The Small-Molecule Inhibitor MRIA9 Reveals Novel Insights into the Cell Cycle Roles of SIK2 in Ovarian Cancer Cells
Simple Summary
Abstract
1. Introduction
2. Results
2.1. MRIA9 Inhibits SIK2 Catalytic Activity in Ovarian Cancer Cells
2.2. Inhibition of SIK2 Reduced the Mitotic Index and Interfered with Spindle Assembly
2.3. Inhibition of SIK2 Altered the Positioning of the Mitotic Spindle
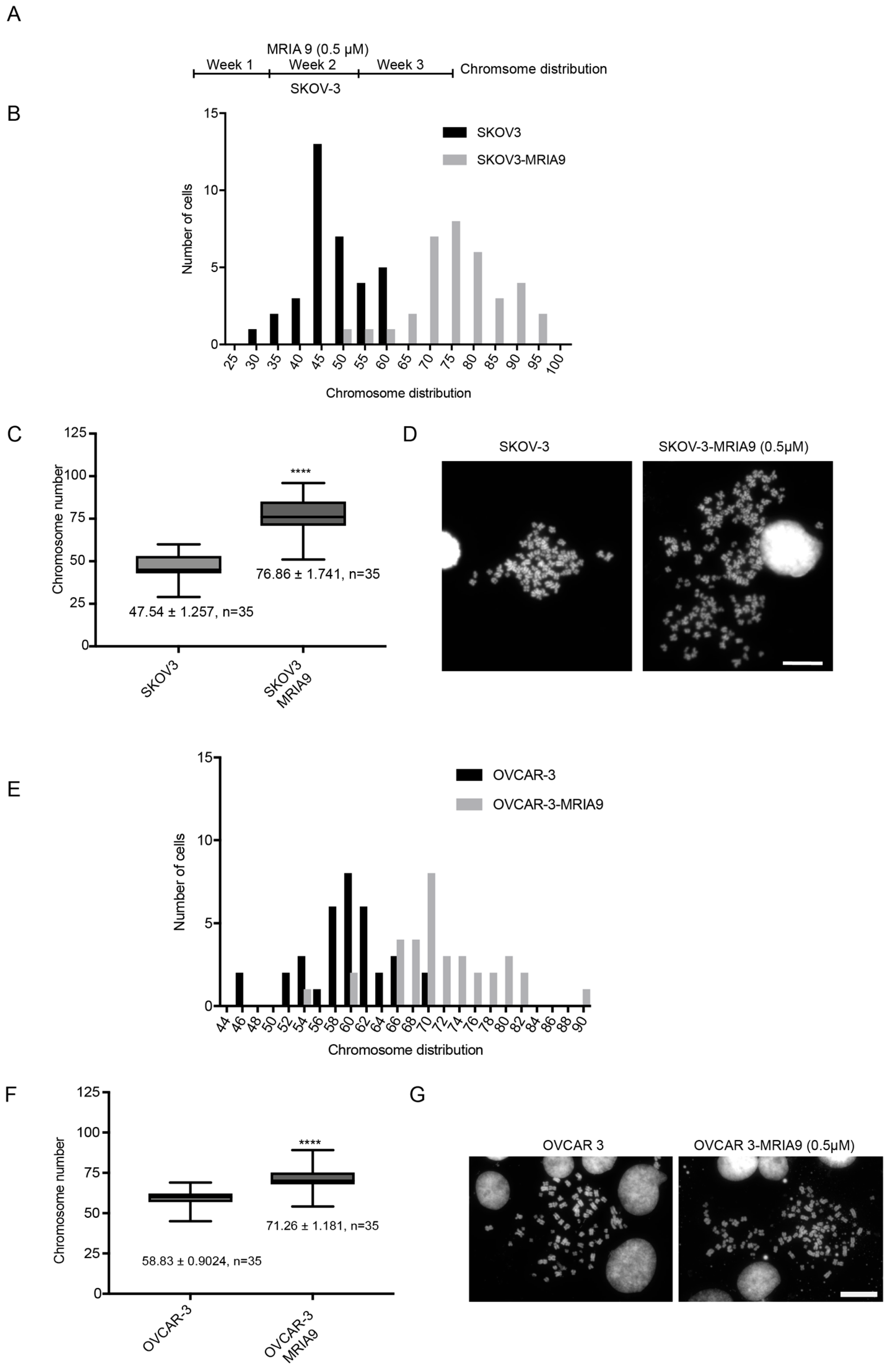
2.4. Long-Term Inhibition of SIK2 Catalytic Activity Enhances Chromosomal Instability
2.5. Long-Term Inhibition of SIK2 Reduced 2D-Colony Forming Ability and Sensitized Ovarian Cancer Cells to Paclitaxel Treatment
2.6. The Combination of Inhibition of SIK2 with Paclitaxel Increases Apoptosis in Ovarian Cancer Cells 3D-Spheroids
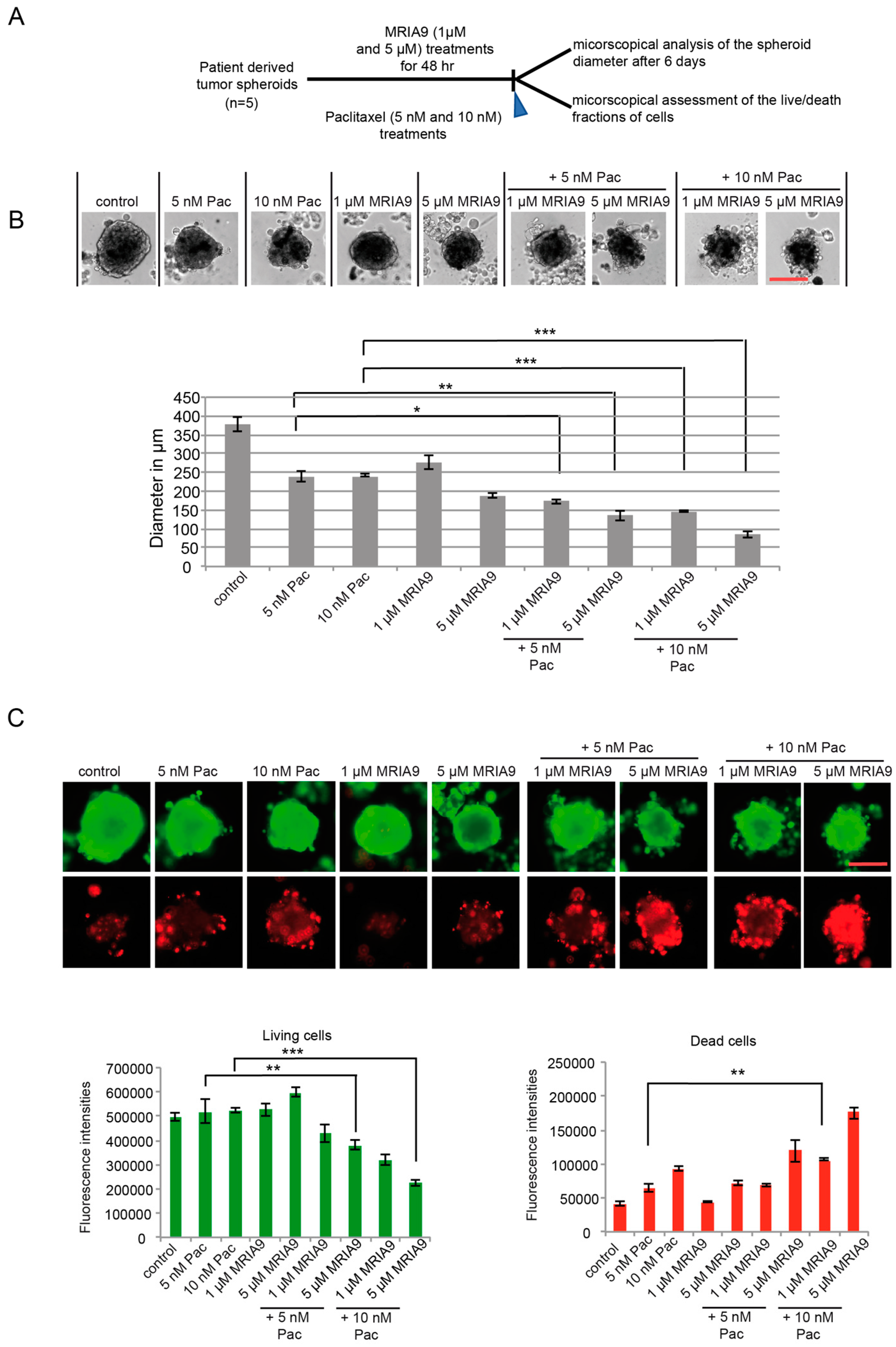
2.7. MRIA9-Induced Inhibition of SIK2 Improves the Paclitaxel Response and Increased Cell Death in Ovarian Patient derived Spheroid Cultures
3. Discussion
4. Material and Methods
4.1. Cell Culture
4.2. Patients and Samples (Primary Cell Culture)
4.3. Colony Formation Assay
4.4. Three-Dimensional (3D) Cultures
4.5. Chromosome Spreads
4.6. Cell Cycle and Apoptosis Assay
4.7. Synchronization
4.8. Cold Kinase Assay In Vitro
4.9. Western Blot Analysis, Antibodies and Chemicals
4.10. Immunofluorescence Assays
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Z.; Takemori, H.; Halder, S.K.; Nonaka, Y.; Okamoto, M. Cloning of a novel kinase (SIK) of the SNF1/AMPK family from high salt diet-treated rat adrenal. FEBS Lett. 1999, 453, 135–139. [Google Scholar] [CrossRef]
- Sun, Z.; Jiang, Q.; Li, J.; Guo, J. The potent roles of salt-inducible kinases (SIKs) in metabolic homeostasis and tumorigenesis. Signal Transduct. Target. Ther. 2020, 5, 150. [Google Scholar] [CrossRef]
- Taub, M.; Springate, J.E.; Cutuli, F. Targeting of renal proximal tubule Na,K-ATPase by salt-inducible kinase. Biochem. Biophys. Res. Commun. 2010, 393, 339–344. [Google Scholar] [CrossRef]
- Chen, F.; Chen, L.; Qin, Q.; Sun, X. Salt-inducible kinase 2: An oncogenic signal transmitter and potential target for cancer therapy. Front Oncol. 2019, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.D.; Vician, L.; Crispino, M.; Hoe, W.; Baudry, M.; Herschman, H.R. The salt-inducible kinase, SIK, is induced by depolarization in brain. J. Neurochem. 2000, 74, 2227–2238. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Chen, Q.; Takemori, H.; Xu, H. SIK2 can be activated by deprivation of nutrition and it inhibits expression of lipogenic genes in adipocytes. Obes. Res. J. 2008, 16, 531–538. [Google Scholar] [CrossRef]
- Zhang, Z.N.; Gong, L.; Lv, S.; Li, J.; Tai, X.; Cao, W.; Peng, B.; Qu, S.; Li, W.; Zhang, C.; et al. SIK2 regulates fasting-induced PPARalpha activity and ketogenesis through p300. Sci. Rep. 2016, 6, 23317. [Google Scholar] [CrossRef]
- Bon, H.; Wadhwa, K.; Schreiner, A.; Osborne, M.; Carroll, T.; Ramos-Montoya, A.; Ross-Adams, H.; Visser, M.; Hoffmann, R.; Ahmed, A.A.; et al. Salt-inducible kinase 2 regulates mitotic progression and transcription in prostate cancer. Mol. Cancer. Res. 2015, 13, 620–635. [Google Scholar] [CrossRef]
- Miranda, F.; Mannion, D.; Liu, S.; Zheng, Y.; Mangala, L.S.; Redondo, C.; Herrero-Gonzalez, S.; Xu, R.; Taylor, C.; Chedom, D.F.; et al. Salt-Inducible kinase 2 couples ovarian cancer cell metabolism with survival at the adipocyte-rich metastatic niche. Cancer Cell 2016, 30, 273–289. [Google Scholar] [CrossRef]
- Zhou, J.; Alfraidi, A.; Zhang, S.; Santiago-O’Farrill, J.M.; Yerramreddy Reddy, V.K.; Alsaadi, A.; Ahmed, A.A.; Yang, H.; Liu, J.; Mao, W.; et al. A novel compound ARN-3236 inhibits salt-inducible kinase 2 and sensitizes ovarian cancer cell lines and xenografts to paclitaxel. Clin. Cancer Res. 2017, 23, 1945–1954. [Google Scholar] [CrossRef]
- Montenegro, R.C.; Howarth, A.; Ceroni, A.; Fedele, V.; Farran, B.; Mesquita, F.P.; Frejno, M.; Berger, B.T.; Heinzlmeir, S.; Sailem, H.Z.; et al. Identification of molecular targets for the targeted treatment of gastric cancer using dasatinib. Oncotarget 2020, 11, 535–549. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Lu, Z.; Jennings, N.B.; Etemadmoghadam, D.; Capalbo, L.; Jacamo, R.O.; Barbosa-Morais, N.; Le, X.-F.; Vivas-Mejia, P.; Lopez-Berestein, G.; et al. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell 2010, 18, 109–121. [Google Scholar] [CrossRef]
- Tarumoto, Y.; Lu, B.; Somerville, T.D.D.; Huang, Y.H.; Milazzo, J.P.; Wu, X.S.; Klingbeil, O.; El Demerdash, O.; Shi, J.; Vakoc, C.R. LKB1, salt-inducible kinases, and MEF2C are linked dependencies in acute myeloid leukemia. Mol. Cell 2018, 69, 1017–1027.e6. [Google Scholar] [CrossRef]
- Patra, K.C.; Kato, Y.; Mizukami, Y.; Widholz, S.; Boukhali, M.; Revenco, I.; Grossman, E.A.; Ji, F.; Sadreyev, R.I.; Liss, A.S.; et al. Mutant GNAS drives pancreatic tumourigenesis by inducing PKA-mediated SIK suppression and reprogramming lipid metabolism. Nat. Cell. Biol. 2018, 20, 811–822. [Google Scholar] [CrossRef]
- Tesch, R.; Rak, M.; Raab, M.; Berger, L.M.; Kronenberger, T.; Joerger, A.C.; Berger, B.T.; Abdi, I.; Hanke, T.; Poso, A.; et al. Structure-based design of selective salt-inducible kinase inhibitors. J. Med. Chem. 2021, 64, 8142–8160. [Google Scholar] [CrossRef]
- Dentin, R.; Liu, Y.; Koo, S.-H.; Hedrick, S.; Vargas, T.; de Heredia, J.M.B.; Yates, J., III; Montminy, M. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nat. Cell Biol. 2007, 449, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Takemori, H.; Horike, N.; Doi, J.; Muraoka, M.; Min, L.; Okamoto, M. Salt-inducible kinase (SIK) isoforms: Their involvement in steroidogenesis and adipogenesis. Mol. Cell Endocrinol. 2004, 217, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Tovar, C.; Chen, S.; Knezevic, D.; Zhao, X.; Sun, H.; Heimbrook, D.C.; Chen, L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA 2006, 103, 10660–10665. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Knapp, S.; Ahmed, A.A. The structural basis of PI3K cancer mutations: From mechanism to therapy. Cancer Res. 2014, 74, 641–646. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, K.E.; Rojo, F.; She, Q.B.; Solit, D.; Mills, G.B.; Smith, D.; Lane, H.; Hofmann, F.; Hicklin, D.J.; Ludwig, D.L.; et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006, 66, 1500–1508. [Google Scholar] [CrossRef]
- Glaubke, E.; Bastians, H. A cell-based assay for mitotic spindle orientation. Methods Mol. Biol. 2018, 1787, 67–75. [Google Scholar] [CrossRef]
- Rogers, S.L.; Rogers, G.C.; Sharp, D.J.; Vale, R.D. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 2002, 158, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Almada, E.; Tonucci, F.M.; Hidalgo, F.; Ferretti, A.; Ibarra, S.; Pariani, A.; Vena, R.; Favre, C.; Girardini, J.; Kierbel, A.; et al. Akap350 recruits Eb1 to the spindle poles, ensuring proper spindle orientation and lumen formation in 3d epithelial cell cultures. Sci. Rep. 2017, 7, 14894. [Google Scholar] [CrossRef]
- Tamura, N.; Draviam, V.M. Microtubule plus-ends within a mitotic cell are ‘moving platforms’ with anchoring, signalling and force-coupling roles. Open Biol. 2012, 2, 120132. [Google Scholar] [CrossRef] [PubMed]
- McHugh, T.; Gluszek, A.A.; Welburn, J.P.I. Microtubule end tethering of a processive kinesin-8 motor Kif18b is required for spindle positioning. J. Cell Biol. 2018, 217, 2403–2416. [Google Scholar] [CrossRef]
- Meraldi, P. Centrosomes in spindle organization and chromosome segregation: A mechanistic view. Chromosome Res. 2016, 24, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Raab, M.; Kobayashi, N.F.; Becker, S.; Kurunci-Csacsko, E.; Kramer, A.; Strebhardt, K.; Sanhaji, M. Boosting the apoptotic response of high-grade serous ovarian cancers with CCNE1 amplification to paclitaxel in vitro by targeting APC/C and the pro-survival protein MCL-1. Int. J. Cancer 2020, 146, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- Thakuri, P.S.; Gupta, M.; Plaster, M.; Tavana, H. Quantitative size-based analysis of tumor spheroids and responses to therapeutics. Assay Drug Dev. Technol. 2019, 17, 140–149. [Google Scholar] [CrossRef]
- Bricambert, J.; Miranda, J.; Benhamed, F.; Girard, J.; Postic, C.; Dentin, R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Investig. 2010, 120, 4316–4331. [Google Scholar] [CrossRef] [PubMed]
- Horike, N.; Takemori, H.; Katoh, Y.; Doi, J.; Min, L.; Asano, T.; Sun, X.J.; Yamamoto, H.; Kasayama, S.; Muraoka, M.; et al. Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J. Biol. Chem. 2003, 278, 18440–18447. [Google Scholar] [CrossRef]
- Horike, N.; Kumagai, A.; Shimono, Y.; Onishi, T.; Itoh, Y.; Sasaki, T.; Kitagawa, K.; Hatano, O.; Takagi, H.; Susumu, T.; et al. Downregulation of SIK2 expression promotes the melanogenic program in mice. Pigment Cell Melanoma Res. 2010, 23, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Takemori, H.; Yagita, Y.; Terasaki, Y.; Uebi, T.; Horike, N.; Takagi, H.; Susumu, T.; Teraoka, H.; Kusano, K.; et al. SIK2 is a key regulator for neuronal survival after ischemia via TORC1-CREB. Neuron 2011, 69, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Ndubaku, C.O.; Crawford, J.J.; Drobnick, J.; Aliagas, I.; Campbell, D.; Dong, P.; Dornan, L.M.; Duron, S.; Epler, J.; Gazzard, L.; et al. Design of selective PAK1 inhibitor G-5555: Improving properties by employing an unorthodox low-pK a polar moiety. ACS Med. Chem. Lett. 2015, 6, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, F.; Echard, A.; Morin, X. Regulation of mitotic spindle orientation: An integrated view. EMBO Rep. 2016, 17, 1106–1130. [Google Scholar] [CrossRef]
- Chen, M.; Cao, Y.; Dong, D.; Zhang, Z.; Zhang, Y.; Chen, J.; Luo, Y.; Chen, Q.; Xiao, X.; Zhou, J.; et al. Regulation of mitotic spindle orientation by phosphorylation of end binding protein. Exp. Cell Res. 2019, 384, 111618. [Google Scholar] [CrossRef]
- Nehlig, A.; Molina, A.; Rodrigues-Ferreira, S.; Honore, S.; Nahmias, C. Regulation of end-binding protein EB1 in the control of microtubule dynamics. Cell Mol. Life Sci. 2017, 74, 2381–2393. [Google Scholar] [CrossRef]
- Stout, J.R.; Yount, A.L.; Powers, J.A.; Leblanc, C.; Ems-McClung, S.C.; Walczak, C.E. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol. Biol. Cell 2011, 22, 3070–3080. [Google Scholar] [CrossRef]
- Pease, J.C.; Tirnauer, J.S. Mitotic spindle misorientation in cancer--out of alignment and into the fire. J. Cell Sci. 2011, 124, 1007–1016. [Google Scholar] [CrossRef]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res. Treat. 2006, 100, 229–235. [Google Scholar] [CrossRef]
- Raab, M.; Kappel, S.; Kramer, A.; Sanhaji, M.; Matthess, Y.; Kurunci-Csacsko, E.; Calzada-Wack, J.; Rathkolb, B.; Rozman, J.; Adler, T.; et al. Toxicity modelling of Plk1-targeted therapies in genetically engineered mice and cultured primary mammalian cells. Nat. Commun. 2011, 2, 395. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raab, M.; Rak, M.; Tesch, R.; Gasimli, K.; Becker, S.; Knapp, S.; Strebhardt, K.; Sanhaji, M. The Small-Molecule Inhibitor MRIA9 Reveals Novel Insights into the Cell Cycle Roles of SIK2 in Ovarian Cancer Cells. Cancers 2021, 13, 3658. https://doi.org/10.3390/cancers13153658
Raab M, Rak M, Tesch R, Gasimli K, Becker S, Knapp S, Strebhardt K, Sanhaji M. The Small-Molecule Inhibitor MRIA9 Reveals Novel Insights into the Cell Cycle Roles of SIK2 in Ovarian Cancer Cells. Cancers. 2021; 13(15):3658. https://doi.org/10.3390/cancers13153658
Chicago/Turabian StyleRaab, Monika, Marcel Rak, Roberta Tesch, Khayal Gasimli, Sven Becker, Stefan Knapp, Klaus Strebhardt, and Mourad Sanhaji. 2021. "The Small-Molecule Inhibitor MRIA9 Reveals Novel Insights into the Cell Cycle Roles of SIK2 in Ovarian Cancer Cells" Cancers 13, no. 15: 3658. https://doi.org/10.3390/cancers13153658
APA StyleRaab, M., Rak, M., Tesch, R., Gasimli, K., Becker, S., Knapp, S., Strebhardt, K., & Sanhaji, M. (2021). The Small-Molecule Inhibitor MRIA9 Reveals Novel Insights into the Cell Cycle Roles of SIK2 in Ovarian Cancer Cells. Cancers, 13(15), 3658. https://doi.org/10.3390/cancers13153658






