Pathophysiological Implications of Urinary Peptides in Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Study Design
2.3. Sample Preparation
2.4. CE-MS Analysis
2.5. CE-MS Data Processing
2.6. Support Vector Machine Model Generation and Classification
2.7. Peptide Sequencing
2.8. In Silico Protease Prediction
2.9. Immunohistochemistry
2.10. Statistics
3. Results
3.1. Identification of Urinary Peptides as HCC Progression Markers by CE-MS Analysis
3.2. Development of the 31 HCC Progression Markers to a Multivariate Classification Model
3.3. CE-MS and Peptide Sequence Characteristics of the Peptide Marker Candidates
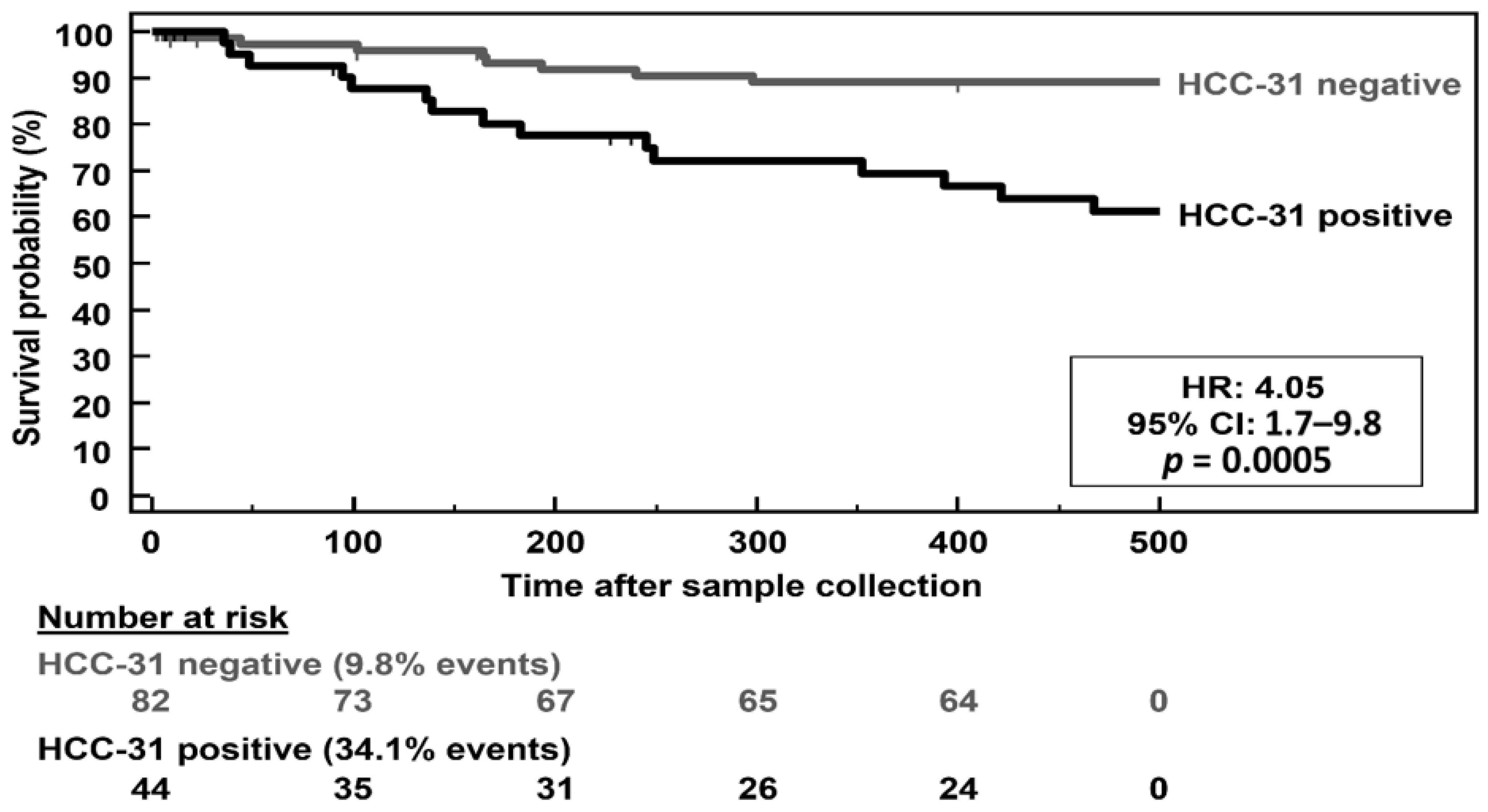
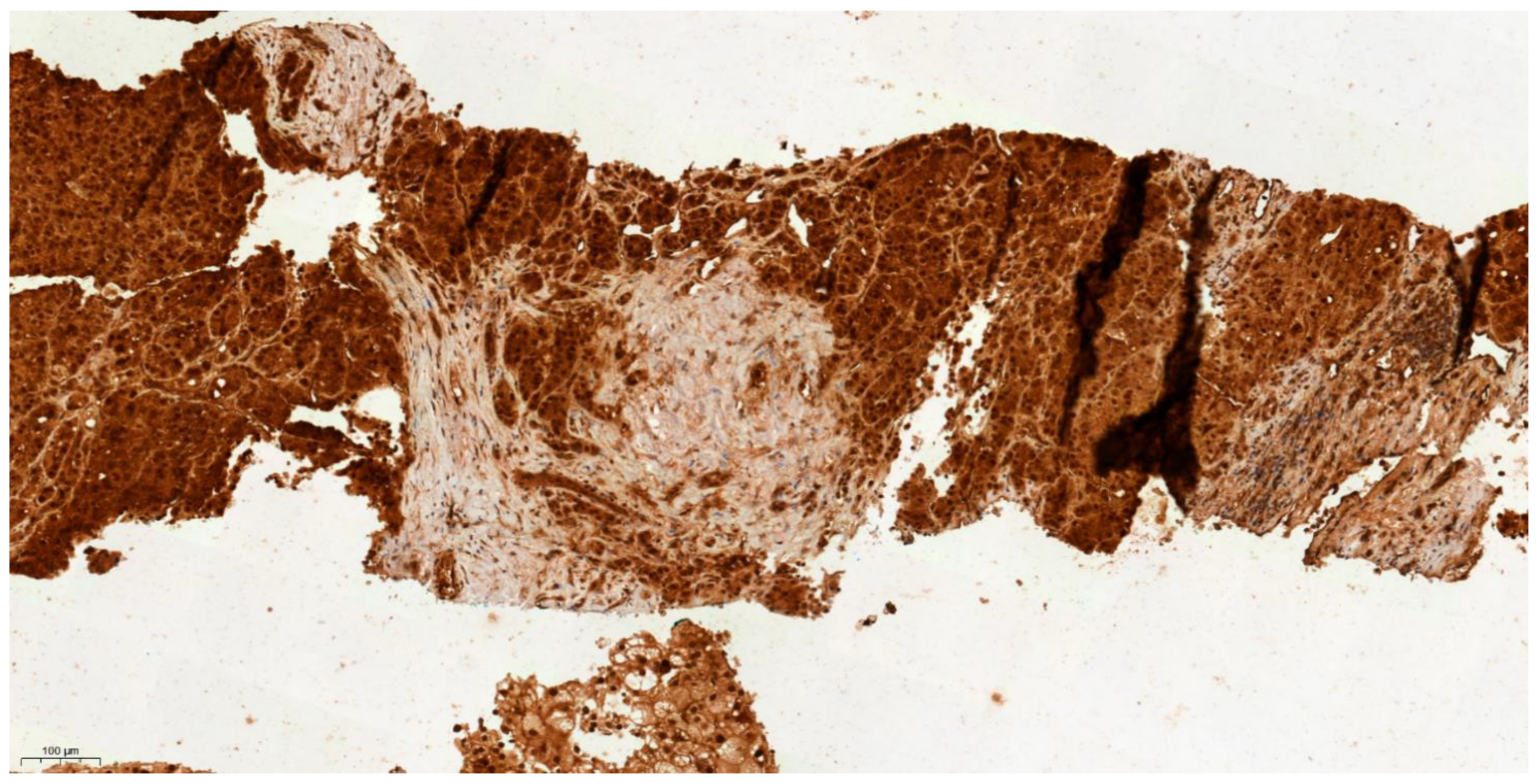
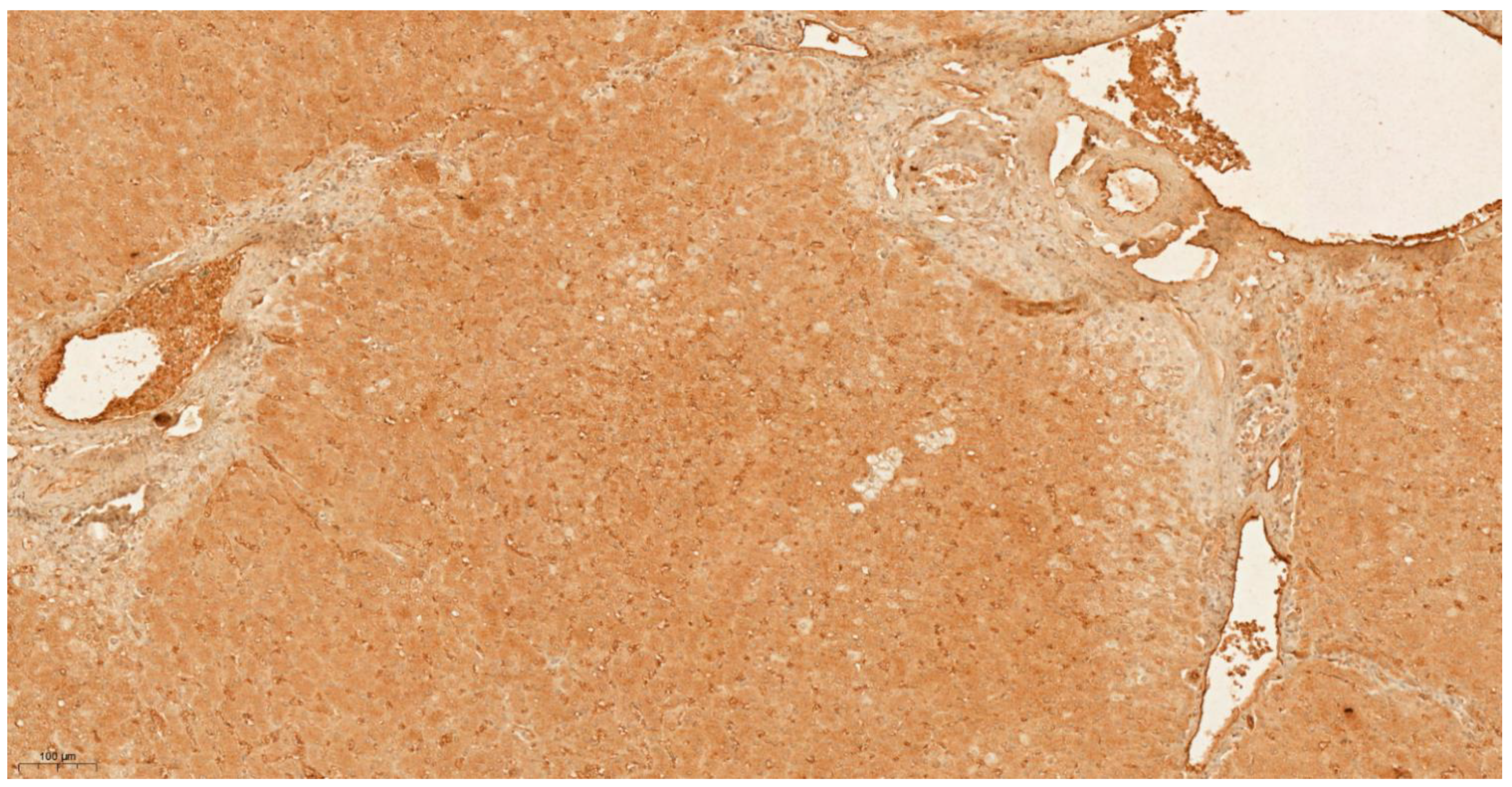
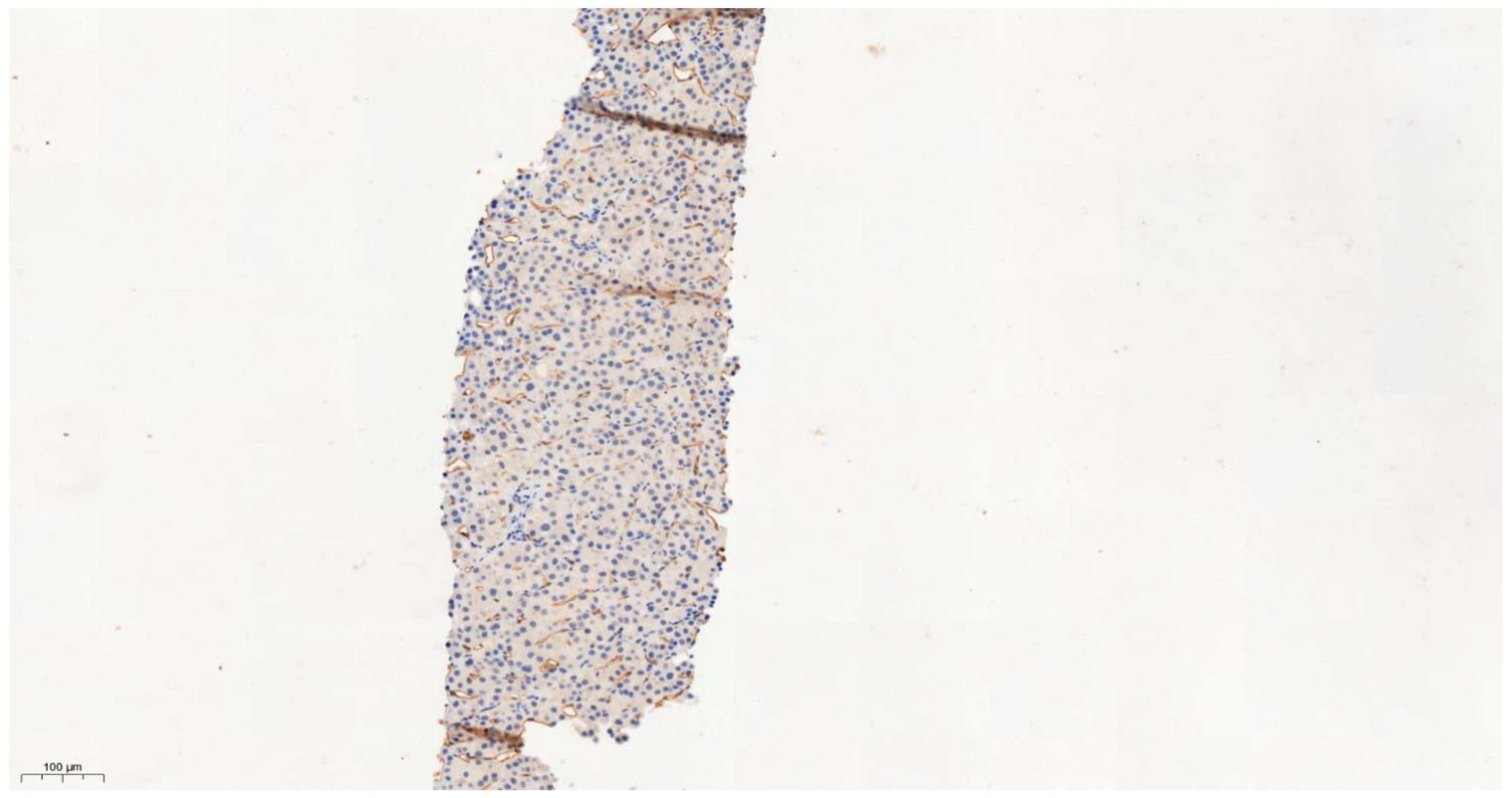
3.4. Differential Expression of KLK6 and MEP1A in HCC, Cirrhosis and Normal Liver Tissue
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [PubMed]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef]
- Schuppan, D.; Afdhal, N.H. Liver cirrhosis. Lancet 2008, 371, 838–851. [Google Scholar] [CrossRef]
- Silantyev, A.S.; Falzone, L.; Libra, M.; Gurina, O.I.; Kardashova, K.S.; Nikolouzakis, T.K.; Nosyrev, A.E.; Sutton, C.W.; Mitsias, P.D.; Tsatsakis, A. Current and Future Trends on Diagnosis and Prognosis of Glioblastoma: From Molecular Biology to Proteomics. Cells 2019, 8, 863. [Google Scholar] [CrossRef]
- Mittal, P.; Klingler-Hoffmann, M.; Arentz, G.; Zhang, C.; Kaur, G.; Oehler, M.K.; Hoffmann, P. Proteomics of endometrial cancer diagnosis, treatment, and prognosis. Proteom. Clin. Appl. 2016, 10, 217–229. [Google Scholar] [CrossRef]
- Li, B.; Li, B.; Guo, T.; Sun, Z.; Li, X.; Li, X.; Wang, H.; Chen, W.; Chen, P.; Qiao, M.; et al. Application Value of Mass Spectrometry in the Differentiation of Benign and Malignant Liver Tumors. Med. Sci. Monit. 2017, 23, 1636–1644. [Google Scholar] [CrossRef]
- Martins, A.M.A.; Garcia, J.H.P.; Eberlin, M.N. Mass Spectrometry as a Clinical Integrative Tool to Evaluate Hepatocellular Carcinoma: Moving to the Mainstream. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Latosinska, A.; Siwy, J.; Mischak, H.; Frantzi, M. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: The past, the present, and the future. Electrophoresis 2019, 40, 2294–2308. [Google Scholar] [CrossRef]
- Lindhardt, M.; Persson, F.; Oxlund, C.; Jacobsen, I.A.; Zürbig, P.; Mischak, H.; Rossing, P.; Heerspink, H.J.L. Predicting albuminuria response to spironolactone treatment with urinary proteomics in patients with type 2 diabetes and hypertension. Nephrol. Dial. Transplant. 2018, 33, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Weissinger, E.M.; Metzger, J.; Schleuning, M.; Schmid, C.; Messinger, D.; Beutel, G.; Wagner-Drouet, E.M.; Schetelig, J.; Baurmann, H.; Rank, A.; et al. A multicenter prospective, randomized, placebo-controlled phase II/III trial for preemptive acute graft-versus-host disease therapy. Leukemia 2021, 35, 1763–1772. [Google Scholar] [CrossRef] [PubMed]
- Tofte, N.; Lindhardt, M.; Adamova, K.; Bakker, S.J.L.; Beige, J.; Beulens, J.W.J.; Birkenfeld, A.L.; Currie, G.; Delles, C.; Dimos, I.; et al. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): A prospective observational study and embedded randomised placebo-controlled trial. Lancet Diabetes Endocrinol. 2020, 8, 301–312. [Google Scholar] [CrossRef]
- Bannaga, A.S.; Metzger, J.; Kyrou, I.; Voigtländer, T.; Book, T.; Melgarejo, J.; Latosinska, A.; Pejchinovski, M.; Staessen, J.A.; Mischak, H.; et al. Discovery, validation and sequencing of urinary peptides for diagnosis of liver fibrosis-A multicentre study. EBioMedicine 2020, 62, 103083. [Google Scholar] [CrossRef]
- Voigtländer, T.; Metzger, J.; Husi, H.; Kirstein, M.M.; Pejchinovski, M.; Latosinska, A.; Frantzi, M.; Mullen, W.; Book, T.; Mischak, H.; et al. Bile and urine peptide marker profiles: Access keys to molecular pathways and biological processes in cholangiocarcinoma. J. Biomed. Sci. 2020, 27, 13. [Google Scholar] [CrossRef] [PubMed]
- Lankisch, T.O.; Metzger, J.; Negm, A.A.; Vosskuhl, K.; Schiffer, E.; Siwy, J.; Weismüller, T.J.; Schneider, A.S.; Thedieck, K.; Baumeister, R.; et al. Bile proteomic profiles differentiate cholangiocarcinoma from primary sclerosing cholangitis and choledocholithiasis. Hepatology 2011, 53, 875–884. [Google Scholar] [CrossRef]
- Klein, J.; Eales, J.; Zurbig, P.; Vlahou, A.; Mischak, H.; Stevens, R. Proteasix: A tool for automated and large-scale prediction of proteases involved in naturally occurring peptide generation. Proteomics 2013, 13, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Metzger, J.; Kirsch, T.; Schiffer, E.; Ulger, P.; Mentes, E.; Brand, K.; Weissinger, E.M.; Haubitz, M.; Mischak, H.; Herget-Rosenthal, S. Urinary excretion of twenty peptides forms an early and accurate diagnostic pattern of acute kidney injury. Kidney Int. 2010, 78, 1252–1262. [Google Scholar] [CrossRef] [PubMed]
- Theodorescu, D.; Wittke, S.; Ross, M.M.; Walden, M.; Conaway, M.; Just, I.; Mischak, H.; Frierson, H.F. Discovery and validation of new protein biomarkers for urothelial cancer: A prospective analysis. Lancet Oncol. 2006, 7, 230–240. [Google Scholar] [CrossRef]
- Mischak, H.; Vlahou, A.; Ioannidis, J.P. Technical aspects and inter-laboratory variability in native peptide profiling: The CE-MS experience. Clin. Biochem. 2013, 46, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Neuhoff, N.; Kaiser, T.; Wittke, S.; Krebs, R.; Pitt, A.; Burchard, A.; Sundmacher, A.; Schlegelberger, B.; Kolch, W.; Mischak, H. Mass spectrometry for the detection of differentially expressed proteins: A comparison of surface-enhanced laser desorption/ionization and capillary electrophoresis/mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Jantos-Siwy, J.; Schiffer, E.; Brand, K.; Schumann, G.; Rossing, K.; Delles, C.; Mischak, H.; Metzger, J. Quantitative urinary proteome analysis for biomarker evaluation in kidney disease. J. Proteome Res. 2009, 8, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Papadopoulos, T.; Mischak, H.; Mullen, W. Comparison of CE-MS/MS and LCMS/ MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis 2014, 35, 1060–1064. [Google Scholar] [CrossRef]
- Pejchinovski, M.; Klein, J.; Ramírez-Torres, A.; Bitsika, V.; Mermelekas, G.; Vlahou, A.; Mullen, W.; Mischak, H.; Jankowski, V. Comparison of higher energy collisional dissociation and collision-induced dissociation MS/MS sequencing methods for identification of naturally occurring peptides in human urine. Proteom. Clin. Appl. 2015, 9, 531–542. [Google Scholar] [CrossRef]
- Zurbig, P.; Renfrow, M.B.; Schiffer, E.; Novak, J.; Walden, M.; Wittke, S.; Just, I.; Pelzing, M.; Neusüss, C.; Theodorescu, D.; et al. Biomarker discovery by CE-MS enables sequence analysis via MS/MS with platform-independent separation. Electrophoresis 2006, 27, 2111–2125. [Google Scholar] [CrossRef]
- Igarashi, Y.; Eroshkin, A.; Gramatikova, S.; Gramatikoff, K.; Zhang, Y.; Smith, J.W.; Osterman, A.L.; Godzik, A. CutDB: A proteolytic event database. Nucleic Acids Res. 2007, 35, 546–549. [Google Scholar] [CrossRef]
- Allred, D.C.; Harvey, J.M.; Berardo, M.; Clark, G.M. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998, 11, 155–168. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Gleiss, A.; Dakna, M.; Mischak, H.; Heinze, G. Two-group comparisons of zero-inflated intensity values: The choice of test statistic matters. Bioinformatics 2015, 14, 2310–2317. [Google Scholar] [CrossRef] [PubMed]
- Siwy, J.; Mullen, W.; Golovko, I.; Franke, J.; Zürbig, P. Human urinary peptide database for multiple disease biomarker discovery. Proteom. Clin. Appl. 2011, 5–6, 367–374. [Google Scholar] [CrossRef]
- Schönemeier, B.; Metzger, J.; Klein, J.; Husi, H.; Bremer, B.; Armbrecht, N.; Dakna, M.; Schanstra, J.; Rosendahl, J.; Wiegand, J.; et al. Urinary Peptide Analysis Differentiates Pancreatic Cancer from Chronic Pancreatitis. Pancreas 2016, 45, 1018–1026. [Google Scholar] [CrossRef]
- Debruyne, E.N.; Delanghe, J.R. Diagnosing and monitoring hepatocellular carcinoma with alpha-fetoprotein: New aspects and applications. Clin. Chim. Acta 2008, 395, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Marcu, A.; Bichmann, L.; Kuchenbecker, L.; Kowalewski, D.J.; Freudenmann, L.K.; Backert, L.; Mühlenbruch, L.; Szolek, A.; Lübke, M.; Wagner, P.; et al. HLA Ligand Atlas: A benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J. Immunother. Cancer. 2021, 9, e002071. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.Q.; Peng, L.H.; Ma, L.J.; Liu, D.B.; Zhang, S.; Luo, S.Z.; Rao, J.H.; Zhu, H.W.; Yang, S.X.; Xi, S.J.; et al. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J. Hepatol. 2020, 72, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, N.V.; Shornikova, A.Y.; Bugrova, A.E.; Baybakova, V.V.; Indeykina, M.I.; Kononikhin, A.S.; Popov, I.A.; Kechko, O.I.; Makarov, A.A.; Nikolaev, E.N. Evaluation of plasma peptides extraction methods by high-resolution mass spectrometry. Eur. J. Mass Spectrom. 2017, 23, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Saichi, N.; Takami, S.; Kang, D.; Toyama, A.; Daigo, Y.; Ishikawa, N.; Kohno, N.; Tamura, K.; Shuin, T.; et al. A comprehensive peptidome profiling technology for the identification of early detection biomarkers for lung adenocarcinoma. PLoS ONE 2011, 12, e18567. [Google Scholar] [CrossRef]
- Koomen, J.M.; Li, D.; Xiao, L.C.; Liu, T.C.; Coombes, K.R.; Abbruzzese, J.; Kobayashi, R. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J. Proteome Res. 2005, 4, 972–981. [Google Scholar] [CrossRef]
- Belogurov, A.A., Jr.; Ivanova, O.M.; Lomakin, Y.A.; Ziganshin, R.H.; Vaskina, M.I.; Knorre, V.D.; Klimova, E.A.; Gabibov, A.G.; Ivanov, V.T.; Govorun, V.M. Mediators and Biomarkers of Inflammation in Meningitis: Cytokine and Peptidome Profiling of Cerebrospinal Fluid. Biochemistry 2016, 81, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Mozdiak, E.; Metzger, J.; Pejchinovski, M.; Gopalakrishnan, K.; James, S.; Arasaradnam, R.P. P308 Exploratory study of urinary peptide markers in colorectal cancer and linkage to histopathology. Gut 2021, 70, A200. [Google Scholar]
- Wei, Y.; Van Nhieu, J.T.; Prigent, S.; Srivatanakul, P.; Tiollais, P.; Buendia, M.A. Altered expression of E-cadherin in hepa- tocellular carcinoma: Correlations with genetic alterations, beta-catenin expression, and clinical features. Hepatology 2002, 36, 692–701. [Google Scholar] [CrossRef]
- Peng, R.; Zhang, P.F.; Yang, X.; Wei, C.Y.; Huang, X.Y.; Cai, J.B.; Lu, J.C.; Gao, C.; Sun, H.X.; Gao, Q.; et al. Overexpression of RNF38 facilitates TGF-β signaling by Ubiquitinating and degrading AHNAK in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 113. [Google Scholar] [CrossRef]
- Htun, N.M.; Magliano, D.J.; Zhang, Z.Y.; Lyons, J.; Petit, T.; Nkuipou-Kenfack, E.; Ramirez-Torres, A.; von Zur Muhlen, C.; Maahs, D.; Schanstra, J.P.; et al. Prediction of acute coronary syndromes by urinary proteome analysis. PLoS ONE 2017, 12, e0172036. [Google Scholar] [CrossRef]
- Good, D.M.; Zürbig, P.; Argilés, A.; Bauer, H.W.; Behrens, G.; Coon, J.J.; Dakna, M.; Decramer, S.; Delles, C.; Dominiczak, A.F.; et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol. Cell Proteom. 2010, 9, 2424–2437. [Google Scholar] [CrossRef]
- Mischak, H.; Kolch, W.; Aivaliotis, M.; Bouyssié, D.; Court, M.; Dihazi, H.; Dihazi, G.H.; Franke, J.; Garin, J.; Gonzalez de Peredo, A.; et al. Comprehensive human urine standards for comparability and standardization in clinical proteome analysis. Proteom. Clin. Appl. 2010, 4, 464–478. [Google Scholar] [CrossRef]
- Amar, S.; Smith, L.; Fields, G.B. Matrix metalloproteinase collagenolysis in health and disease. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1940–1951. [Google Scholar] [CrossRef]
- Metzger, J.; Negm, A.A.; Plentz, R.R.; Weismüller, T.J.; Wedemeyer, J.; Karlsen, T.H.; Dakna, M.; Mullen, W.; Mischak, H.; Manns, M.P.; et al. Urine proteomic analysis differentiates cholangiocarcinoma from primary sclerosing cholangitis and other benign biliary disorders. Gut 2013, 62, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Frantzi, M.; Metzger, J.; Banks, R.E.; Husi, H.; Klein, J.; Dakna, M.; Mullen, W.; Cartledge, J.J.; Schanstra, J.P.; Brand, K.; et al. Discovery and validation of urinary biomarkers for detection of renal cell carcinoma. J. Proteom. 2014, 98, 44–58. [Google Scholar] [CrossRef] [PubMed]
- Belczacka, I.; Latosinska, A.; Metzger, J.; Marx, D.; Vlahou, A.; Mischak, H.; Frantzi, M. Proteomics biomarkers for solid tumors: Current status and future prospects. Mass Spectrom. Rev. 2019, 38, 49–78. [Google Scholar] [CrossRef]
- Frantzi, M.; van Kessel, K.; Zwarthoff, E.; Marquez, M.; Rava, M.; Malats, N.; Merseburger, A.; Katafigiotis, I.; Stravodimos, K.; Mullen, W.; et al. Development and Validation of Urine-based Peptide Biomarker Panels for Detecting Bladder Cancer in a Multicenter Study. Clin. Cancer Res. 2016, 22, 4077–4086. [Google Scholar] [CrossRef]
- Theodorescu, D.; Schiffer, E.; Bauer, H.W.; Douwes, F.; Eichhorn, F.; Polley, R.; Schmidt, T.; Schöfer, W.; Zürbig, P.; Good, D.M.; et al. Discovery and validation of urinary biomarkers for prostate cancer. Proteom. Clin. Appl. 2008, 2, 556–570. [Google Scholar] [CrossRef]
- Emami, N.; Diamandis, E.P. New insights into the functional mechanisms and clinical applications of the kallikrein-related peptidase family. Mol. Oncol. 2007, 1, 269–287. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Zhang, Y.; Fang, J.; Wilson, J.J.; Luo, S.; Huang, R.P. Development of Monoclonal Antibodies and Characterization of an ELISA Platform Against Kallikrein-Related Peptidase 6 as a Tumor Biomarker. Monoclon. Antibodies Immunodiagn. Immunother. 2015, 34, 346–353. [Google Scholar] [CrossRef]
- Petraki, C.; Dubinski, W.; Scorilas, A.; Saleh, C.; Pasic, M.D.; Komborozos, V.; Khalil, B.; Gabril, M.Y.; Streutker, C.; Diamandis, E.P.; et al. Evaluation and prognostic significance of human tissue kallikrein-related peptidase 6 (KLK6) in colorectal cancer. Pathol. Res. Pract. 2012, 208, 104–108. [Google Scholar] [CrossRef]
- Seiz, L.; Dorn, J.; Kotzsch, M.; Walch, A.; Grebenchtchikov, N.I.; Gkazepis, A.; Schmalfeldt, B.; Kiechle, M.; Bayani, J.; Diamandis, E.P.; et al. Stromal cell-associated expression of kallikrein-related peptidase 6 (KLK6) indicates poor prognosis of ovarian cancer patients. Biol. Chem. 2012, 393, 391–401. [Google Scholar] [CrossRef]
- Laouirem, S.; Le Faouder, J.; Alexandrov, T.; Mestivier, D.; Léger, T.; Baudin, X.; Mebarki, M.; Paradis, V.; Camadro, J.M.; Bedossa, P. Progression from cirrhosis to cancer is associated with early ubiquitin post-translational modifications: Identification of new biomarkers of cirrhosis at risk of malignancy. J. Pathol. 2014, 234, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, H.; Zhang, B.; Pan, X.; Zhu, X.; Ding, Y.; Li, G. Peptide network for detection of tissue-remodeling enzyme in the prognosis of hepatocellular carcinoma. ACS Appl. Mater. Interfaces 2015, 7, 4401–4405. [Google Scholar] [CrossRef]
- Sterchi, E.E.; Stocker, W.; Bond, J.S. Meprins, membrane-bound and secreted astacin metalloproteinases. Mol. Asp. Med. 2008, 29, 309–328. [Google Scholar] [CrossRef] [PubMed]
- Bond, J.S.; Matters, G.L.; Banerjee, S.; Dusheck, R.E. Meprin metalloprotease expression and regulation in kidney, intestine, urinary tract infections and cancer. FEBS Lett. 2005, 579, 3317–3322. [Google Scholar] [CrossRef]
- Lottaz, D.; Maurer, C.A.; Hahn, D.; Büchler, M.W.; Sterchi, E.E. Nonpolarized secretion of human meprin alpha in colorectal cancer generates an increased proteolytic potential in the stroma. Cancer Res. 1999, 59, 1127–1133. [Google Scholar] [PubMed]
- Breig, O.; Yates, M.; Neaud, V.; Couchy, G.; Grigoletto, A.; Lucchesi, C.; Prox, J.; Zucman-Rossi, J.; Becker-Pauly, C.; Rosenbaum, J. Metalloproteinase meprin α regulates migration and invasion of human hepatocarcinoma cells and is a mediator of the oncoprotein Reptin. Oncotarget 2017, 8, 7839–7851. [Google Scholar] [CrossRef][Green Version]
- OuYang, H.Y.; Xu, J.; Luo, J.; Zou, R.H.; Chen, K.; Le, Y.; Zhang, Y.F.; Wei, W.; Guo, R.P.; Shi, M. MEP1A contributes to tumor progression and predicts poor clinical outcome in human hepatocellular carcinoma. Hepatology 2016, 63, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Peters, F.; Scharfenberg, F.; Colmorgen, C.; Armbrust, F.; Wichert, R.; Arnold, P.; Potempa, B.; Potempa, J.; Pietrzik, C.U.; Häsler, R.; et al. Tethering soluble meprin α in an enzyme complex to the cell surface affects IBD-associated genes. FASEB J. 2019, 33, 7490–7504. [Google Scholar] [CrossRef]
- Tran, P.L.; Vigneron, J.P.; Pericat, D.; Dubois, S.; Cazals, D.; Hervy, M.; DeClerck, Y.A.; Degott, C.; Auclair, C. Gene therapy for hepatocellular carcinoma using non-viral vectors composed of bis guanidinium-tren-cholesterol and plasmids encoding the tissue inhibitors of metalloproteinases TIMP-2 and TIMP-3. Cancer Gene Ther. 2003, 10, 435–444. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Droga-Mazovec, G.; Bojic, L.; Petelin, A.; Ivanova, S.; Romih, R.; Repnik, U.; Salvesen, G.S.; Stoka, V.; Turk, V.; Turk, B. Cysteine cathepsins trigger caspase-dependent cell death through cleavage of bid and antiapoptotic Bcl-2 homologues. J. Biol. Chem. 2008, 283, 19140–19150. [Google Scholar] [CrossRef] [PubMed]


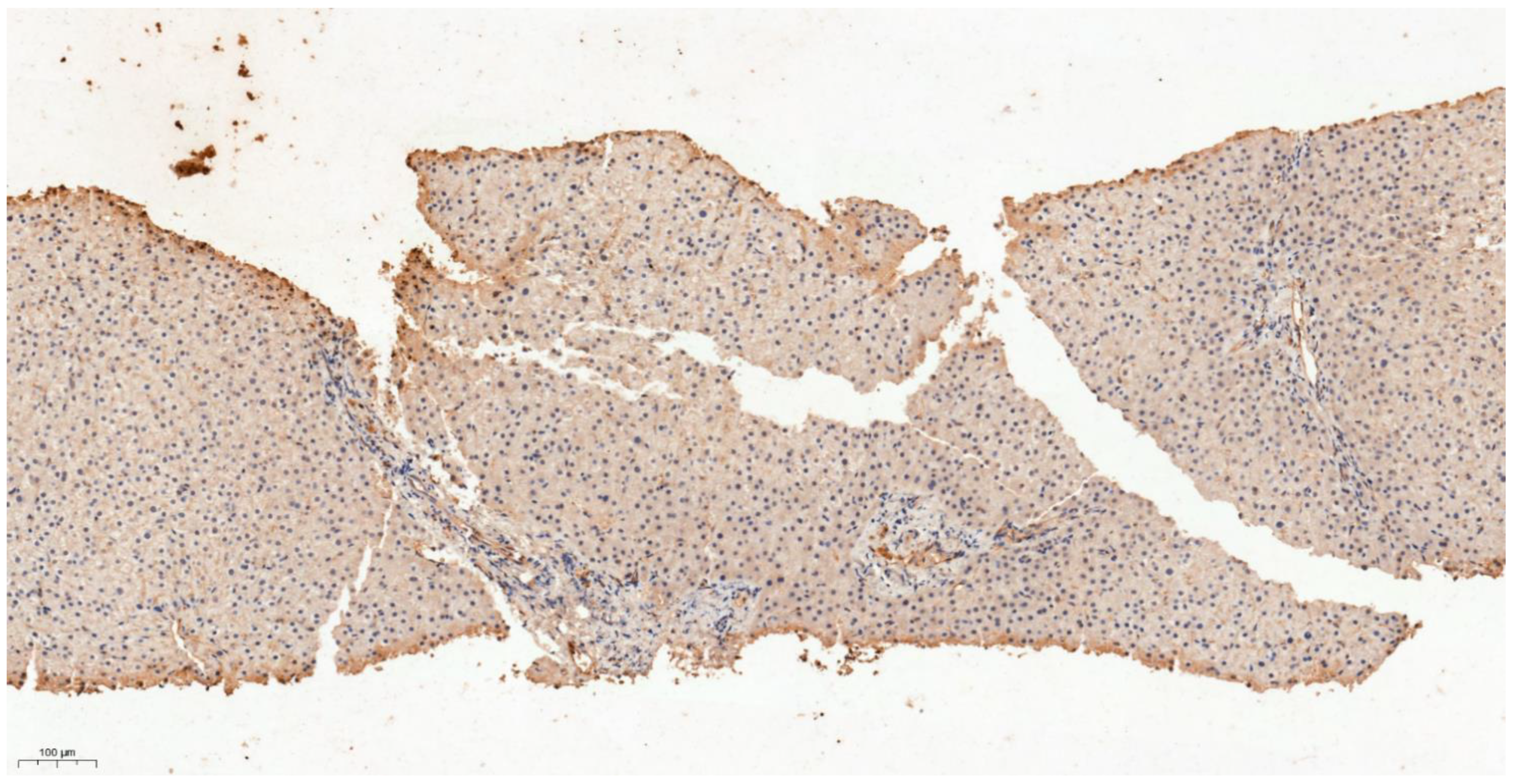
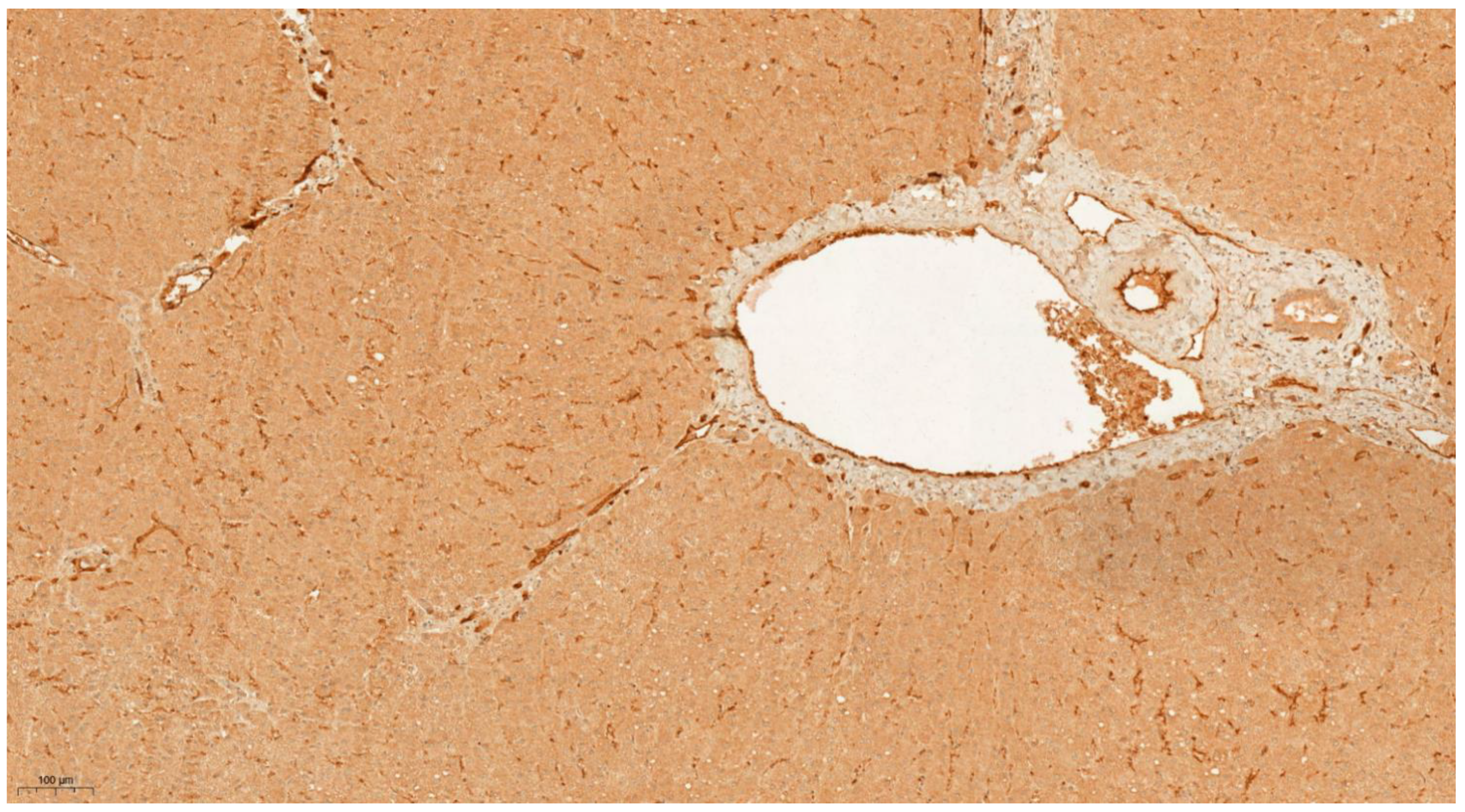
| Study Phase | Discovery | Validation | |||||
|---|---|---|---|---|---|---|---|
| Patient Group | HCC | Non-HCC | p * | HCC | Non-HCC | p * | |
| Patients/samples, n | 18/18 | 51/51 | n.a. | 39/39 | 87/87 | n.a. | |
| Age, years, mean/range | 58/28–76 | 52/18–82 | 0.08 | 67/38–87 | 56/20–85 | 0.0001 | |
| Female/male, n | 3/15 | 20/31 | 0.14 | 9/30 | 35/52 | 0.07 | |
| No. detected peptides, mean/range | 1753/623–2965 | 2329/920–4488 | 0.02 | 2872/1073–4617 | 2461/811–4057 | 0.008 | |
| Liver cirrhosis, n (%) | 18 (100) | 25 (49) | <0.0001 | 28 (72) | 47 (54) | 0.08 | |
| Diabetes mellitus, n (%) | 3 (17) | 13 (25) | 0.53 | 16 (41) | 23 (26) | 0.14 | |
| Body mass index, mean/range | 26.8/18.3–32.1 | 27.8/20.3–41.7 | 0.76 | 27.7/20.2–41.0 | 27.5/16.4–46.6 | 0.52 | |
| Platelet count, ×109/L, mean/range | 99/26–310 | 162/25–391 | 0.02 | 195/28–595 | 234/44–961 | 0.04 | |
| a-Fetoprotein (AFP), µg/L, mean/range | 1821/3–22,826 | 4/1–50 | <0.0001 | 6669/1–107,202 | 39/1–1493 | <0.0001 | |
| Alkaline phosphatase, U/L, mean/range | 151/60–380 | 125/45–797 | 0.008 | 291/83–1781 | 196/37–693 | 0.02 | |
| Aspartate aminotransferase (AST), U/L, mean/range | 74/24–284 | 49/13–120 | 0.03 | 112/26–457 | 69/13–1606 | <0.0001 | |
| Alanine aminotransferase (ALT), U/L, mean/range | 61/14–288 | 53/10–304 | 0.84 | 69/12–242 | 72/7–1038 | 0.37 | |
| AST:ALT ratio, mean/range | 1.48/0.89–3.41 | 1.17/0.20–3.20 | 0.05 | 2.11/0.55–7.46 | 1.13/0.18–3.06 | 0.0004 | |
| Approx. Ishak Fibrosis Score as per FIB-4 index, n, 0–1/2–3/4–6 | 0/1/17 | 14/12/17 | 0.0004 | 2/9/28 | 36/28/23 | <0.0001 | |
| Albumin, g/L, mean/range | 32/19–48 | 40/26–51 | 0.0003 | 34/16–48 | 38/15–70 | 0.01 | |
| Bilirubin, µmol/L, mean/range | 27/9–86 | 26/3–163 | 0.05 | 43/6–254 | 34/3–390 | 0.03 | |
| ALBI stage, n, 1/2/3 | 2/11/5 | 26/15/4 | 0.003 | 11/20/8 | 47/22/18 | 0.009 | |
| Liver Disease Etiology, n | |||||||
| Primary HCC | 0 | 0 | 1 | 0 | |||
| Alcoholic cirrhosis (C2) | 7 | 4 | 7 | 6 | |||
| Virus-related cirrhosis (HBV/HCV/HDV) | 3/4/0 | 0/4/1 | 3/4/0 | 2/2/0 | |||
| Cryptogenic/Biliary cirrhosis | 0/0 | 3/1 | 1/0 | 2/0 | |||
| Hereditary (Mucoviscidosis/Hemochromatosis/AATD) | 1/1/0 | 0/0/0 | 1/0/1 | 0/0/0 | |||
| Cholestasis (PBC/PSC/SSC/PFIC) | 2/0/0/0 | 2/2/1/1 | 2/5/0/0 | 2/14/1/0 | |||
| Autoimmune hepatitis (AIH) | 0 | 2 | 0 | 0 | |||
| NAFLD/NASH/NASH-LC | 0/0/0 | 9/8/4 | 0/2/12 | 18/6/15 | |||
| GI cancer (CCA/PCA) with LC | 0/0 | 0/0 | 0/0 | 7/2 | |||
| Other benign liver diseases (CHP/CDL) | 0/0 | 0/0 | 0/0 | 7/3 | |||
| No liver disease | 0 | 9 | 0 | 0 | |||
| Peptide ID † | Group-Wise Comparison § of Peptide Distributions HCC (n = 18) vs. Non-HCC Liver Disease (n = 51) | Rank Correlation of Peptide Amplitudes to a Grading Score 0 = Non-LC #, 1 = LC & 2 = HCC | Peptide Distribution in the Discovery Patient Groups | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Non-LC # (n = 26) | LC (n = 25) | HCC (n = 18) | ||||||||
| p-Value ‡ for Group Differences | AUC for Group Differences | Spearman Rho Coef. | p-Value ‡ for Rank Differences | Mean Amp (SD) | Freq. | Mean Amp (SD) | Freq. | Mean Amp (SD) | Freq. | |
| 54 | 8.34 × 10−03 | 0.70 | 0.353 | 1.43 × 10−02 | 2 (10) | 4 | 7 (28) | 12 | 53 (85) | 56 |
| 1059 | 4.57 × 10−02 | 0.63 | 0.303 | 4.06 × 10−02 | 4 (18) | 8 | 3 (13) | 12 | 30 (55) | 33 |
| 1160 | 4.23 × 10−02 | 0.63 | 0.431 | 1.83 × 10−03 | 1 (4) | 8 | 34 (46) | 48 | 61 (81) | 67 |
| 1778 | 1.50 × 10−02 | 0.65 | 0.407 | 3.66 × 10−03 | 5 (26) | 4 | 18 (34) | 32 | 29 (40) | 50 |
| 2314 | 2.92 × 10−02 | 0.65 | 0.385 | 6.52 × 10−03 | 1911 (3077) | 42 | 2696 (2346) | 80 | 3017 (2222) | 94 |
| 3559 | 1.30 × 10−02 | 0.63 | 0.396 | 4.97 × 10−03 | 47 (88) | 31 | 184 (223) | 60 | 201 (224) | 67 |
| 3662 | 8.27 × 10−04 | 0.76 | 0.489 | 3.08 × 10−04 | 76 (91) | 62 | 161 (188) | 76 | 256 (177) | 89 |
| 4564 | 6.81 × 10−03 | 0.72 | 0.479 | 4.18 × 10−04 | 270 (130) | 92 | 639 (579) | 96 | 591 (449) | 94 |
| 4610 | 2.50 × 10−03 | 0.65 | 0.398 | 4.74 × 10−03 | 0 (0) | 0 | 64 (157) | 16 | 166 (272) | 39 |
| 4829 | 1.54 × 10−02 | 0.66 | 0.409 | 3.49 × 10−03 | 59 (142) | 27 | 277 (358) | 76 | 245 (286) | 67 |
| 5284 | 5.61 × 10−03 | 0.70 | 0.362 | 1.15 × 10−02 | 63 (102) | 42 | 81 (137) | 44 | 172 (174) | 67 |
| 6601 | 3.57 × 10−06 | 0.72 | 0.470 | 5.54 × 10−04 | 0 (0) | 0 | 1 (4) | 4 | 199 (286) | 44 |
| 6607 | 8.67 × 10−04 | 0.65 | 0.380 | 7.39 × 10−03 | 0 (0) | 0 | 19 (83) | 8 | 259 (737) | 33 |
| 8720 | 6.37 × 10−04 | 0.75 | −0.587 | 8.05 × 10−06 | 658 (664) | 73 | 231 (641) | 48 | 31 (98) | 11 |
| 9080 | 3.34 × 10−02 | 0.65 | 0.391 | 5.69 × 10−03 | 813 (2125) | 15 | 893 (1236) | 48 | 1618 (1815) | 61 |
| 9510 | 5.03 × 10−03 | 0.72 | 0.370 | 9.56 × 10−03 | 2227 (4587) | 31 | 2847 (4477) | 68 | 4563 (3380) | 83 |
| 9728 | 1.90 × 10−03 | 0.73 | 0.511 | 1.51 × 10−04 | 345 (745) | 23 | 902 (1038) | 56 | 1359 (916) | 89 |
| 10177 | 3.24 × 10−03 | 0.71 | 0.358 | 1.29 × 10−02 | 162 (288) | 38 | 170 (224) | 48 | 476 (376) | 89 |
| 11725 | 6.63 × 10−04 | 0.65 | 0.385 | 6.64 × 10−03 | 0 (0) | 0 | 4 (12) | 8 | 28 (50) | 33 |
| 12459 | 8.13 × 10−06 | 0.86 | −0.526 | 8.68 × 10−05 | 373 (152) | 96 | 335 (236) | 92 | 119 (124) | 61 |
| 13134 | 7.85 × 10−04 | 0.73 | 0.311 | 3.45 × 10−02 | 32 (73) | 27 | 170 (375) | 24 | 364 (411) | 67 |
| 13176 | 1.20 × 10−03 | 0.73 | 0.410 | 3.47 × 10−03 | 27 (54) | 27 | 103 (190) | 40 | 171 (155) | 72 |
| 14389 | 5.32 × 10−04 | 0.74 | −0.400 | 4.53 × 10−03 | 53 (64) | 54 | 20 (37) | 40 | 0 (0) | 0 |
| 14925 | 7.69 × 10−04 | 0.73 | −0.567 | 1.82 × 10−05 | 689 (1120) | 58 | 237 (627) | 32 | 0 (0) | 0 |
| 15342 | 1.38 × 10−04 | 0.80 | −0.604 | 3.66 × 10−06 | 766 (308) | 100 | 420 (323) | 76 | 224 (219) | 78 |
| 17066 | 1.33 × 10−04 | 0.66 | 0.397 | 4.89 × 10−03 | 0 (3) | 4 | 8 (53) | 4 | 257 (693) | 33 |
| 17805 | 8.75 × 10−04 | 0.75 | −0.514 | 1.37 × 10−04 | 394 (478) | 69 | 235 (349) | 52 | 16 (36) | 22 |
| 19681 | 5.57 × 10−04 | 0.76 | −0.630 | 1.14 × 10−06 | 127 (129) | 85 | 34 (53) | 40 | 9 (21) | 17 |
| 20237 | 1.82 × 10−04 | 0.79 | 0.483 | 3.69 × 10−04 | 292 (480) | 35 | 316 (635) | 56 | 940 (1064) | 94 |
| 24328 | 2.07 × 10−05 | 0.84 | −0.636 | 8.84 × 10−07 | 5557 (3207) | 100 | 2341 (2674) | 100 | 890 (1187) | 72 |
| 29919 | 1.20 × 10−02 | 0.69 | 0.465 | 6.63 × 10−04 | 199 (505) | 23 | 525 (661) | 60 | 857 (1223) | 78 |
| Peptide ID † | Exp. Mass [Da] | CE Time [min] | Protein | AA | Proteases for N-Terminal Cleavage | Peptide Sequence (Black) with Flanking Regions (Grey) ‡ | Proteases for C-Terminal Cleavage |
|---|---|---|---|---|---|---|---|
| 54 | 807.39 | 23.2 | CLU | 366–371 | --- | LNEQ|FNWVSR|LANL | --- |
| 1059 | 920.34 | 21.2 | --- | --- | --- | --- | --- |
| 1160 | 928.51 | 24.6 | UMOD | 592–599 | MEP1A, MMP3 | RSGS|VIDQSRVL|NLGP | MEP1A, KLK6, CTS [B,D,E] |
| 1778 | 981.49 | 24.4 | ACTB | 107–115 | MEP1A, MMP [3,13], CTS [B,D,E] | HPVL|LTEAPLNPK|ANRE | --- |
| 2314 | 1032.45 | 25.9 | ACTB | 95–103 | --- | YNEL|RVAPEEHPV|LLTE | MEP1A, MMP [3,13], CTS [B,D,E] |
| 3559 | 1134.59 | 24.0 | COL3A1 | 755–766 | --- | ADGV|PGKDGPRGPTGP|IGPP | --- |
| 3662 | 1142.55 | 22.0 | ADGRF3 | 56–64 | --- | DKAW|NERIDRPFP|ACPI | --- |
| 4564 | 1199.55 | 20.8 | TKT | 343–352 | --- | DGDT|KNSTFSEIFK|KEHP | --- |
| 4610 | 1201.53 | 24.9 | GAGE12H | 58–70 | MEP1A, MMP3, CTSB | AAAQ|KGEDEGASAGQGP|KPEA | MMP3 |
| 4829 | 1217.48 | 27.7 | COL3A1 | 179–191 | --- | PAGP|pGPpGPpGTSGHp|GSPG | KLK6, CTSB |
| 5284 | 1250.64 | 20.6 | HBB | 136–147 | --- | QKVV|AGVANALAHKYH | C-terminal end |
| 6601 | 1352.78 | 22.0 | AHNAK | 772–784 | --- | EVDV|NLPKADVDISGPK|IDVT | MEP1A, KLK6 |
| 6607 | 1353.53 | 23.8 | FGA | 605–617 | MEP1A | YKMA|DEAGSEADHEGTH|STKR | --- |
| 8720 | 1513.62 | 29.5 | CDH1 | 397–410 | --- | ITTL|KVTDADAPNTPAWE|AVYT | --- |
| 9080 | 1539.74 | 40.4 | COL18A1 | 1400–1416 | MEP1A, CTSB | EGRQ|GPpGPpGPPGPPSFPGP|HRQT | MMP3 |
| 9510 | 1576.68 | 44.9 | --- | --- | --- | --- | --- |
| 9728 | 1594.77 | 40.3 | COL1A1 | 1177–1194 | MMP3 | DAGP|VGPpGPpGPpGPpGPPSA|GFDF | MMP [3,13], CTSB |
| 10177 | 1624.73 | 25.1 | COL2A1 | 1150–1167 | --- | GPSG|DQGASGpAGpSGpRGPpG|PVGP | --- |
| 11725 | 1733.73 | 29.8 | GSN | 605–621 | CTSD | AAYL|WVGTGASEAEKTGAQEL|LRVL | MEP1A, MMP3, CTS [D,E] |
| 12459 | 1782.85 | 26.0 | --- | --- | --- | --- | --- |
| 13134 | 1836.79 | 31.1 | COL1A2 | 918–937 | MMP3 | SPGV|NGApGEAGRDGNPGNDGPpG|RDGQ | MEP1A, MMP [3,13], CTSB |
| 13176 | 1840.83 | 41.9 | COL2A1 | 1193–1213 | MEP1A, KLK6 | PRGR|SGETGPAGppGNPGPPGPpGP|PGPG | KLK6 |
| 14389 | 1930.89 | 31.6 | COL3A1 | 618–639 | MEP1A, CTSB | TGPQ|GPpGPTGPGGDKGDTGPpGPQG|LQGL | MEP1A, MMP [3,13], CTSB |
| 14925 | 1972.96 | 25.0 | PCSK1N | 223–241 | MEP1A | RRAA|DHDVGSELPPEGVLGALLR|VKRL | MEP1A, MMP [3,13] |
| 15342 | 2009.88 | 32.4 | COL5A2 | 136–157 | MEP1A, MMP [3,13], CTSB | GAPG|SKGEAGpTGPMGDpGTVGPPGP|VGER | MMP3 |
| 17066 | 2169.98 | 33.7 | COL16A1 | 1145–1166 | MEP1A, MMP [3,13], CTSB | GPQG|NSGEKGDQGFQGQPGFpGPPGP|PGFP | --- |
| 17805 | 2232.00 | 33.6 | COL5A1 | 999–1021 | --- | PPGV|VGpQGpTGETGpMGERGHPGPpG|PPGE | MEP1A, CTSB |
| 19681 | 2380.08 | 35.8 | COL3A1 | 201–228 | MEP1A, CTSB | PGYQ|GPPGEPGQAGpSGpPGppGAIGPSGPAG|KDGE | MEP1A, MMP [3,13], CTSB |
| 20237 | 2430.60 | 35.7 | --- | --- | --- | --- | --- |
| 24328 | 2854.37 | 34.6 | COL3A1 | 616–646 | CTSB | GETG|PQGPpGPTGpGGDKGDTGPpGPQGLQGLpGT|GGPP | MEP1A, CTSB |
| 29919 | 3524.75 | 32.4 | CLU | 390–423 | --- | VTTV|ASHTSDSDVPSGVTEVVVKLFDSDPITVTVPVEV|SRKN | MMP3 |
| ACTB, Actin, cytoplasmic 1; ADGRF3, Adhesion G-protein coupled receptor F3; AHNAK, Neuroblast differentiation-associated protein; CDH1, Cadherin-1; CLU, Clusterin; COL1A1, Collagen α-1(I) chain; COL1A2, Collagen α-2(I) chain; COL2A1, Collagen α-1(II) chain; COL3A1, Collagen α-1(III) chain; COL5A1, Collagen α-1(V) chain COL5A2, Collagen α-2(V) chain; COL16A1, Collagen α-1(XVI) chain; COL18A1, Collagen α-1(XVIII) chain; CTS, Cathepsin; FGA, Fibrinogen α chain; GAGE12H, G antigen 12H; GSN, Gelsolin; HBB, Hemoglobin subunit β; KLK6, Kallikrein-6; MEP1A, Meprin A subunit α; MMP, Matrix metallopeptidase; PCSK1N, ProSAAS; TKT, Transketolase; UMOD, uromodulin. | |||||||
| Protease | Peptide Substrate Distribution [Avg. Ion Counts ± SD] | Fold-Change Case/Control | p | |
|---|---|---|---|---|
| HCC Case Group (n = 18) | Non-HCC Liver Disease Control Group (n = 51) | |||
| MEP1A | 196.23 ± 93.19 | 365.65 ± 231.12 | 0.54 | 0.003 |
| MMP3 | 632.99 ± 317.56 | 393.63 ± 331.29 | 1.60 | 0.007 |
| MMP13 | 729.36 ± 402.76 | 495.03 ± 539.78 | 1.47 | 0.012 |
| KLK6 | 166.40 ± 79.87 | 67.09 ± 88.51 | 2.47 | <0.0001 |
| CTSB | 347.63 ± 173.67 | 643.77 ± 399.10 | 0.53 | 0.004 |
| CTSD | 32.90 ± 33.44 | 17.35 ± 31.21 | 1.89 | 0.015 |
| CTSE | 34.41 ± 31.99 | 22.56 ± 41.25 | 1.52 | 0.031 |
| Tissue Type | Allred IHC Score | |
|---|---|---|
| KLK6 | MEP1A | |
| HCC 1 | 3 | 0 |
| HCC 2 | 3 | 1 |
| HCC 3 | 3 | 0 |
| HCC 4 | 3 | 0 |
| HCC 5 | 3 | 0 |
| Cirrhosis 1 | 2 | 0 |
| Cirrhosis 2 | 2 | 0 |
| Cirrhosis 3 | 3 | 0 |
| Cirrhosis 4 | 3 | 0 |
| Normal liver 1 | 1 | 1 |
| Normal liver 2 | 1 | 0 |
| Normal liver 3 | 1 | 1 |
| Normal liver 4 | 1 | 1 |
| Normal liver 5 | 1 | 1 |
| HCC-31 Peptide Marker ID | Amino Acid Sequence | Protein Name | Protein Symbol | Biological Source of Identification (Other than Urine) | Reference |
|---|---|---|---|---|---|
| 11354 | 107-LTEAPLNPK-115 | Actin; α skeletal muscle | ACTA1 | Cerebellum tissue ‡ | Marcu et al. [33] |
| HCC tumor tissue ‡ | Liang-Qing et al. [34] | ||||
| 14071 | 95-RVAPEEHPV-103 | Actin, cytoplasmic 1 | ACTB | Lung tissue ‡ | Marcu et al. [33] |
| 25411 | 179-PGPPGPPGTSGHP-191 | Collagen α-1(III) chain | COL3A1 | Plasma | Zakharova et al. [35] |
| 33901 | 605-DEAGSEADHEGTH-617 | Fibrinogen α chain | FGA | Serum | Ueda et al. [36] |
| Plasma | Koomen et al. [37] | ||||
| 135817 | 390-ASHTSDSDVPSGVTEV VVKLFDSDPITVTVPVEV-423 | Clusterin | CLU | Cerebrospinal fluid | Belogurov et al. [38] |
| 57312 | 605-WVGTGASEAEK TGAQEL-621 | Gelsolin | GSN | Plasma | Modzdiak et al. [39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bannaga, A.; Metzger, J.; Voigtländer, T.; Pejchinovski, M.; Frantzi, M.; Book, T.; James, S.; Gopalakrishnan, K.; Mischak, H.; Manns, M.P.; et al. Pathophysiological Implications of Urinary Peptides in Hepatocellular Carcinoma. Cancers 2021, 13, 3786. https://doi.org/10.3390/cancers13153786
Bannaga A, Metzger J, Voigtländer T, Pejchinovski M, Frantzi M, Book T, James S, Gopalakrishnan K, Mischak H, Manns MP, et al. Pathophysiological Implications of Urinary Peptides in Hepatocellular Carcinoma. Cancers. 2021; 13(15):3786. https://doi.org/10.3390/cancers13153786
Chicago/Turabian StyleBannaga, Ayman, Jochen Metzger, Torsten Voigtländer, Martin Pejchinovski, Maria Frantzi, Thorsten Book, Sean James, Kishore Gopalakrishnan, Harald Mischak, Michael P. Manns, and et al. 2021. "Pathophysiological Implications of Urinary Peptides in Hepatocellular Carcinoma" Cancers 13, no. 15: 3786. https://doi.org/10.3390/cancers13153786
APA StyleBannaga, A., Metzger, J., Voigtländer, T., Pejchinovski, M., Frantzi, M., Book, T., James, S., Gopalakrishnan, K., Mischak, H., Manns, M. P., & Arasaradnam, R. P. (2021). Pathophysiological Implications of Urinary Peptides in Hepatocellular Carcinoma. Cancers, 13(15), 3786. https://doi.org/10.3390/cancers13153786







