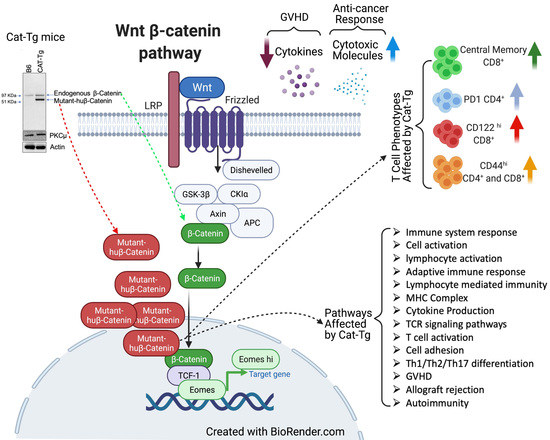
Human Wnt/β-Catenin Regulates Alloimmune Signaling during Allogeneic Transplantation
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
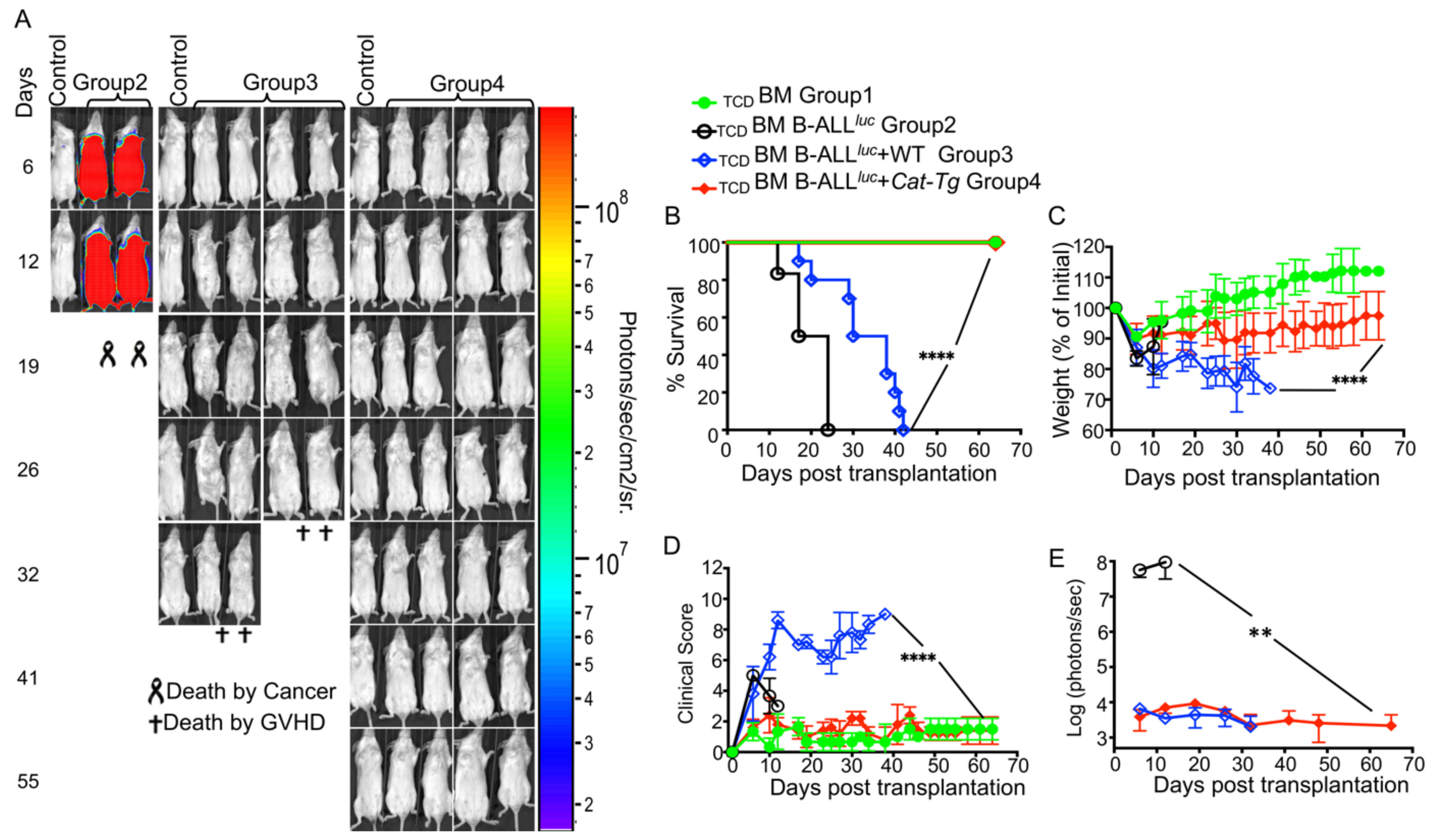
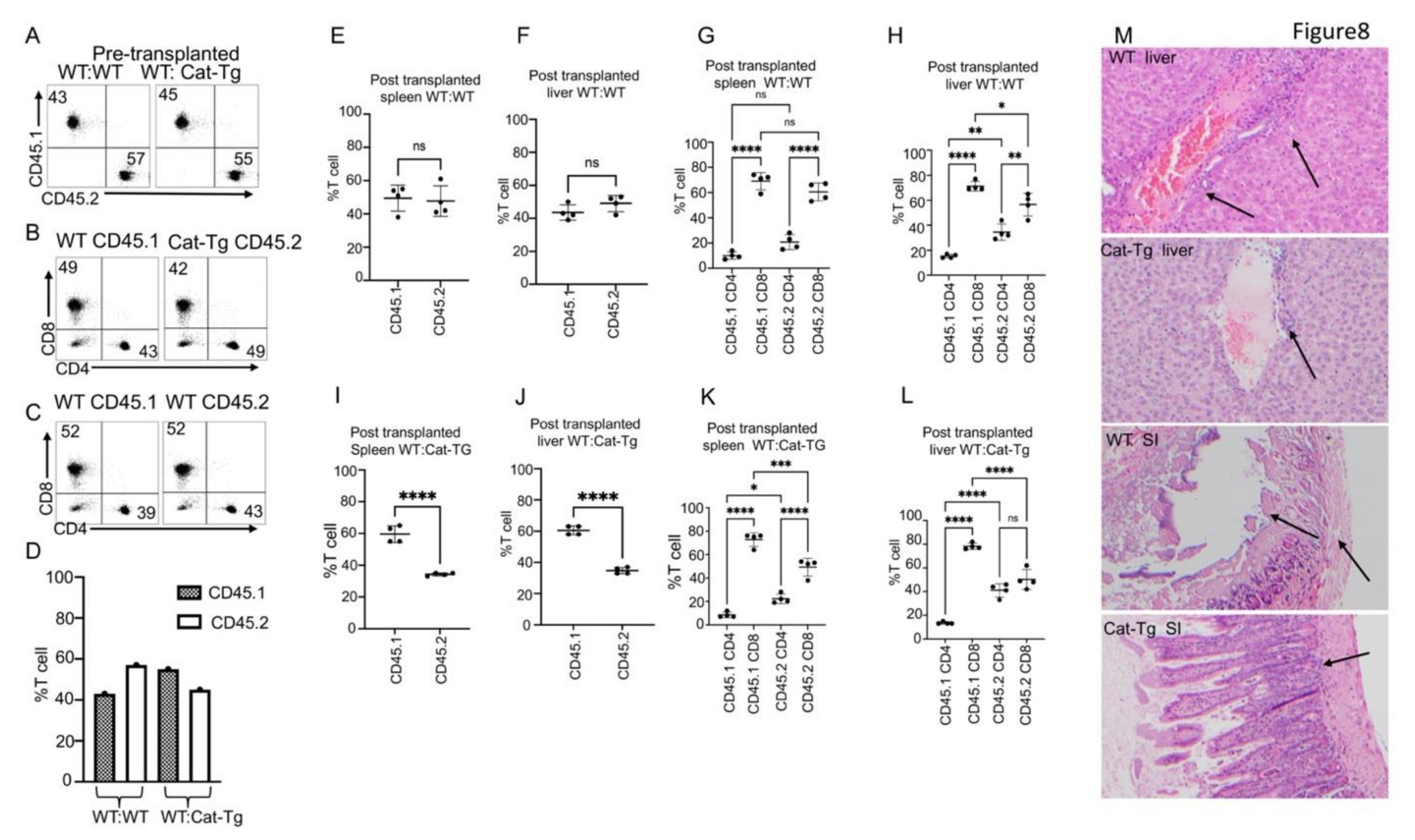
2.1. Donor T Cells from Cat-Tg Mice Do Not Induce GVHD but Maintain GVL Function
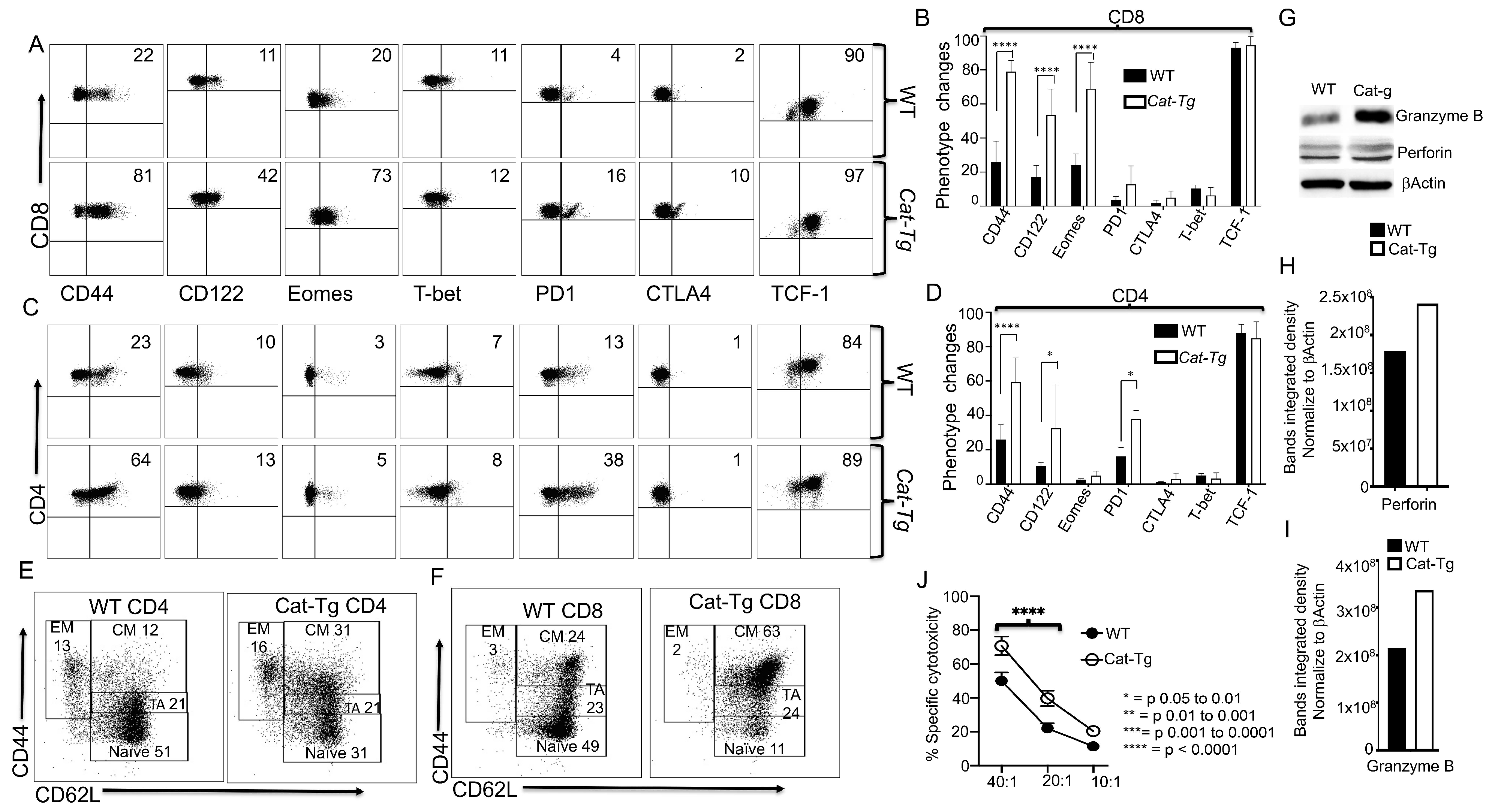
2.2. Wnt/β-Catenin Affects T Cell Phenotype and Cytotoxic Function
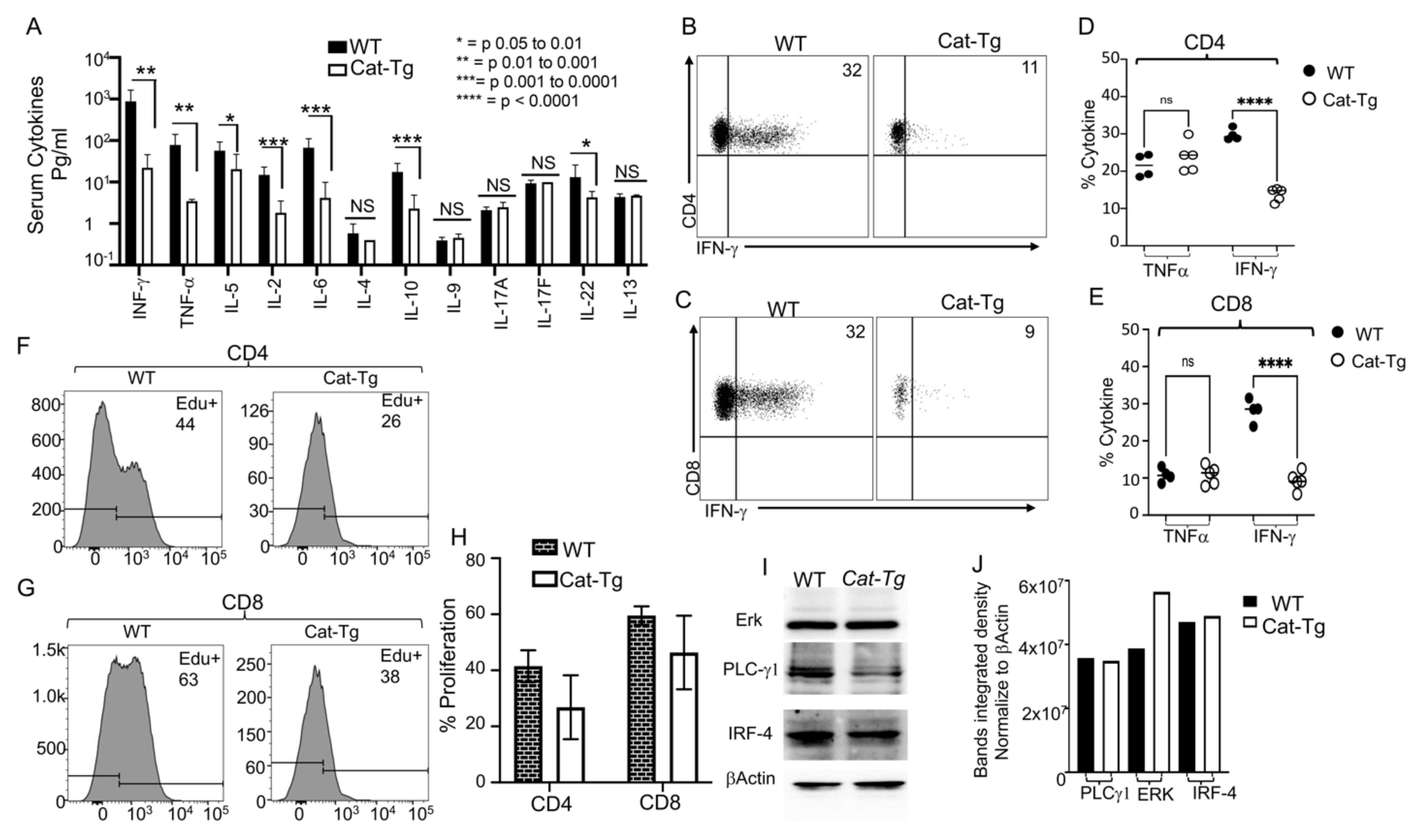
2.3. Wnt/β-Catenin Overexpression Results in Reduced Cytokine Production and Donor T Cell Proliferation without Affecting TCR Signaling
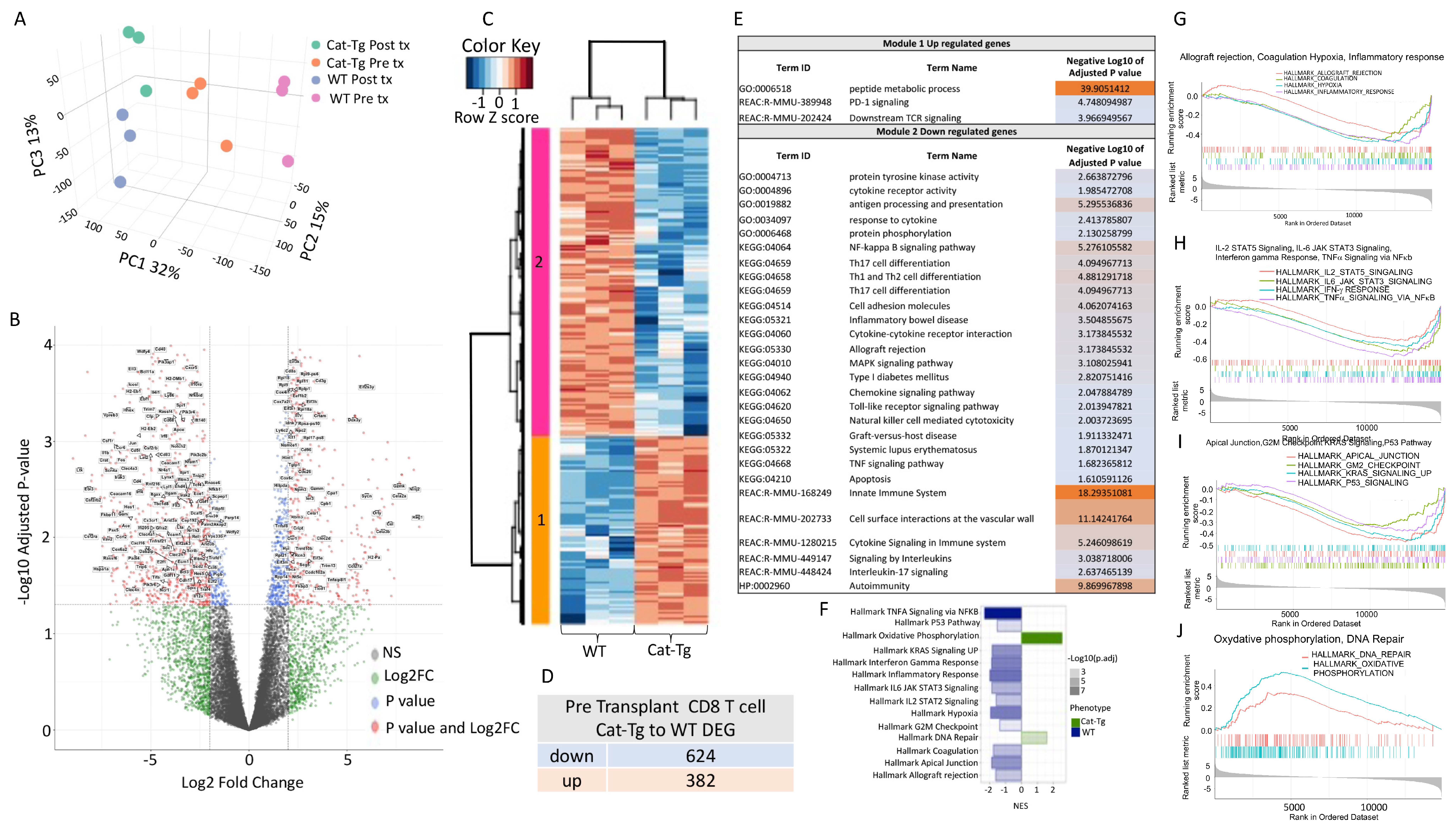
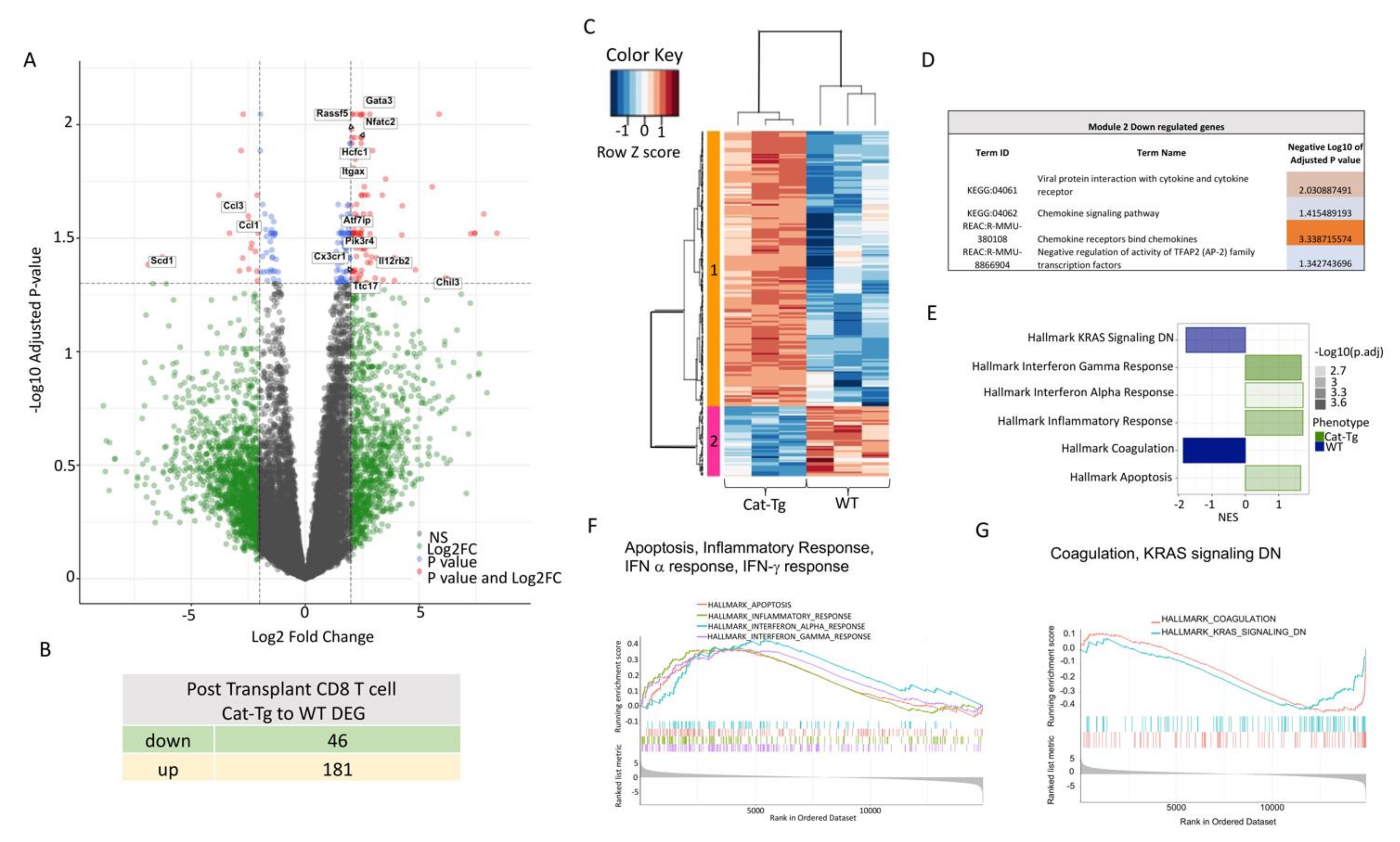
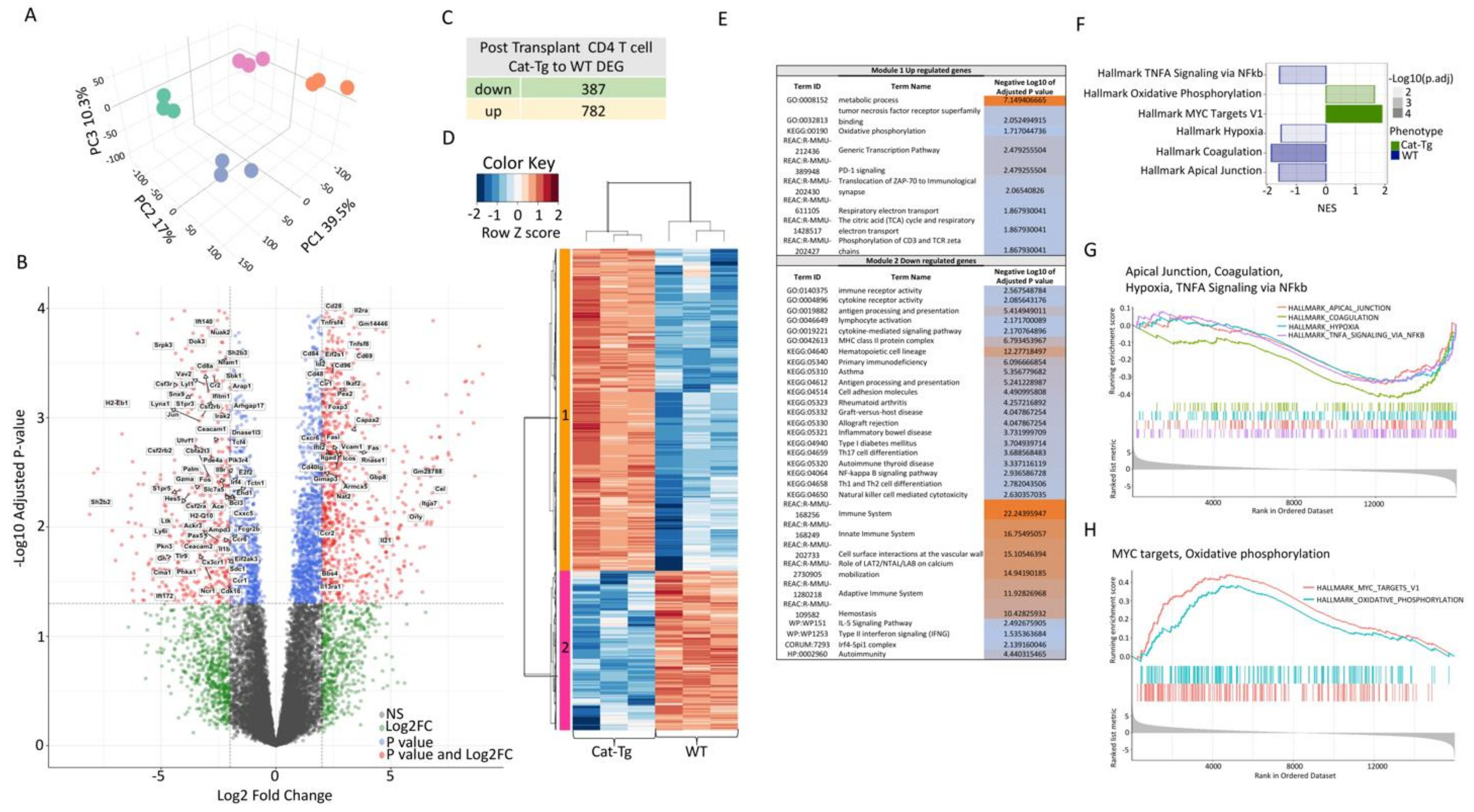
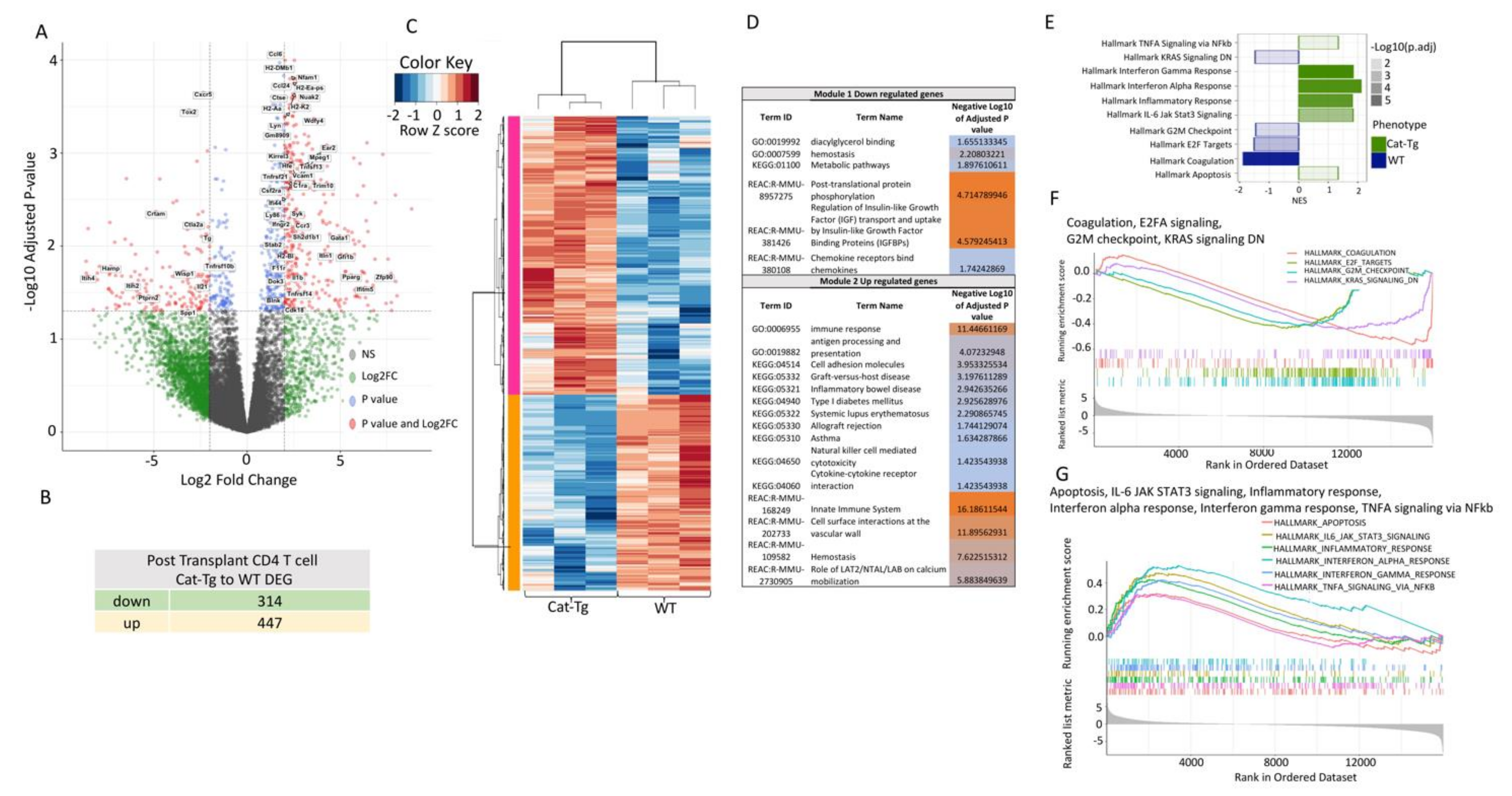
2.4. Wnt/β-Catenin Expression Regulates Gene Expression in T Cells during GVHD
2.5. Wnt/β-Catenin Overexpression Specifically Affects CD8+ T Cell Functions
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Reagents, Cell Lines, Flow Cytometry
4.3. Allo-HSCT and GVL Studies
4.4. Cytokine Production, Cytotoxicity, and EdU Incorporation Assays
4.5. Migration Assays
4.6. RNA Sequencing
4.7. Western Blotting
4.8. Histopathological Evaluation
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bleakley, M.; Turtle, C.J.; Riddell, S.R. Augmentation of anti-tumor immunity by adoptive T-cell transfer after allogeneic hematopoietic stem cell transplantation. Expert. Rev. Hematol. 2012, 5, 409–425. [Google Scholar] [CrossRef]
- Breems, D.A.; Lowenberg, B. Autologous stem cell transplantation in the treatment of adults with acute myeloid leukaemia. Br. J. Haematol. 2005, 130, 825–833. [Google Scholar] [CrossRef]
- Ghimire, S.; Weber, D.; Mavin, E.; Wang, X.N.; Dickinson, A.M.; Holler, E. Pathophysiology of GvHD and Other HSCT-Related Major Complications. Front. Immunol. 2017, 8, 79. [Google Scholar] [CrossRef]
- Zeiser, R. Introduction to a review series on pathophysiology and treatment of acute GVHD. Blood 2020, 136, 375–376. [Google Scholar] [CrossRef]
- Li, X.; Xiang, Y.; Li, F.; Yin, C.; Li, B.; Ke, X. WNT/beta-Catenin Signaling Pathway Regulating T Cell-Inflammation in the Tumor Microenvironment. Front. Immunol. 2019, 10, 2293. [Google Scholar] [CrossRef]
- Haseeb, M.; Pirzada, R.H.; Ain, Q.U.; Choi, S. Wnt Signaling in the Regulation of Immune Cell and Cancer Therapeutics. Cells 2019, 8, 1380. [Google Scholar] [CrossRef]
- Driessens, G.; Zheng, Y.; Locke, F.; Cannon, J.L.; Gounari, F.; Gajewski, T.F. Beta-catenin inhibits T cell activation by selective interference with linker for activation of T cells-phospholipase C-gamma1 phosphorylation. J. Immunol. 2011, 186, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Sharma, A.; Sen, J.M. TCF1 and beta-catenin regulate T cell development and function. Immunol. Res. 2010, 47, 45–55. [Google Scholar] [CrossRef]
- Willinger, T.; Freeman, T.; Herbert, M.; Hasegawa, H.; McMichael, A.J.; Callan, M.F.C. Human naive CD8 T cells down-regulate expression of the WNT pathway transcription factors lymphoid enhancer binding factor 1 and transcription factor 7 (T cell factor-1) following antigen encounter in vitro and in vivo. J. Immunol. 2006, 176, 1439–1446. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Hottiger, M.O. Crosstalk between Wnt/beta-Catenin and NF-kappaB Signaling Pathway during Inflammation. Front. Immunol. 2016, 7, 378. [Google Scholar] [CrossRef] [PubMed]
- Gattinoni, L.; Ji, Y.; Restifo, N.P. Wnt/beta-catenin signaling in T-cell immunity and cancer immunotherapy. Clin. Cancer Res. 2010, 16, 4695–4701. [Google Scholar] [CrossRef]
- Van Loosdregt, J.; Coffer, P.J. The Role of WNT Signaling in Mature T Cells: T Cell Factor Is Coming Home. J. Immunol. 2018, 201, 2193–2200. [Google Scholar] [CrossRef] [PubMed]
- Boudousquié, C.; Danilo, M.; Pousse, L.; Raj, B.J.; Angelov, G.S.; Chennupati, V.; Zehn, D.; Held, W. Differences in the transduction of canonical Wnt signals demarcate effector and memory CD8 T cells with distinct recall proliferation capacity. J. Immunol. 2014, 193, 2784–2791. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Chikwava, K.; Wu, C.; Zhang, H.; Bhagat, A.; Pei, D.; Choi, J.K.; Tong, W. LNK/SH2B3 regulates IL-7 receptor signaling in normal and malignant B-progenitors. J. Clin. Investig. 2016, 126, 1267–1281. [Google Scholar] [CrossRef]
- Mammadli, M.; Huang, W.; Harris, R.; Sultana, A.; Cheng, Y.; Tong, W.; Pu, J.; Gentile, T.; Dsouza, S.; Yang, Q.; et al. Targeting Interleukin-2-Inducible T-Cell Kinase (ITK) Differentiates GVL and GVHD in Allo-HSCT. Front. Immunol. 2020, 11, 593863. [Google Scholar] [CrossRef] [PubMed]
- Socie, G.; Blazar, B.R. Acute graft-versus-host disease: From the bench to the bedside. Blood 2009, 114, 4327–4336. [Google Scholar] [CrossRef]
- Mammadli, M.; Huang, W.; Harris, R.; Xiong, H.; Weeks, S.; May, A.; Gentile, T.; Ridilla, J.H.; Waickman, A.T.; August, A.; et al. Targeting SLP76: ITK interaction separates GVHD from GVL in allo-HSCT. iScience 2021, 24, 102286. [Google Scholar] [CrossRef] [PubMed]
- Cooke, K.R.; Kobzik, L.; Martin, T.R.; Brewer, J.; Delmonte, J., Jr.; Crawford, J.M.; Ferrara, J.L. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. Blood 1996, 88, 3230–3239. [Google Scholar] [CrossRef]
- Huang, W.; Hu, J.; August, A. Cutting edge: Innate memory CD8+ T cells are distinct from homeostatic expanded CD8+ T cells and rapidly respond to primary antigenic stimuli. J. Immunol. 2013, 190, 2490–2494. [Google Scholar] [CrossRef] [PubMed]
- Henden, A.S.; Hill, G.R. Cytokines in Graft-versus-Host Disease. J. Immunol. 2015, 194, 4604–4612. [Google Scholar] [CrossRef]
- Holler, E. Cytokines, viruses, and graft-versus-host disease. Curr. Opin. Hematol. 2002, 9, 479–484. [Google Scholar] [CrossRef]
- Ju, X.P.; Xu, B.; Xiao, Z.P.; Li, J.Y.; Chen, L.; Lu, S.Q.; Huang, Z.X. Cytokine expression during acute graft-versus-host disease after allogeneic peripheral stem cell transplantation. Bone Marrow Transpl. 2005, 35, 1179–1186. [Google Scholar] [CrossRef][Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Wysocki, C.A.; Panoskaltsis-Mortari, A.; Blazar, B.R.; Serody, J.S. Leukocyte migration and graft-versus-host disease. Blood 2005, 105, 4191–4199. [Google Scholar] [CrossRef]
- Gooptu, M.; Koreth, J. Translational and clinical advances in acute graft-versus-host disease. Haematologica 2020, 105, 2550–2560. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, J.L.; Cooke, K.R.; Pan, L.; Krenger, W. The immunopathophysiology of acute graft-versus-host-disease. Stem Cells 1996, 14, 473–489. [Google Scholar] [CrossRef] [PubMed]
- Dutt, S.; Baker, J.; Kohrt, H.E.; Kambham, N.; Sanyal, M.; Negrin, R.S.; Strober, S. CD8+CD44(hi) but not CD4+CD44(hi) memory T cells mediate potent graft antilymphoma activity without GVHD. Blood 2011, 117, 3230–3239. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.E.; McNiff, J.; Yan, J.; Doyle, H.; Mamula, M.; Shlomchik, M.J.; Shlomchik, W.D. Memory CD4+ T cells do not induce graft-versus-host disease. J. Clin. Investig. 2003, 112, 101–108. [Google Scholar] [CrossRef]
- Huang, W.; Mo, W.; Jiang, J.; Chao, N.J.; Chen, B.J. Donor Allospecific CD44(high) Central Memory T Cells Have Decreased Ability to Mediate Graft-vs.-Host Disease. Front. Immunol. 2019, 10, 624. [Google Scholar] [CrossRef]
- Carty, S.A.; Koretzky, G.A.; Jordan, M.S. Interleukin-4 regulates eomesodermin in CD8+ T cell development and differentiation. PLoS ONE 2014, 9, e106659. [Google Scholar] [CrossRef]
- Ma, J.; Wang, R.; Fang, X.; Sun, Z. Beta-catenin/TCF-1 pathway in T cell development and differentiation. J. Neuroimmune Pharmacol. 2012, 7, 750–762. [Google Scholar] [CrossRef]
- Yu, Q.; Sen, J.M. Beta-catenin regulates positive selection of thymocytes but not lineage commitment. J. Immunol. 2007, 178, 5028–5034. [Google Scholar] [CrossRef] [PubMed]
- Lovatt, M.; Bijlmakers, M.J. Stabilisation of beta-catenin downstream of T cell receptor signalling. PLoS ONE 2010, 5, e12794. [Google Scholar] [CrossRef][Green Version]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]
- Gattinoni, L.; Zhong, X.S.; Palmer, D.C.; Ji, Y.; Hinrichs, C.S.; Yu, Z.; Wrzesinski, C.; Boni, A.; Cassard, L.; Church, L.; et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 2009, 15, 808–813. [Google Scholar] [CrossRef]
- Zhao, D.M.; Yu, S.; Zhou, X.; Haring, J.S.; Held, W.; Badovinac, V.P.; Harty, J.T.; Xue, H.H. Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J. Immunol. 2010, 184, 1191–1199. [Google Scholar] [CrossRef]
- Schroeder, M.A.; DiPersio, J.F. Mouse models of graft-versus-host disease: Advances and limitations. Dis. Model. Mech. 2011, 4, 318–333. [Google Scholar] [CrossRef] [PubMed]
- Bouazzaoui, A.; Spacenko, E.; Mueller, G.; Miklos, S.; Huber, E.; Holler, E.; Andreesen, R.; Hildebrandt, G.C. Chemokine and chemokine receptor expression analysis in target organs of acute graft-versus-host disease. Genes Immun. 2009, 10, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Blazar, B.R.; Taylor, P.A.; Panoskaltsis-Mortari, A.; Gray, G.S.; Vallera, D.A. Coblockade of the LFA1: ICAM and CD28/CTLA4:B7 pathways is a highly effective means of preventing acute lethal graft-versus-host disease induced by fully major histocompatibility complex-disparate donor grafts. Blood 1995, 85, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Saidu, N.E.B.; Bonini, C.; Dickinson, A.; Grce, M.; Inngjerdingen, M.; Koehl, U.; Toubert, A.; Zeiser, R.; Galimberti, S. New Approaches for the Treatment of Chronic Graft-Versus-Host Disease: Current Status and Future Directions. Front. Immunol. 2020, 11, 578314. [Google Scholar] [CrossRef]
- Jankovic, S.V.; Hari, P.; Jacobs, P.; Komorowski, R.; Drobyski, W.R. NF-kappaB as a target for the prevention of graft-versus-host disease: Comparative efficacy of bortezomib and PS-1145. Blood 2006, 107, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Augello, G.; Emma, M.R.; Cusimano, A.; Azzolina, A.; Montalto, G.; McCubrey, J.A.; Cervello, M. The Role of GSK-3 in Cancer Immunotherapy: GSK-3 Inhibitors as a New Frontier in Cancer Treatment. Cells 2020, 9, 1427. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020, 13, 165. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Xu, M.; Sen, J.M. Beta-catenin expression enhances IL-7 receptor signaling in thymocytes during positive selection. J. Immunol. 2007, 179, 126–131. [Google Scholar] [CrossRef]
- Gat, U.; DasGupta, R.; Degenstein, L.; Fuchs, E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell 1998, 95, 605–614. [Google Scholar] [CrossRef]
- Garvin, A.M.; Abraham, K.M.; Forbush, K.A.; Farr, A.G.; Davison, B.L.; Perlmutter, R.M. Disruption of thymocyte development and lymphomagenesis induced by SV40 T-antigen. Int. Immunol. 1990, 2, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Mulroy, T.; Xu, Y.; Sen, J.M. Beta-Catenin expression enhances generation of mature thymocytes. Int. Immunol. 2003, 15, 1485–1494. [Google Scholar] [CrossRef]
- Contag, C.H.; Bachmann, M.H. Advances in in vivo bioluminescence imaging of gene expression. Annu. Rev. Biomed. Eng. 2002, 4, 235–260. [Google Scholar] [CrossRef]
- Mancusi, A.; Piccinelli, S.; Velardi, A.; Pierini, A. The Effect of TNF-alpha on Regulatory T Cell Function in Graft-versus-Host Disease. Front. Immunol. 2018, 9, 356. [Google Scholar] [CrossRef]
- Karimi, M.A.; Lee, E.; Bachmann, M.H.; Salicioni, A.M.; Behrens, E.M.; Kambayashi, T.; Baldwin, C.L. Measuring cytotoxicity by bioluminescence imaging outperforms the standard chromium-51 release assay. PLoS ONE 2014, 9, e89357. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Greene, A.E.; Charney, J.; Nichols, W.W.; Coriell, L.L. Species identity of insect cell lines. In Vitro 1972, 7, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Reiner, A.; Yekutieli, D.; Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003, 19, 368–375. [Google Scholar] [CrossRef]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g: Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.; Mammadli, M.; May, A.; Yang, Q.; Fung, I.T.H.; Sen, J.M.; Karimi, M. TCF-1 in CD4 T cells regulates GVHD severity and persistence. bioRxiv 2021. [Google Scholar] [CrossRef]
- Proffitt, K.D.; Virshup, D.M. Precise regulation of porcupine activity is required for physiological Wnt signaling. J. Biol. Chem. 2012, 287, 34167–34178. [Google Scholar] [CrossRef]
- Ye, Z.; Gould, T.M.; Zhang, H.; Jin, J.; Weyand, C.M.; Goronzy, J.J. The GSK3beta-beta-catenin-TCF1 pathway improves naive T cell activation in old adults by upregulating miR-181a. NPJ Aging Mech. Dis. 2021, 7, 4. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mammadli, M.; Harris, R.; Mahmudlu, S.; Verma, A.; May, A.; Dhawan, R.; Waickman, A.T.; Sen, J.M.; August, A.; Karimi, M. Human Wnt/β-Catenin Regulates Alloimmune Signaling during Allogeneic Transplantation. Cancers 2021, 13, 3798. https://doi.org/10.3390/cancers13153798
Mammadli M, Harris R, Mahmudlu S, Verma A, May A, Dhawan R, Waickman AT, Sen JM, August A, Karimi M. Human Wnt/β-Catenin Regulates Alloimmune Signaling during Allogeneic Transplantation. Cancers. 2021; 13(15):3798. https://doi.org/10.3390/cancers13153798
Chicago/Turabian StyleMammadli, Mahinbanu, Rebecca Harris, Sara Mahmudlu, Anjali Verma, Adriana May, Rohan Dhawan, Adam T. Waickman, Jyoti Misra Sen, Avery August, and Mobin Karimi. 2021. "Human Wnt/β-Catenin Regulates Alloimmune Signaling during Allogeneic Transplantation" Cancers 13, no. 15: 3798. https://doi.org/10.3390/cancers13153798
APA StyleMammadli, M., Harris, R., Mahmudlu, S., Verma, A., May, A., Dhawan, R., Waickman, A. T., Sen, J. M., August, A., & Karimi, M. (2021). Human Wnt/β-Catenin Regulates Alloimmune Signaling during Allogeneic Transplantation. Cancers, 13(15), 3798. https://doi.org/10.3390/cancers13153798