.; Azar, G.; Sidhu, S.S.; Piaggio, E. Humanized mice for the study of immuno-oncology. Trends Immunol. 2018, 39, 748–763. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.J.; Bird, G.; Refaeli, Y.; Jimeno, A. Humanized mouse xenograft models: Narrowing the tumor-microenvironment gap. Cancer Res. 2016, 76, 6153–6158. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Facciponte, J.; Jin, M.; Shen, Q.; Lin, Q. Humanized NOD-SCID IL2rg-/- mice as a preclinical model for cancer research and its potential use for individualized cancer therapies. Cancer Lett. 2014, 344, 13–19. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Fridman, R.; Kibbey, M.C.; Royce, L.S.; Zain, M.; Sweeney, T.M.; Jicha, D.L.; Yannelli, J.R.; Martin, G.R.; Kleinman, H.K. Enhanced tumor growth of both primary and established human and murine tumor cells in athymic mice after coinjection with matrigel. J. Natl. Cancer Inst. 1991, 83, 769–774. [Google Scholar] [CrossRef]
- Igarashi, T.; Iwasaki, N.; Kasahara, Y.; Minami, A. A cellular implantation system using an injectable ultra-purified alginate gel for repair of osteochondral defects in a rabbit model. J. Biomed. Mater. Res. Part A 2010, 94, 844–855. [Google Scholar] [CrossRef]
- Rabbany, S.Y.; Pastore, J.; Yamamoto, M.; Miller, T.; Rafii, S.; Aras, R.; Penn, M. Continuous delivery of stromal cell-derived factor-1 from alginate scaffolds accelerates wound healing. Cell Transplant. 2010, 19, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, C.; Heinze, T.; Hipler, U.C. Comparative in vitro study on cytotoxicity, antimicrobial activity, and binding capacity for pathophysiological factors in chronic wounds of alginate and silver-containing alginat. Wound Repair Regen. 2009, 17, 511–521. [Google Scholar] [CrossRef]
- Murakami, K.; Aoki, H.; Nakamura, S.; Nakamura, S.-i.; Takikawa, M.; Hanzawa, M.; Kishimoto, S.; Hattori, H.; Tanaka, Y.; Kiyosawa, T.; et al. Hydrogel blends of chitin/chitosan, fucoidan and alginate as healing-impaired wound dressings. Biomaterials 2010, 31, 83–90. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, R.; Murata, S.; Cheng, C.; Mori, A.; Nie, Y.; Mikami, S.; Hasegawa, S.; Tadokoro, T.; Okamoto, S.; Taniguchi, H. A Novel Orthotopic Liver Cancer Model for Creating a Human-like Tumor Microenvironment. Cancers 2021, 13, 3997. https://doi.org/10.3390/cancers13163997
Qiu R, Murata S, Cheng C, Mori A, Nie Y, Mikami S, Hasegawa S, Tadokoro T, Okamoto S, Taniguchi H. A Novel Orthotopic Liver Cancer Model for Creating a Human-like Tumor Microenvironment. Cancers. 2021; 13(16):3997. https://doi.org/10.3390/cancers13163997
Chicago/Turabian StyleQiu, Rong, Soichiro Murata, Chao Cheng, Akihiro Mori, Yunzhong Nie, Satoshi Mikami, Shunsuke Hasegawa, Tomomi Tadokoro, Satoshi Okamoto, and Hideki Taniguchi. 2021. "A Novel Orthotopic Liver Cancer Model for Creating a Human-like Tumor Microenvironment" Cancers 13, no. 16: 3997. https://doi.org/10.3390/cancers13163997
APA StyleQiu, R., Murata, S., Cheng, C., Mori, A., Nie, Y., Mikami, S., Hasegawa, S., Tadokoro, T., Okamoto, S., & Taniguchi, H. (2021). A Novel Orthotopic Liver Cancer Model for Creating a Human-like Tumor Microenvironment. Cancers, 13(16), 3997. https://doi.org/10.3390/cancers13163997





