Is Mammographic Breast Density an Endophenotype for Breast Cancer?
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.1.1. kConFab
2.1.2. Busselton Health Study (BHS)
2.2. Status Allocation
2.3. Mammogram Selection
2.4. Data Analysis
2.4.1. Test of Criterion (i)—The Endophenotype Is Associated with Illness in the Population
2.4.2. Test of Criterion (ii)—The Endophenotype Is Heritable
2.4.3. Test of Criteria (iii)—Within Families, Endophenotype and Illness Co-Segregate and (iv)—The Endophenotype Found in Affected Family Members Is Found in Non-Affected Family Members at a Higher Rate than in the General Population
3. Results
3.1. Test of Criterion (i)—MBD Is Significantly Associated with Breast Cancer
3.2. Test of Criterion (ii)—MBD Is Heritable
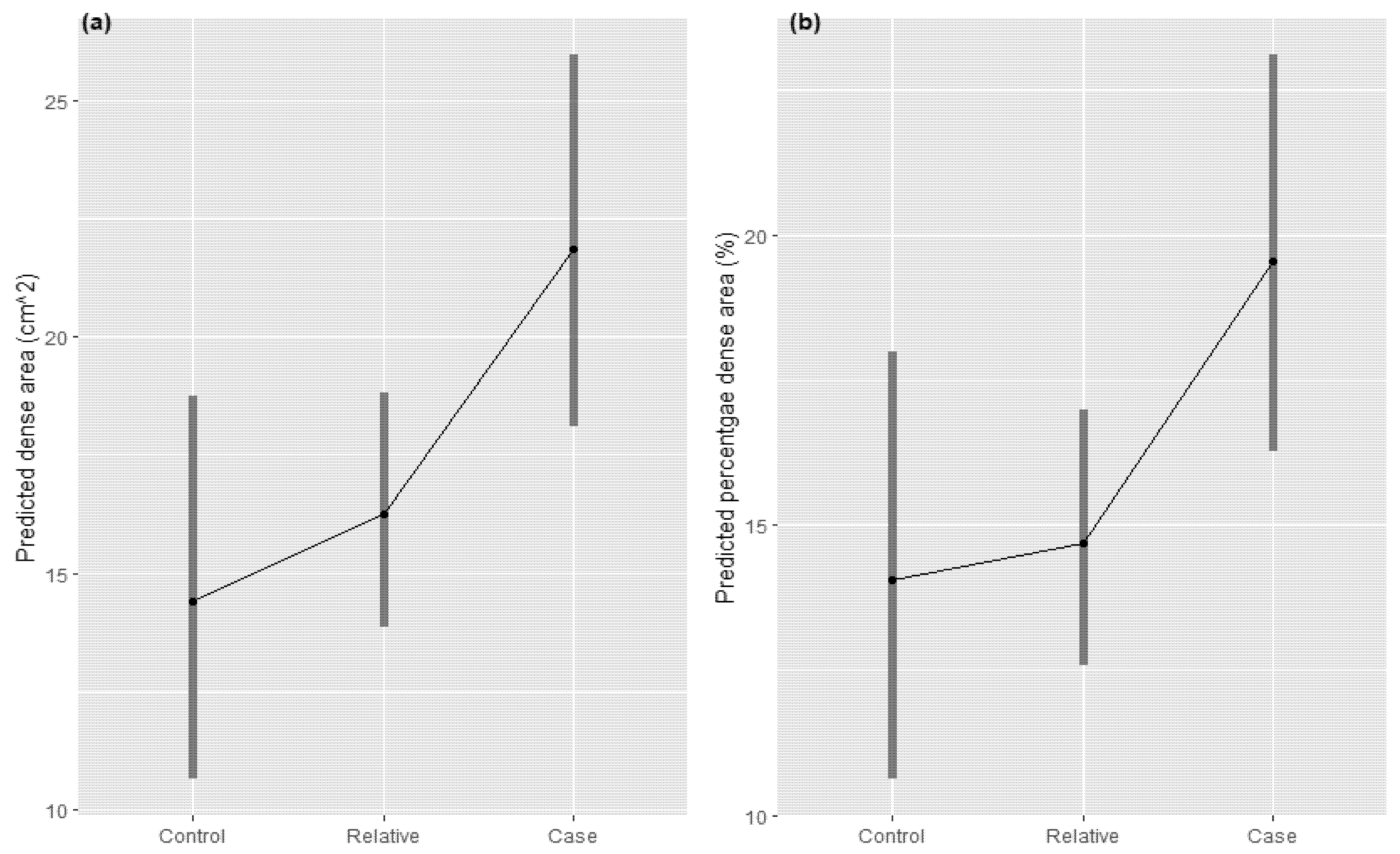
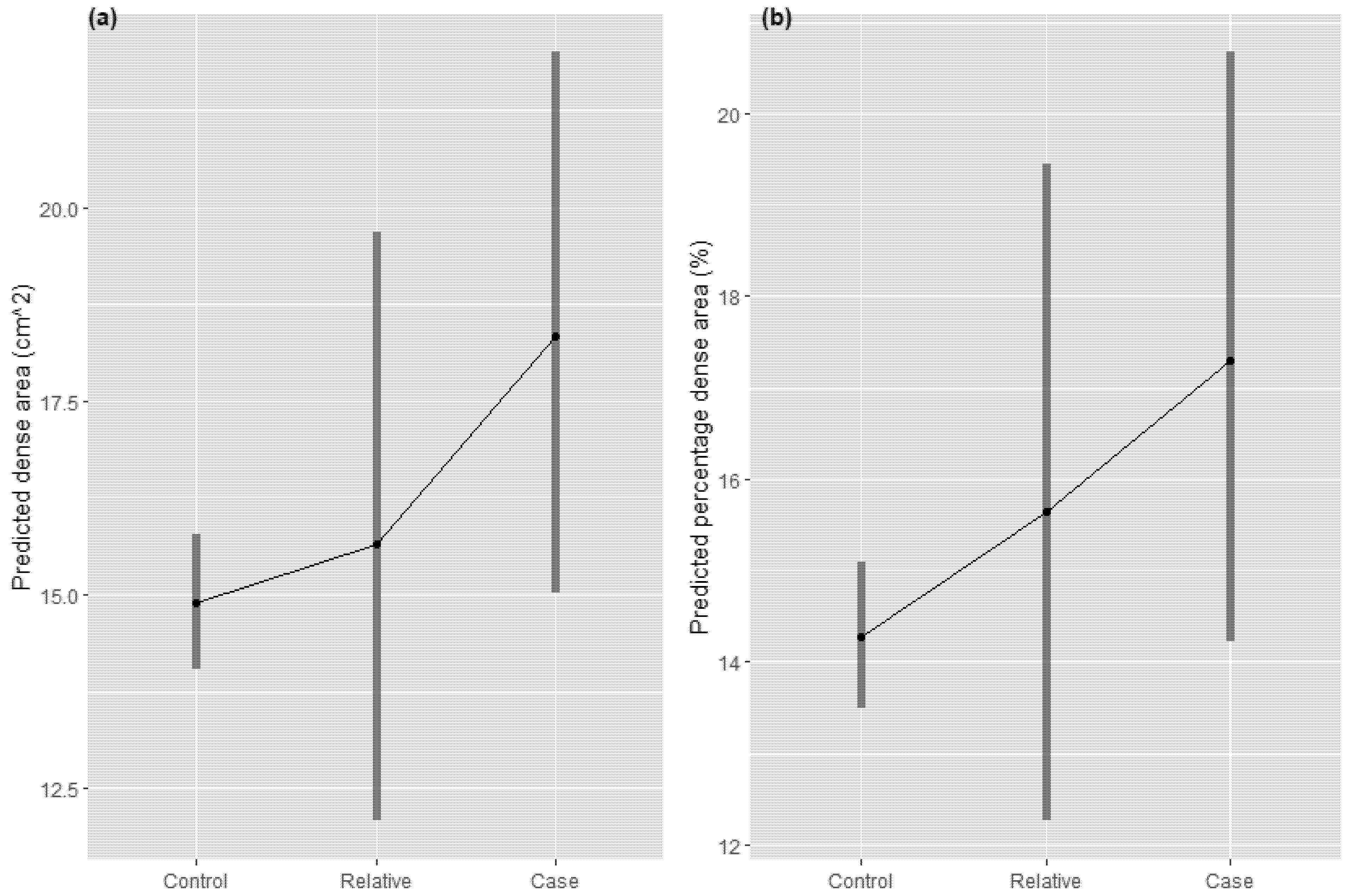
3.3. Test of Criterion (iii)—Within Families, Endophenotype and Illness Co-Segregate and (iv)—The Endophenotype Found in Affected Family Members Is Found in Non-Affected Family Members at a Higher Rate than in the General Population
4. Discussion
4.1. Criteria 1: MBD Is Associated with Breast Cancer Risk
4.2. Criteria 2: MBD Is Heritable
4.3. Criteria 3 and 4: Breast Cancer Segregates with Breast Density within Families, and Non-Affected Family Members Have an Intermediate (between Cases and Unrelated Controls) Breast Density
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Breast Cancer Foundation. About Breast Cancer. Available online: https://www.nationalbreastcancer.org/about-breast-cancer/ (accessed on 9 June 2021).
- Michailidou, K.; Hall, P.; Gonzalez-Neira, A.; Ghoussaini, M.; Dennis, J.; Milne, R.L.; Schmidt, M.K.; Chang-Claude, J.; Bojesen, S.E.; Bolla, M.K.; et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013, 45, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, K.; Lindstrom, S.; Dennis, J.; Beesley, J.; Hui, S.; Kar, S.; Lemacon, A.; Soucy, P.; Glubb, D.; Rostamianfar, A.; et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017, 551, 92–94. [Google Scholar] [CrossRef] [Green Version]
- Bond-Smith, D.; Stone, J. Methodological challenges and updated findings from a meta-analysis of the association between mammographic density and breast cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 22–31. [Google Scholar] [CrossRef] [Green Version]
- McCormack, V.A.; dos Santos Silva, I. Breast density and parenchymal patterns as markers of breast cancer risk: A meta-analysis. Cancer Epidemiol. Biomark. Prev 2006, 15, 1159–1169. [Google Scholar] [CrossRef] [Green Version]
- Boyd, N.F.; Dite, G.S.; Stone, J.; Gunasekara, A.; English, D.R.; McCredie, M.R.; Giles, G.G.; Tritchler, D.; Chiarelli, A.; Yaffe, M.J.; et al. Heritability of mammographic density, a risk factor for breast cancer. N. Engl. J. Med. 2002, 347, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.; Dite, G.S.; Gunasekara, A.; English, D.R.; McCredie, M.R.; Giles, G.G.; Cawson, J.N.; Hegele, R.A.; Chiarelli, A.M.; Yaffe, M.J.; et al. The heritability of mammographically dense and nondense breast tissue. Cancer Epidemiol. Biomark. Prev. 2006, 15, 612–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprague, B.L.; Gangnon, R.E.; Burt, V.; Trentham-Dietz, A.; Hampton, J.M.; Wellman, R.D.; Kerlikowske, K.; Miglioretti, D.L. Prevalence of mammographically dense breasts in the United States. J. Natl. Cancer Inst. 2014, 106, dju255. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J.; Warwick, J.; Pinney, E.; Warren, R.M.; Duffy, S.W. Tamoxifen and breast density in women at increased risk of breast cancer. J. Natl. Cancer Inst. 2004, 96, 621–628. [Google Scholar] [CrossRef] [Green Version]
- Cuzick, J.; Warwick, J.; Pinney, E.; Duffy, S.W.; Cawthorn, S.; Howell, A.; Forbes, J.F.; Warren, R.M. Tamoxifen-induced reduction in mammographic density and breast cancer risk reduction: A nested case-control study. J. Natl. Cancer Inst. 2011, 103, 744–752. [Google Scholar] [CrossRef]
- Stone, J.; Thompson, D.J.; Dos Santos Silva, I.; Scott, C.; Tamimi, R.M.; Lindstrom, S.; Kraft, P.; Hazra, A.; Li, J.; Eriksson, L.; et al. Novel Associations between Common Breast Cancer Susceptibility Variants and Risk-Predicting Mammographic Density Measures. Cancer Res. 2015, 75, 2457–2467. [Google Scholar] [CrossRef] [Green Version]
- Gottesman, I.I.; Gould, T.D. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am. J. Psychiatry 2003, 160, 636–645. [Google Scholar] [CrossRef]
- Kerlikowske, K.; Zhu, W.; Tosteson, A.N.; Sprague, B.L.; Tice, J.A.; Lehman, C.D.; Miglioretti, D.L.; Breast Cancer Surveillance, C. Identifying women with dense breasts at high risk for interval cancer: A cohort study. Ann. Intern. Med. 2015, 162, 673–681. [Google Scholar] [CrossRef] [Green Version]
- Mann, G.J.; Thorne, H.; Balleine, R.L.; Butow, P.N.; Clarke, C.L.; Edkins, E.; Evans, G.M.; Fereday, S.; Haan, E.; Gattas, M.; et al. Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res. 2006, 8, R12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, A.T.; Armstrong, R.M.; Grassi, T.D.; Gaut, B.; Van Der Weyden, M.B. On our selection: Australian longitudinal research studies. Med. J. Aust. 2008, 189, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Cadby, G.; Melton, P.E.; McCarthy, N.S.; Almeida, M.; Williams-Blangero, S.; Curran, J.E.; VandeBerg, J.L.; Hui, J.; Beilby, J.; Musk, A.W.; et al. Pleiotropy of cardiometabolic syndrome with obesity-related anthropometric traits determined using empirically derived kinships from the Busselton Health Study. Hum. Genet. 2018, 137, 45–53. [Google Scholar] [CrossRef]
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Speed, D.; Hemani, G.; Johnson, M.R.; Balding, D.J. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 2012, 91, 1011–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaitlen, N.; Kraft, P.; Patterson, N.; Pasaniuc, B.; Bhatia, G.; Pollack, S.; Price, A.L. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 2013, 9, e1003520. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Conomos, M.P. GMMAT: Generalized Linear Mixed Model Association Tests. 2020. Available online: https://cran.r-project.org/web/packages/GMMAT/GMMAT.pdf (accessed on 9 June 2021).
- Boyd, N.F.; Byng, J.W.; Jong, R.A.; Fishell, E.K.; Little, L.E.; Miller, A.B.; Lockwood, G.A.; Tritchler, D.L.; Yaffe, M.J. Quantitative classification of mammographic densities and breast cancer risk: Results from the Canadian National Breast Screening Study. J. Natl. Cancer Inst. 1995, 87, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Byrne, C.; Schairer, C.; Wolfe, J.; Parekh, N.; Salane, M.; Brinton, L.A.; Hoover, R.; Haile, R. Mammographic features and breast cancer risk: Effects with time, age, and menopause status. J. Natl. Cancer Inst. 1995, 87, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Risch, H.A.; McLaughlin, J.R.; Cole, D.E.; Rosen, B.; Bradley, L.; Fan, I.; Tang, J.; Li, S.; Zhang, S.; Shaw, P.A.; et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: A kin-cohort study in Ontario, Canada. J. Natl. Cancer Inst. 2006, 98, 1694–1706. [Google Scholar] [CrossRef] [PubMed]
- Darcey, E.; Lloyd, R.; Cadby, G.; Pilkington, L.; Redfern, A.; Thompson, S.C.; Saunders, C.; Wylie, E.; Stone, J. The association between mammographic density and breast cancer risk in Western Australian Aboriginal women. Breast Cancer Res. Treat. 2019, 176, 235–242. [Google Scholar] [CrossRef]
- Maskarinec, G.; Pagano, I.; Lurie, G.; Wilkens, L.R.; Kolonel, L.N. Mammographic density and breast cancer risk: The multiethnic cohort study. Am. J. Epidemiol. 2005, 162, 743–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| kConFab | BHS | |||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | Total (n = 323) | Controls (n = 56, 17.3%) | Relatives (n = 179, 55.4%) | Cases (n = 88, 27.2%) | Total (n = 1587) | Controls (n = 1423, 89.7%) | Relatives (n = 72, 4.5%) | Cases (n = 92, 5.8%) |
| Median age (years) at mammogram (IQR) | 51.0 (13.0) | 51.0 (8.5) | 50.0 (14.5) | 56.0 (12.3) | 52.6 (15.9) | 52.8 (15.7) | 49.5 (11.9) | 54.0 (16.9) |
| Median BMI at interview (IQR) a | 26.0 (7.5) | 26.7 (6.4) | 24.9 (7.6) | 26.4 (7.0) | 25.1 (6.0) | 25.1 (6.0) | 24.8 (6.0) | 25.9 (5.6) |
| Median time (days) between mammogram and interview (IQR) a | −1084 (2644) | −2990 (3452) | −658 (2316) | −1200 (2230) | 305 (3282) | 331 (3278) | 1043 (3371) | 24.5 (2207) |
| Median dense area cm2 (IQR) | 17.6 (26.3) | 15.2 (28.6) | 15.4 (25.3) | 22.0 (26.3) | 15.3 (24.4) | 14.7 (23.8) | 22.0 (28.6) | 19.7 (24.1) |
| Median percentage dense area b % (IQR) | 16.6 (26.1) | 16.8 (33.8) | 14.8 (24.4) | 20.9 (25.8) | 15.0 (28.0) | 14.0 (27.5) | 23.0 (32.5) | 19.0 (26.0) |
| kConFab 1 | BHS 2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n = 88)/ Relatives + Controls (n = 235 3) | Cases (n = 88)/ Relatives (n = 179 3) | Cases (n = 88)/ Controls (n = 56) | Cases (n = 92)/ Relatives + Controls (n = 1495) | Cases (n = 92)/ Relatives (n = 72) | Cases (n = 92)/ Controls (n = 1423) | |||||||
| OR (95% CI) | p-Value 4 | OR (95% CI) | p-Value 4 | OR (95%CI) | p-Value 4 | OR (95% CI) | p-Value 4 | OR (95% CI) | p-Value 4 | OR (95%CI) | p-Value 4 | |
| Dense area (cm2) | 1.015 (1.002, 1.029) | 0.029 | 1.015 (1.000, 1.030) | 0.043 | 1.023 (0.9998, 1.046) | 0.050 | 1.009 (0.998, 1.019) | 0.094 | 1.004 (0.984, 1.025) | 0.692 | 1.009 (0.998, 1.019) | 0.101 |
| Percentage dense area (%) | 1.019 (1.002, 1.037) | 0.030 | 1.020 (1.002, 1.039) | 0.031 | 1.022 (0.996, 1.049) | 0.098 | 1.011 (0.998, 1.025) | 0.102 | 0.999 (0.978, 1.021) | 0.934 | 1.012 (0.998, 1.025) | 0.097 |
| βcases vs. relatives (CI) | βcases vs. controls (CI) | βrelatives vs. controls (CI) | |
|---|---|---|---|
| kConFab | |||
| Dense area (cm2) | 0.649 (0.165, 1.13) | 6.93 (0.174, 13.68) 1 | 0.253 (−0.404, 0.909) |
| Percentage dense area (%) | 0.593 (0.155, 1.03) | 0.818 (0.177, 1.46) | 0.106 (−0.503, 0.716) |
| BHS | |||
| Dense area (cm2) | 0.197 (−0.428, 0.822 | 0.429 (0.009, 0.850) | 0.106 (−0.394, 0.605) |
| Percentage dense area (%) | 0.186 (−0.508, 0.685) | 0.391 (−0.007, 0.790) | 0.201 (−0.269, 0.672) |
| Meta-analysed 2 | |||
| Dense area (cm2) | 0.478 (0.098, 0.859) | 0.596 (0.240, 0.951) | 0.161 (−0.236, 0.556) |
| Percentage dense area (%) | 0.413 (0.060, 0.7767) | 0.511 (0.173, 0.848) | 0.162 (−0.210, 0.534) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darcey, E.; McCarthy, N.; Moses, E.K.; Saunders, C.; Cadby, G.; Stone, J. Is Mammographic Breast Density an Endophenotype for Breast Cancer? Cancers 2021, 13, 3916. https://doi.org/10.3390/cancers13153916
Darcey E, McCarthy N, Moses EK, Saunders C, Cadby G, Stone J. Is Mammographic Breast Density an Endophenotype for Breast Cancer? Cancers. 2021; 13(15):3916. https://doi.org/10.3390/cancers13153916
Chicago/Turabian StyleDarcey, Ellie, Nina McCarthy, Eric K. Moses, Christobel Saunders, Gemma Cadby, and Jennifer Stone. 2021. "Is Mammographic Breast Density an Endophenotype for Breast Cancer?" Cancers 13, no. 15: 3916. https://doi.org/10.3390/cancers13153916
APA StyleDarcey, E., McCarthy, N., Moses, E. K., Saunders, C., Cadby, G., & Stone, J. (2021). Is Mammographic Breast Density an Endophenotype for Breast Cancer? Cancers, 13(15), 3916. https://doi.org/10.3390/cancers13153916






