Electrochemical Detection of Global DNA Methylation Using Biologically Assembled Polymer Beads
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Chemicals
2.2. DNA Preparation
2.3. Preparation of DNA from Cell Line and Ovarian Cancer Samples
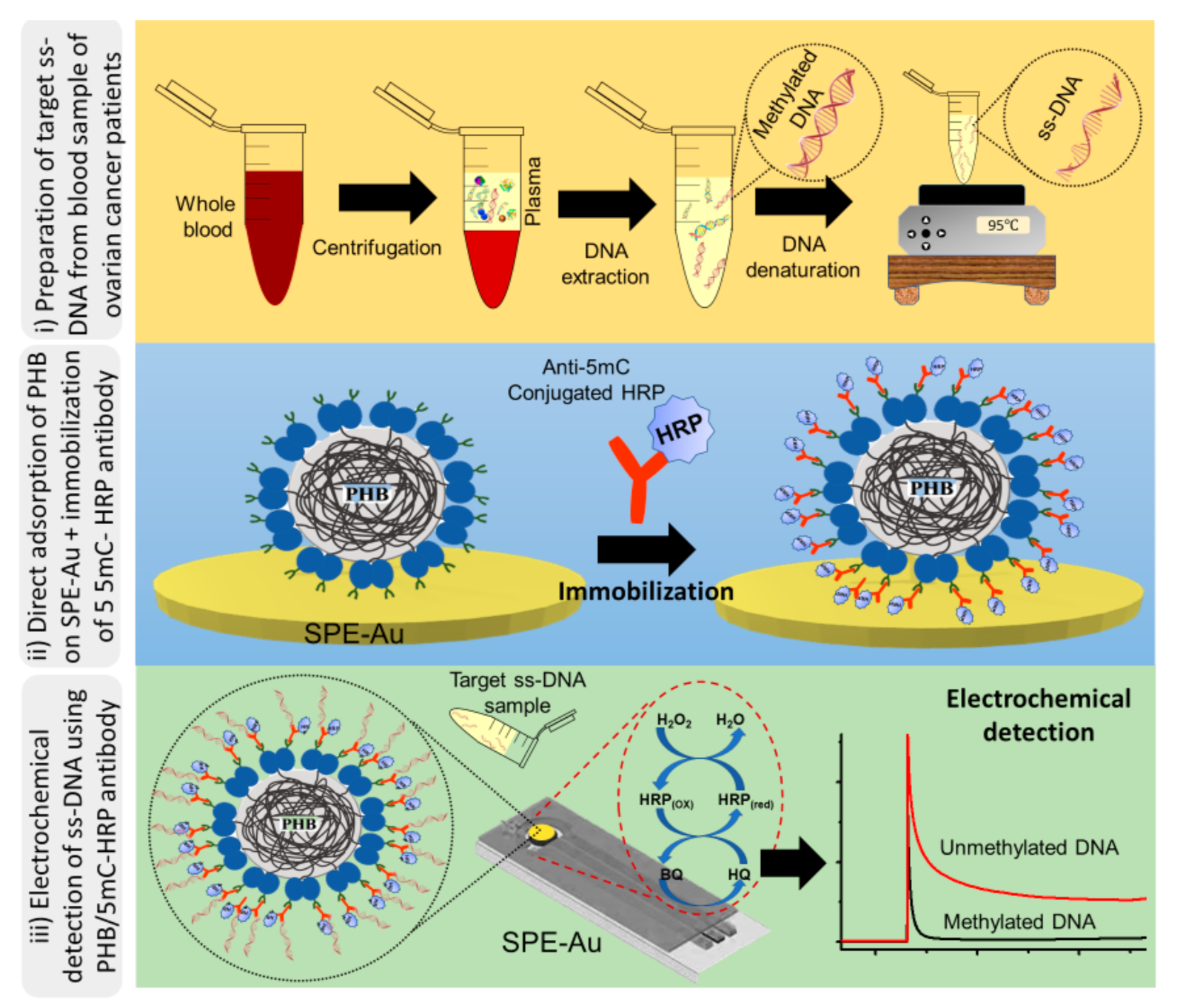
2.4. Electrochemical Detection of Methylated DNA Sequences
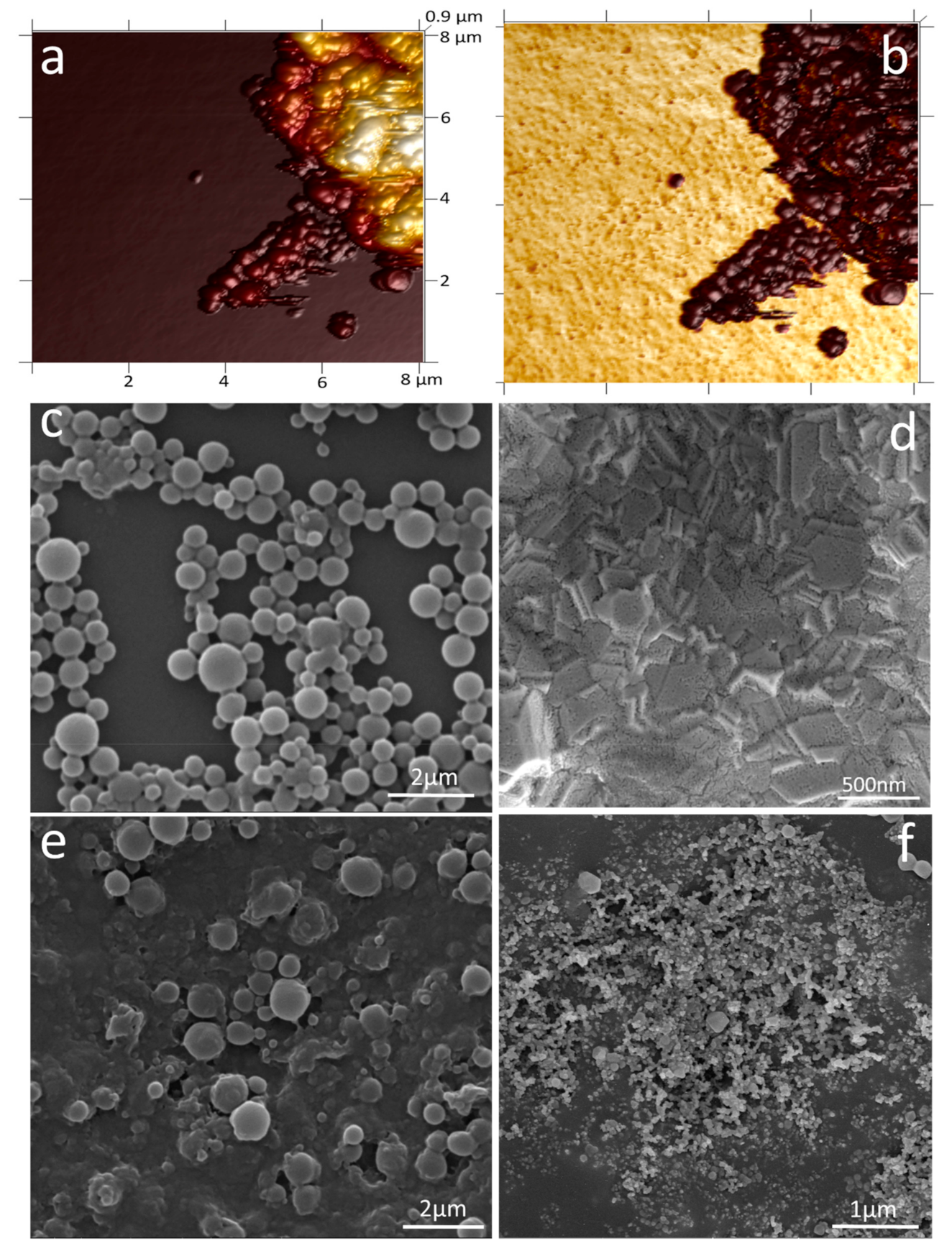
2.5. Preparation and Characterisation of PHB Nanobeads
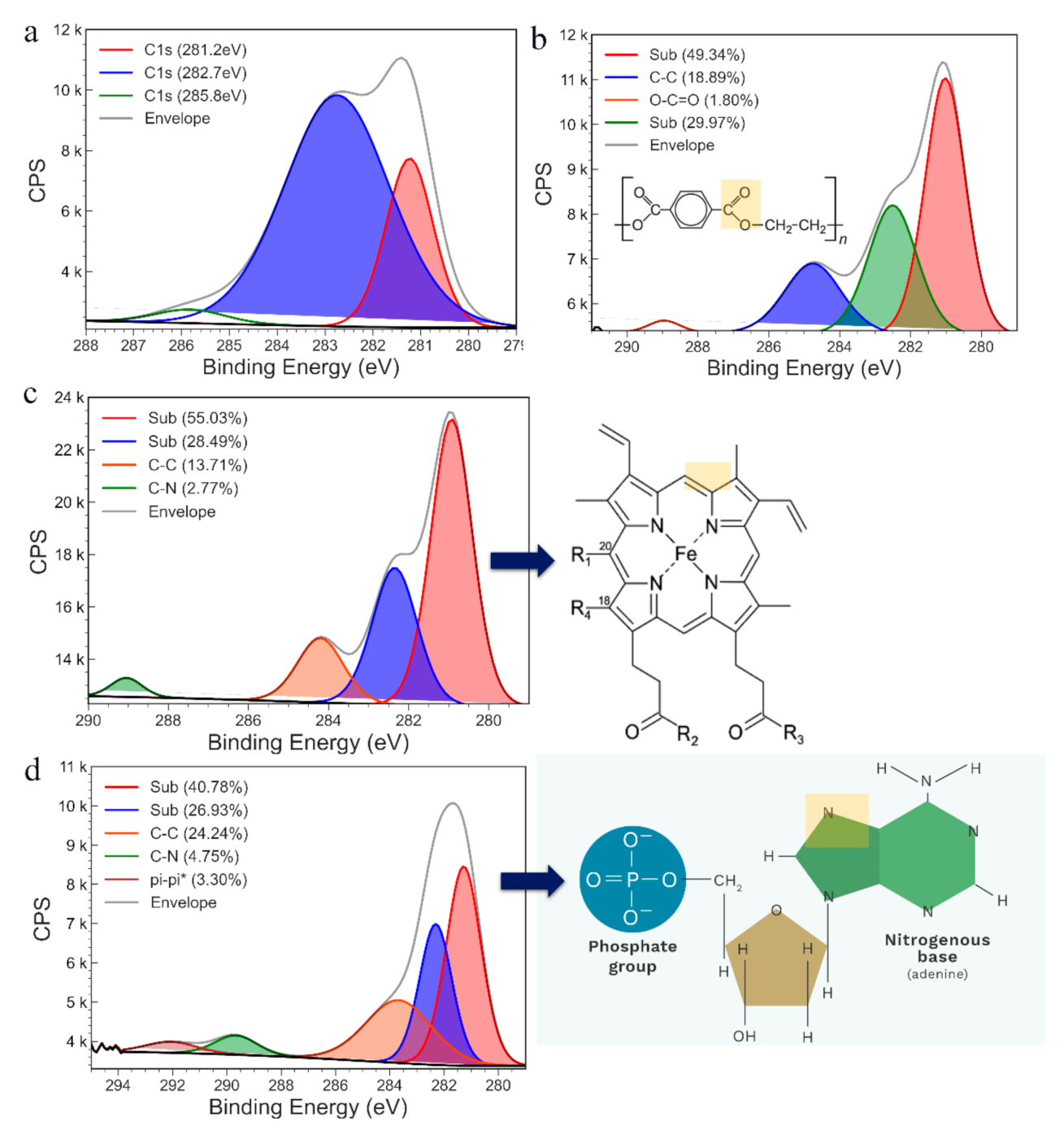
2.6. Electrode Surface Characterisation
2.7. Functionality of PHB Nanobeads Displaying ZZ Domains
3. Results and Discussion
3.1. Characterisation of PHB Nanobeads
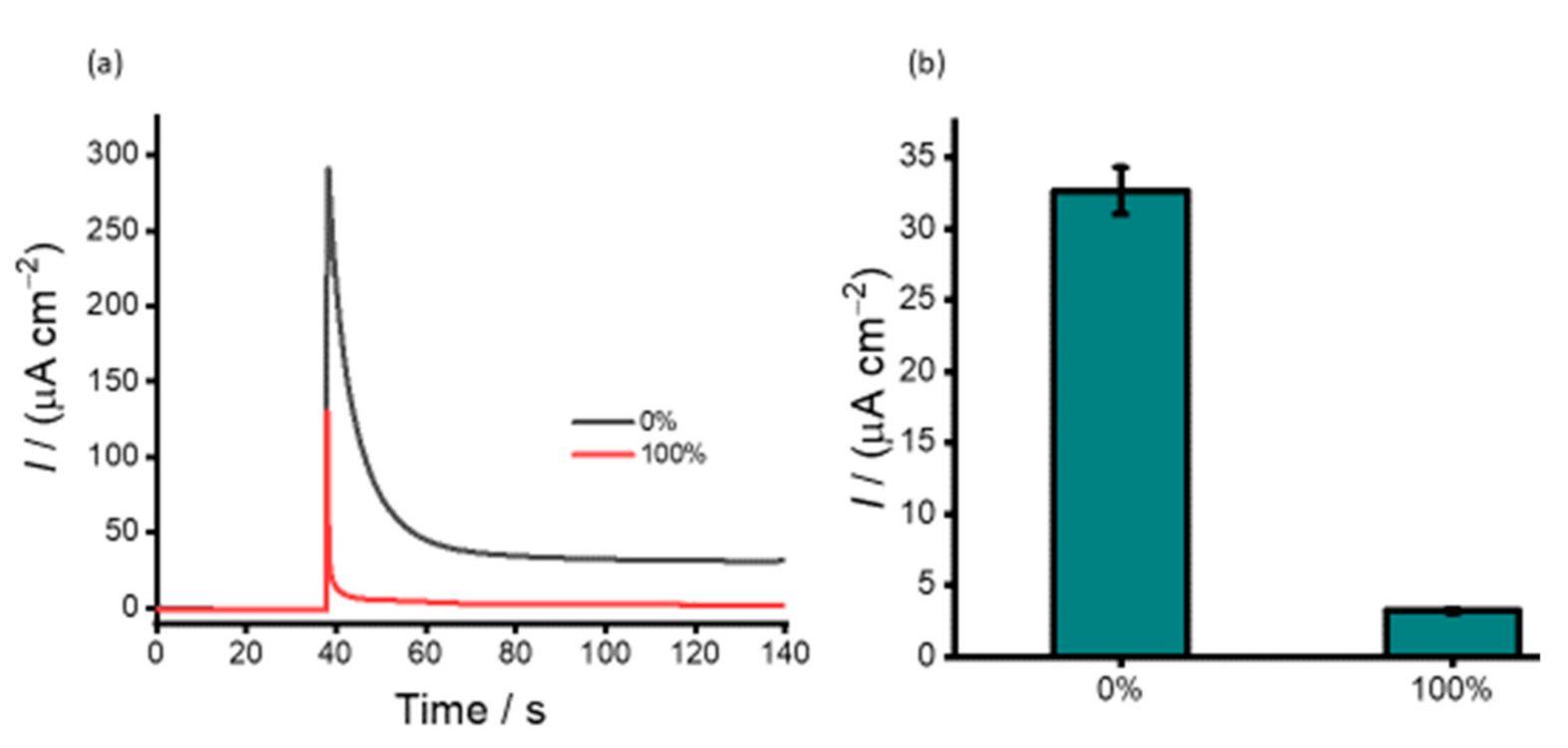
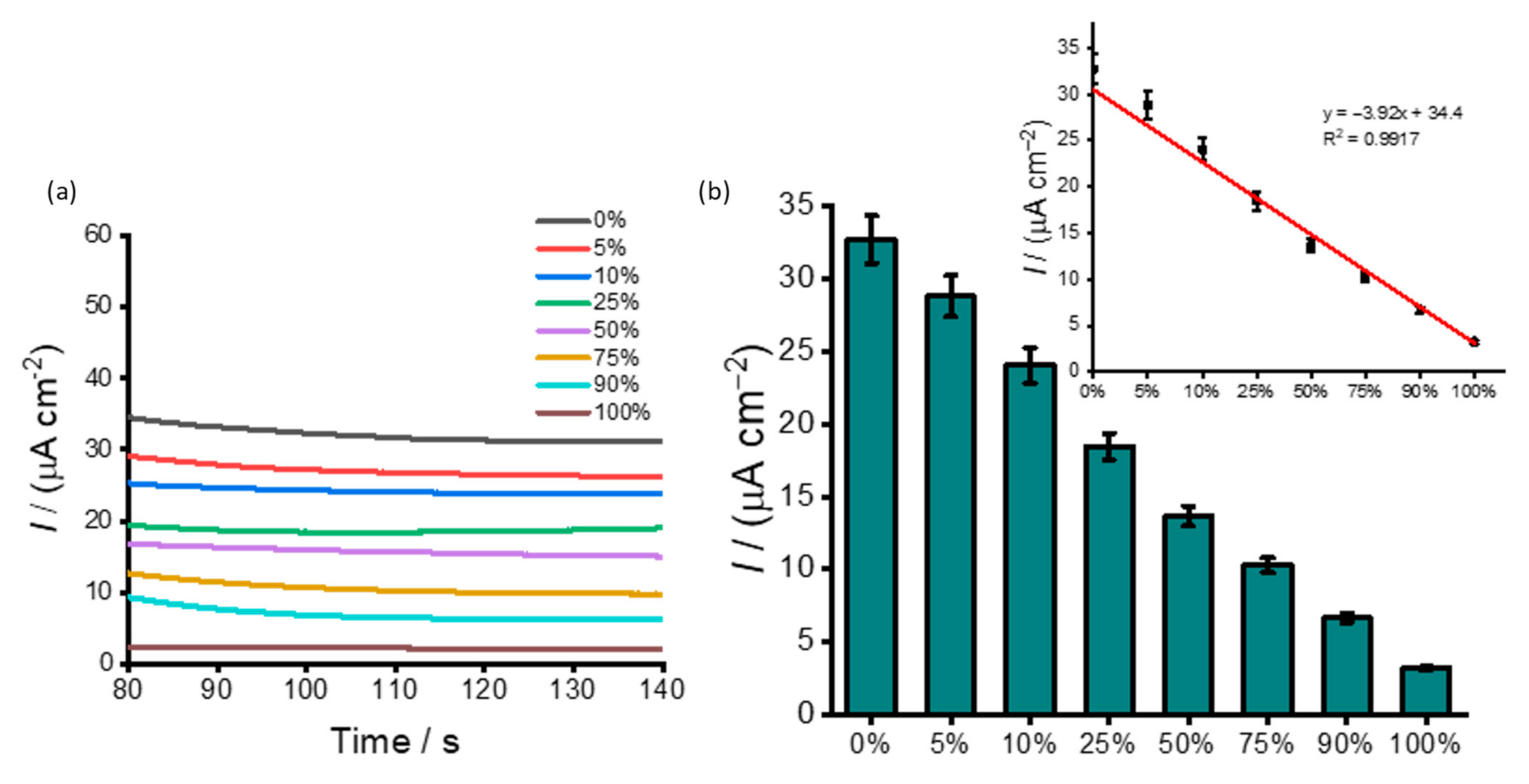
3.2. DNA Methylation Analysis in Heterogeneous Samples
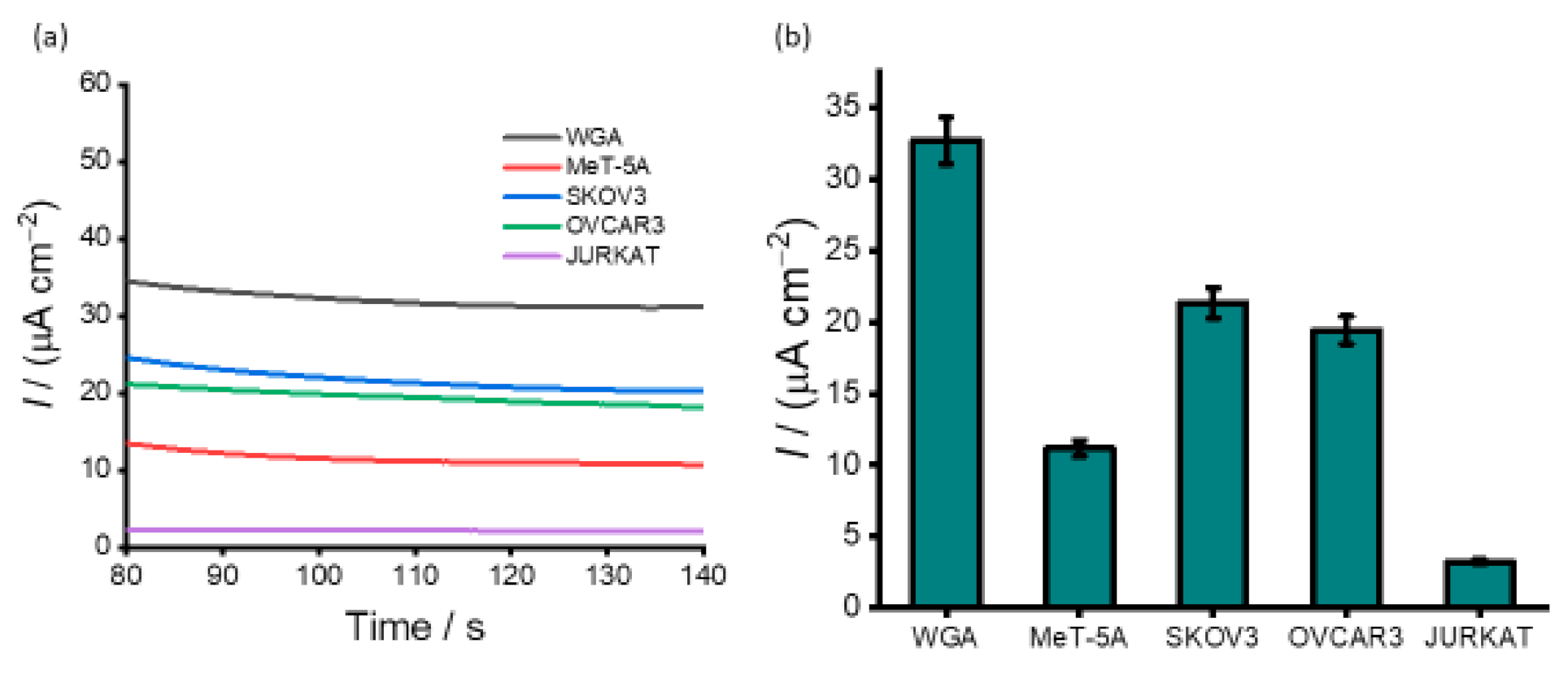
3.3. Detection of DNA Methylation in Cell Line Samples
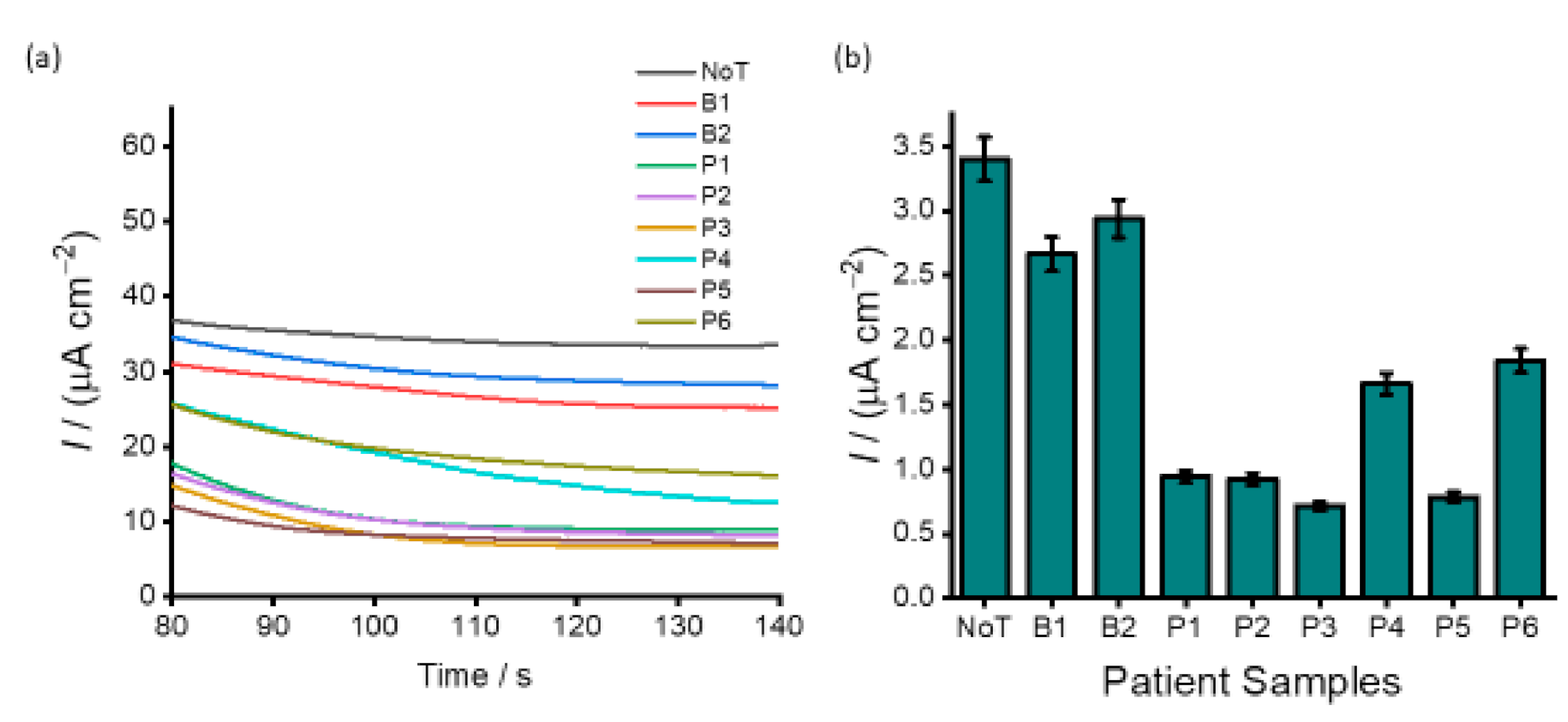
3.4. Analysis of DNA Methylation in Clinical Samples
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef]
- Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Robertson, K.D.; Jones, P.A. DNA methylation: Past, present and future directions. Carcinogenesis 2000, 21, 461–467. [Google Scholar] [CrossRef]
- Kulis, M.; Esteller, M. DNA methylation and cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar]
- Laird, P.W. The power and the promise of DNA methylation markers. Nat. Rev. Cancer 2003, 3, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.S.; Estécio, M.R.; Doshi, K.; Kondo, Y.; Tajara, E.H.; Issa, J.P.J. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004, 32, e38. [Google Scholar] [CrossRef]
- Xiong, Z.; Laird, P.W. COBRA: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997, 25, 2532–2534. [Google Scholar] [CrossRef] [PubMed]
- Kremer, D.; Metzger, S.; Kolb-Bachofen, V. Quantitative measurement of genome-wide DNA methylation by a reliable and cost-efficient enzyme-linked immunosorbent assay technique. Anal. Biochem. 2012, 422, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, K.M.; Bermingham, E.N.; Bassett, S.A.; Treloar, B.P.; Roy, N.C.; Barnett, M.P. Global DNA methylation measurement by HPLC using low amounts of DNA. Biotechnol. J. 2011, 6, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Stach, D.; Schmitz, O.J.; Stilgenbauer, S.; Benner, A.; Döhner, H.; Wiessler, M.; Lyko, F. Capillary electrophoretic analysis of genomic DNA methylation levels. Nucleic Acids Res. 2003, 31, e2. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, R.; Moriam, S.; Nguyen, N.-T.; Shiddiky, M.J. A bisulfite treatment and PCR-free global DNA methylation detection method using electrochemical enzymatic signal engagement. Biosens. Bioelectron. 2019, 126, 102–107. [Google Scholar] [CrossRef]
- Hossain, T.; Mahmudunnabi, R.G.; Masud, M.K.; Islam, N.; Ooi, L.; Konstantinov, K.; Hossain, S.; Martinac, B.; Alici, G.; Nguyen, N.-T.; et al. Electrochemical biosensing strategies for DNA methylation analysis. Biosens. Bioelectron. 2017, 94, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Laird, P.W. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 2010, 11, 191–203. [Google Scholar] [CrossRef]
- Soda, N.; Umer, M.; Kashaninejad, N.; Kasetsirikul, S.; Kline, R.; Salomon, C.; Nguyen, N.-T.; Shiddiky, M.J.A. PCR-free detection of long non-coding HOTAIR RNA in ovarian cancer cell lines and plasma samples. Cancers 2020, 12, 2233. [Google Scholar] [CrossRef]
- Hentze, J.L.; Høgdall, C.; Høgdall, E.V. Methylation and ovarian cancer: Can DNA methylation be of diagnostic use? (Review). Mol. Clin. Oncol. 2019, 10, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.L.; Nemeth, E.; Tran, H.; Shvartsman, H.; Cass, I.; Narod, S.; Karlan, B.Y. BRCA1 promoter region hypermethylation in ovarian carcinoma: A population-based study. Cancer Res. 2000, 60, 5329–5333. [Google Scholar] [PubMed]
- Wu, Q.; Lothe, R.A.; Ahlquist, T.; Silins, I.; Tropé, C.G.; Micci, F.; Nesland, J.M.; Suo, Z.; Lind, G.E. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol. Cancer 2007, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Rehm, B.; Steinbüchel, A. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int. J. Biol. Macromol. 1999, 25, 3–19. [Google Scholar] [CrossRef]
- Moradali, M.F.; Rehm, B.H.A. Bacterial biopolymers: From pathogenesis to advanced materials. Nat. Rev. Genet. 2020, 18, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.W.; Kim, H.W.; Kim, Y.B.; Rhee, Y.H. Poly (ethylene glycol)-grafted poly (3-hydroxyundecenoate) networks for enhanced blood compatibility. Int. J. Biol. Macromol. 2003, 32, 17–22. [Google Scholar] [CrossRef]
- Hazer, B.; Steinbüchel, A. Increased diversification of polyhydroxyalkanoates by modification reactions for industrial and medical applications. Appl. Microbiol. Biotechnol. 2007, 74, 1–12. [Google Scholar] [CrossRef]
- Legat, A.; Gruber, C.; Zangger, K.; Wanner, G.; Stan-Lotter, H. Identification of polyhydroxyalkanoates in Halococcus and other haloarchaeal species. Appl. Microbiol. Biotechnol. 2010, 87, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Parlane, N.A.; Gupta, S.; Rubio-Reyes, P.; Chen, S.; Gonzalez-Miro, M.; Wedlock, N.; Rehm, B.H.A. Self-assembled protein-coated polyhydroxyalkanoate beads: Properties and biomedical applications. ACS Biomater. Sci. Eng. 2016, 3, 3043–3057. [Google Scholar] [CrossRef]
- Lewis, J.D.; Meehan, R.; Henzel, W.; Maurer-Fogy, I.; Jeppesen, P.; Klein, F.; Bird, A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 1992, 69, 905–914. [Google Scholar] [CrossRef]
- Meehan, R.; Lewis, J.D.; McKay, S.; Kleiner, E.L.; Bird, A. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 1989, 58, 499–507. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, X.; Zhang, L.; Chen, D.; Dai, Z.; Zou, X. Simultaneous detection of double gene-specific methylation loci based on hairpin probes tagged with electrochemical quantum dots barcodes. J. Electroanal. Chem. 2016, 781, 356–362. [Google Scholar] [CrossRef]
- Ogura, K.; Rehm, B.H.A. Alginate encapsulation of bioengineered protein-coated polyhydroxybutyrate particles: A new platform for multifunctional composite materials. Adv. Funct. Mater. 2019, 29. [Google Scholar] [CrossRef]
- Grage, K.; McDermott, P.; Rehm, B.H.A. Engineering Bacillus megaterium for production of functional intracellular materials. Microb. Cell Factories 2017, 16, 1–12. [Google Scholar] [CrossRef]
- Lewis, J.G.; Rehm, B.H. ZZ polyester beads: An efficient and simple method for purifying IgG from mouse hybridoma supernatants. J. Immunol. Methods 2009, 346, 71–74. [Google Scholar] [CrossRef]
- Mifune, J.; Grage, K.; Rehm, B.H.A. Production of functionalized biopolyester granules by recombinant Lactococcus lactis. Appl. Environ. Microbiol. 2009, 75, 4668–4675. [Google Scholar] [CrossRef]
- Brockelbank, J.A.; Peters, V.; Rehm, B.H.A. Recombinant Escherichia coli strain produces a ZZ domain displaying biopolyester granules suitable for immunoglobulin G purification. Appl. Environ. Microbiol. 2006, 72, 7394–7397. [Google Scholar] [CrossRef]
- Schlosser, M.J.; Shurina, R.D.; Kalf, G.F. Prostaglandin H synthase catalyzed oxidation of hydroquinone to a sulfhydryl-binding and DNA-damaging metabolite. Chem. Res. Toxicol. 1990, 3, 333–339. [Google Scholar] [CrossRef]
- Puckett-Vaughn, D.L.; Stenken, J.A.; Scott, D.; Lunte, S.M.; Lunte, C.E. Enzymatic formation and electrochemical characterization of multiply substituted glutathione conjugates of hydroquinone. Life Sci. 1993, 52, 1239–1247. [Google Scholar] [CrossRef]
- Lévay, G.; Ross, D.; Bodell, W.J.; György, L. Peroxidase activation of hydroquinone results in the formation of DNA adducts in HL-60 cells, mouse bone marrow macrophages and human bone marrow. Carcinogenesis 1993, 14, 2329–2334. [Google Scholar] [CrossRef] [PubMed]
- Amara, A.A.; Rehm, B.H.A. Replacement of the catalytic nucleophile cysteine-296 by serine in class II polyhydroxyalkanoate synthase from Pseudomonas aeruginosa-mediated synthesis of a new polyester: Identification of catalytic residues. Biochem. J. 2003, 374, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Reyes, P.R.; Parlane, N.A.; Wedlock, N.; Rehm, B.H. Immunogencity of antigens from Mycobacterium tuberculosis self-assembled as particulate vaccines. Int. J. Med. Microbiol. 2016, 306, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Spiekermann, P.; Rehm, B.H.A.; Kalscheuer, R.; Baumeister, D.; Steinbüchel, A.J.A.O.M. A sensitive, viable colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 1999, 171, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Evert, B.; Vezina, B.; Rehm, B.H.A. Catalytically active bioseparation resin utilizing a covalent intermediate for tagless protein purification. ACS Appl. Bio Mater. 2020, 3, 8911–8922. [Google Scholar] [CrossRef]
- Lemgruber, R.D.S.P.; Valgepea, K.; Tappel, R.; Behrendorff, J.B.; Palfreyman, R.W.; Plan, M.; Hodson, M.P.; Simpson, S.D.; Nielsen, L.K.; Köpke, M.; et al. Systems-level engineering and characterisation of Clostridium autoethanogenum through heterologous production of poly-3-hydroxybutyrate (PHB). Metab. Eng. 2019, 53, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Jahns, A.C.; Maspolim, Y.; Chen, S.; Guthrie, J.M.; Blackwell, L.F.; Rehm, B.H.A. In vivo self-assembly of fluorescent protein microparticles displaying specific binding domains. Bioconjug. Chem. 2013, 24, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.-J.; Lin, D.-C.; Dinh, H.; Mayakonda, A.; Jiang, Y.-Y.; Chang, C.; Jiang, Y.; Lu, C.-C.; Shi, Z.-Z.; Xu, X.; et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat. Genet. 2016, 48, 1500–1507. [Google Scholar] [CrossRef]
- Mazor, T.; Pankov, A.; Song, J.; Costello, J.F. Intratumoral heterogeneity of the epigenome. Cancer Cell 2016, 29, 440–451. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Moriam, S.; Umer, M.; Nguyen, N.-T.; Shiddiky, M.J.A. DNA methylation detection: Recent developments in bisulfite free electrochemical and optical approaches. Analyst 2018, 143, 4802–4818. [Google Scholar] [CrossRef]
- Mikeska, T.; Candiloro, I.L.M.; Dobrovic, A. The implications of heterogeneous DNA methylation for the accurate quantification of methylation. Epigenomics 2010, 2, 561–573. [Google Scholar] [CrossRef]
- Candiloro, I.L.; Mikeska, T.; Dobrovic, A. Assessing combined methylation–sensitive high-resolution melting and pyrosequencing for the analysis of heterogeneous DNA methylation. Epigenetics 2011, 6, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Wee, E.J.H.; Ngo, T.H.; Trau, M. Colorimetric detection of both total genomic and loci-specific DNA methylation from limited DNA inputs. Clin. Epigenetics 2015, 7, 65. [Google Scholar] [CrossRef]
- Haque, H.; Bhattacharjee, R.; Islam, N.; Gopalan, V.; Nguyen, N.-T.; Lam, A.K.; Shiddiky, M.J.A. Colorimetric and electrochemical quantification of global DNA methylation using a methyl cytosine-specific antibody. Analyst 2017, 142, 1900–1908. [Google Scholar] [CrossRef]
- Ibn Sina, A.A.; Howell, S.; Carrascosa, L.G.; Rauf, S.; Shiddiky, M.J.A.; Trau, M. eMethylsorb: Electrochemical quantification of DNA methylation at CpG resolution using DNA–gold affinity interactions. Chem. Commun. 2014, 50, 13153–13156. [Google Scholar] [CrossRef] [PubMed]
- Soda, N.; Gonzaga, Z.J.; Chen, S.; Koo, K.M.; Nguyen, N.-T.; Shiddiky, M.J.A.; Rehm, B.H.A. Bioengineered polymer nanobeads for isolation and electrochemical detection of cancer biomarkers. ACS Appl. Mater. Interfaces 2021. [Google Scholar] [CrossRef] [PubMed]
- Kuo, K.C.; McCune, R.A.; Gehrke, C.W.; Midgett, R.; Ehrlich, M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980, 8, 4763–4776. [Google Scholar] [CrossRef]
- Singer, J.; Schnute, W.C.; Shively, J.E.; Todd, C.W.; Riggs, A.D. Sensitive detection of 5-methylcytosine and quantitation of the 5-methylcytosine/cytosine ratio in DNA by gas chromatography-mass spectrometry using multiple specific ion monitoring. Anal. Biochem. 1979, 94, 297–301. [Google Scholar] [CrossRef]
- Jiménez Sánchez, A. Characterisation of the Tumour Microenvironment in Ovarian Cancer. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2019. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soda, N.; Gonzaga, Z.J.; Pannu, A.S.; Kashaninejad, N.; Kline, R.; Salomon, C.; Nguyen, N.-T.; Sonar, P.; Rehm, B.H.A.; Shiddiky, M.J.A. Electrochemical Detection of Global DNA Methylation Using Biologically Assembled Polymer Beads. Cancers 2021, 13, 3787. https://doi.org/10.3390/cancers13153787
Soda N, Gonzaga ZJ, Pannu AS, Kashaninejad N, Kline R, Salomon C, Nguyen N-T, Sonar P, Rehm BHA, Shiddiky MJA. Electrochemical Detection of Global DNA Methylation Using Biologically Assembled Polymer Beads. Cancers. 2021; 13(15):3787. https://doi.org/10.3390/cancers13153787
Chicago/Turabian StyleSoda, Narshone, Zennia Jean Gonzaga, Amandeep Singh Pannu, Navid Kashaninejad, Richard Kline, Carlos Salomon, Nam-Trung Nguyen, Prashant Sonar, Bernd H. A. Rehm, and Muhammad J. A. Shiddiky. 2021. "Electrochemical Detection of Global DNA Methylation Using Biologically Assembled Polymer Beads" Cancers 13, no. 15: 3787. https://doi.org/10.3390/cancers13153787
APA StyleSoda, N., Gonzaga, Z. J., Pannu, A. S., Kashaninejad, N., Kline, R., Salomon, C., Nguyen, N.-T., Sonar, P., Rehm, B. H. A., & Shiddiky, M. J. A. (2021). Electrochemical Detection of Global DNA Methylation Using Biologically Assembled Polymer Beads. Cancers, 13(15), 3787. https://doi.org/10.3390/cancers13153787











