The BET Inhibitor JQ1 Augments the Antitumor Efficacy of Gemcitabine in Preclinical Models of Pancreatic Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cell Lines and Compounds
2.3. In Vitro Cell Viability Assay
2.4. Clonogenic Assay
2.5. Immunoblot Analysis
2.6. In Vivo Drug Efficacy
2.7. Hematoxylin and Eosin Staining
2.8. Immunohistochemistry
2.9. RNA Isolation and RNA-Sequencing
2.10. Bioinformatics
2.11. Real-Time PCR, qRT-PCR
2.12. Statistics
3. Results
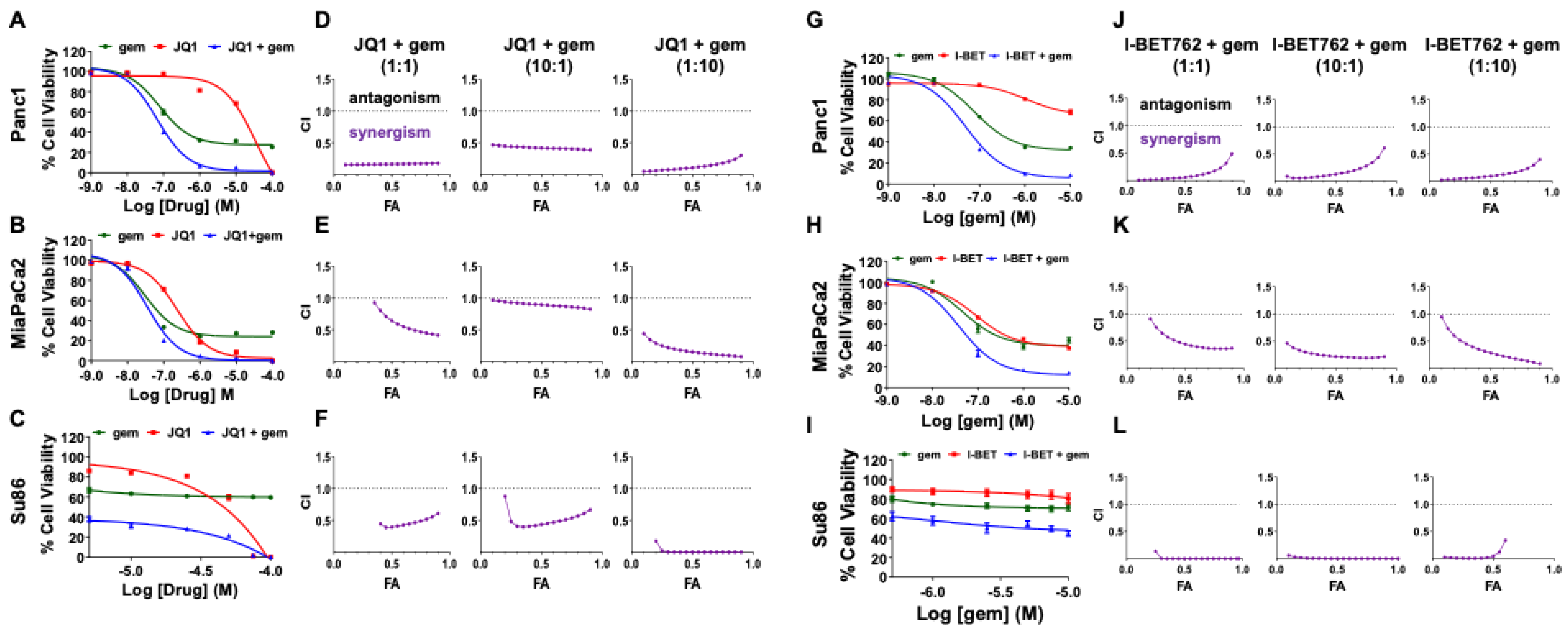
3.1. The Combination of a BET Inhibitor + Gemcitabine Exerts Synergistic Cytotoxicity in PDAC Cell Lines
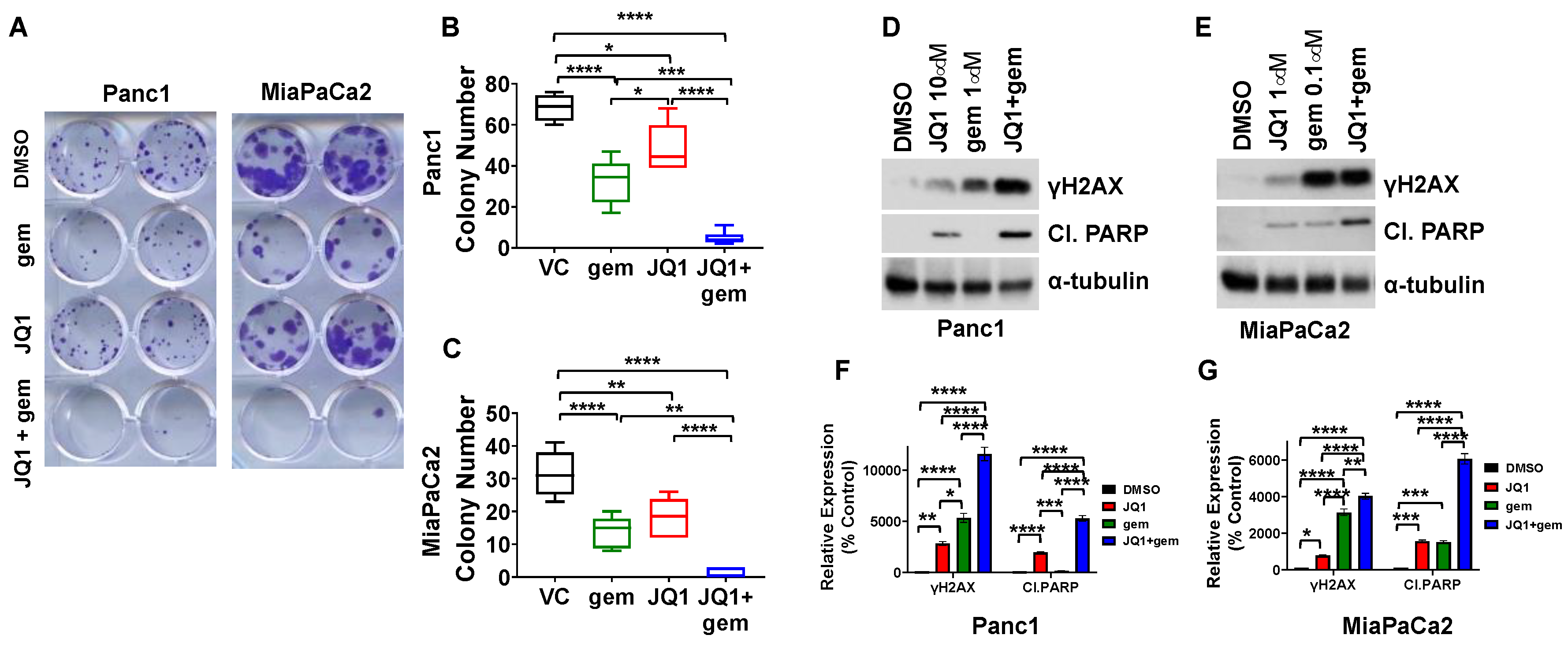
3.2. JQ1 + Gemcitabine Decreases Colony Formation In Vitro, Concomitant with Increases in Levels of the DNA Damage Marker γH2AX and the Apoptosis Marker Cleaved PARP
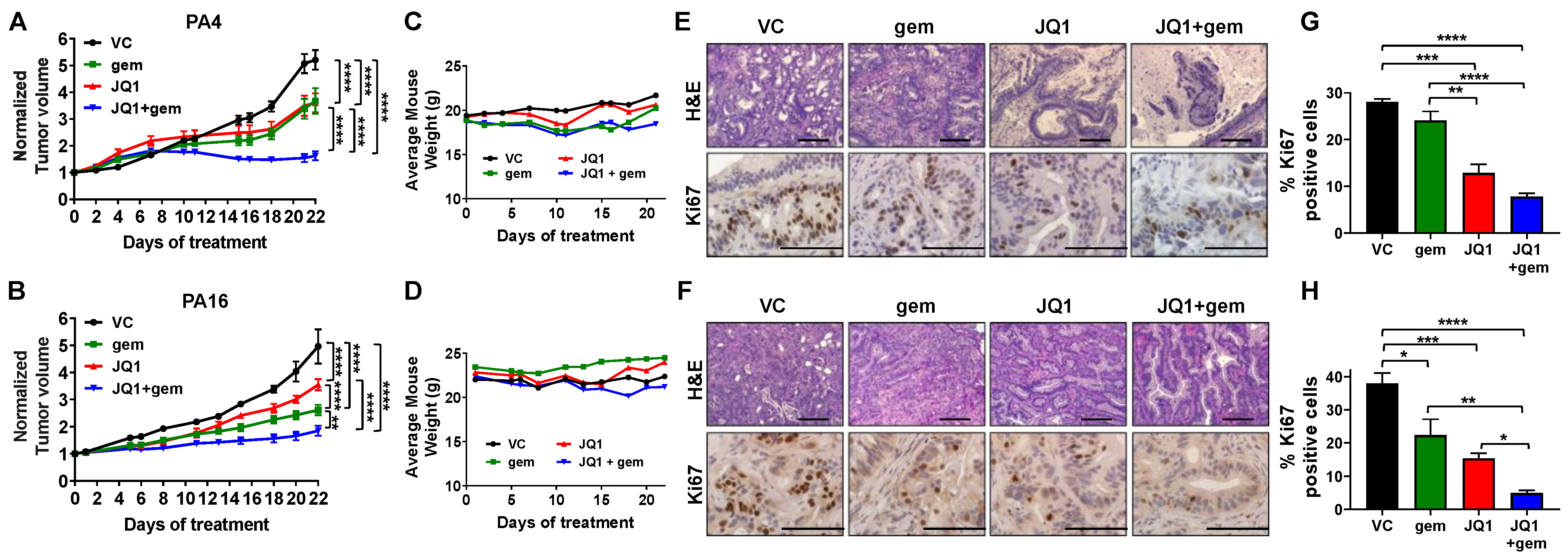
3.3. JQ1 + Gemcitabine Suppresses PDAC Tumor Growth In Vivo
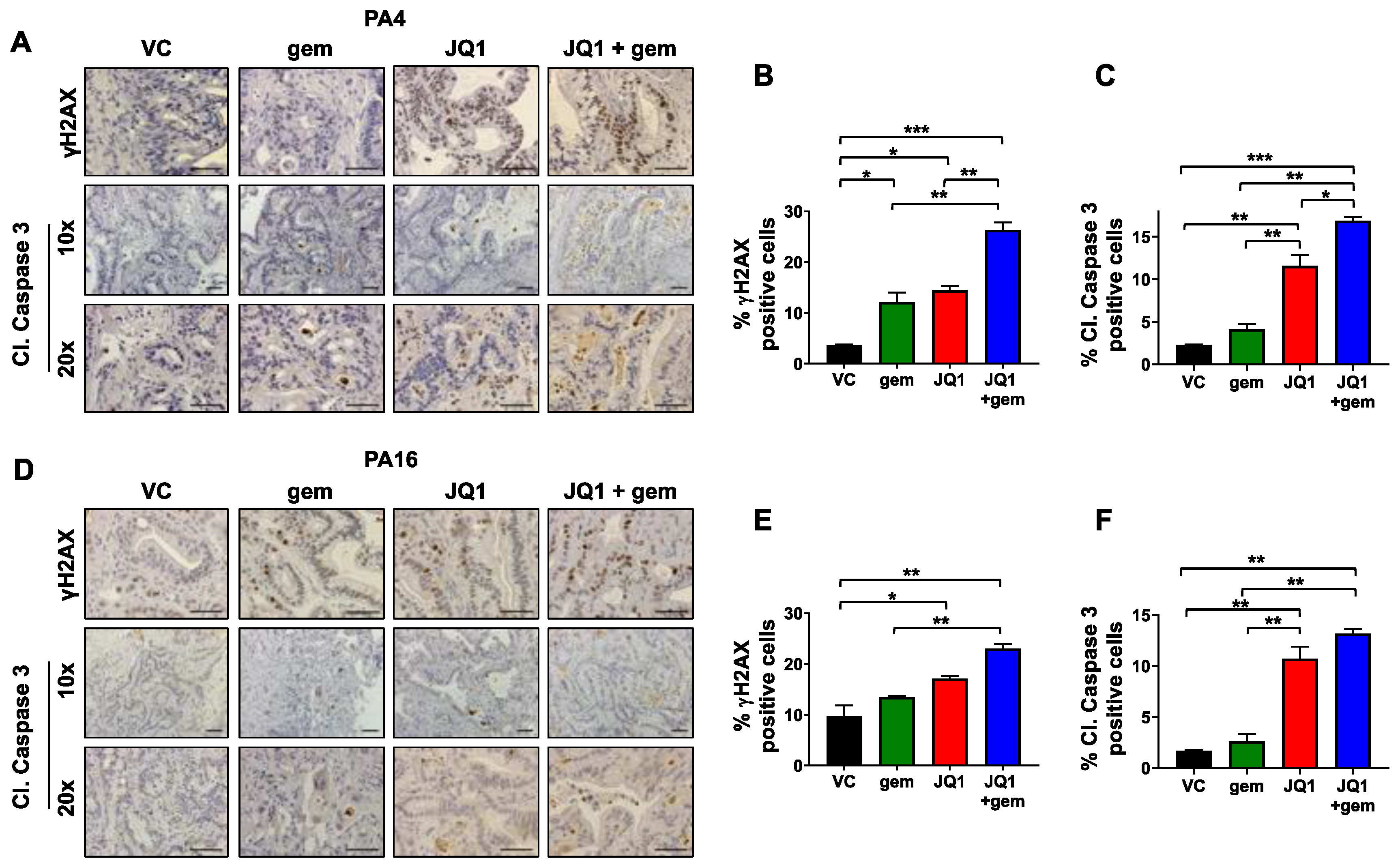
3.4. Tumors Exposed to JQ1 + Gemcitabine In Vivo Have Higher Levels of the DNA Damage Marker γH2AX and the Apoptosis Marker Cleaved Caspase 3 Than Either Drug as a Single Agent
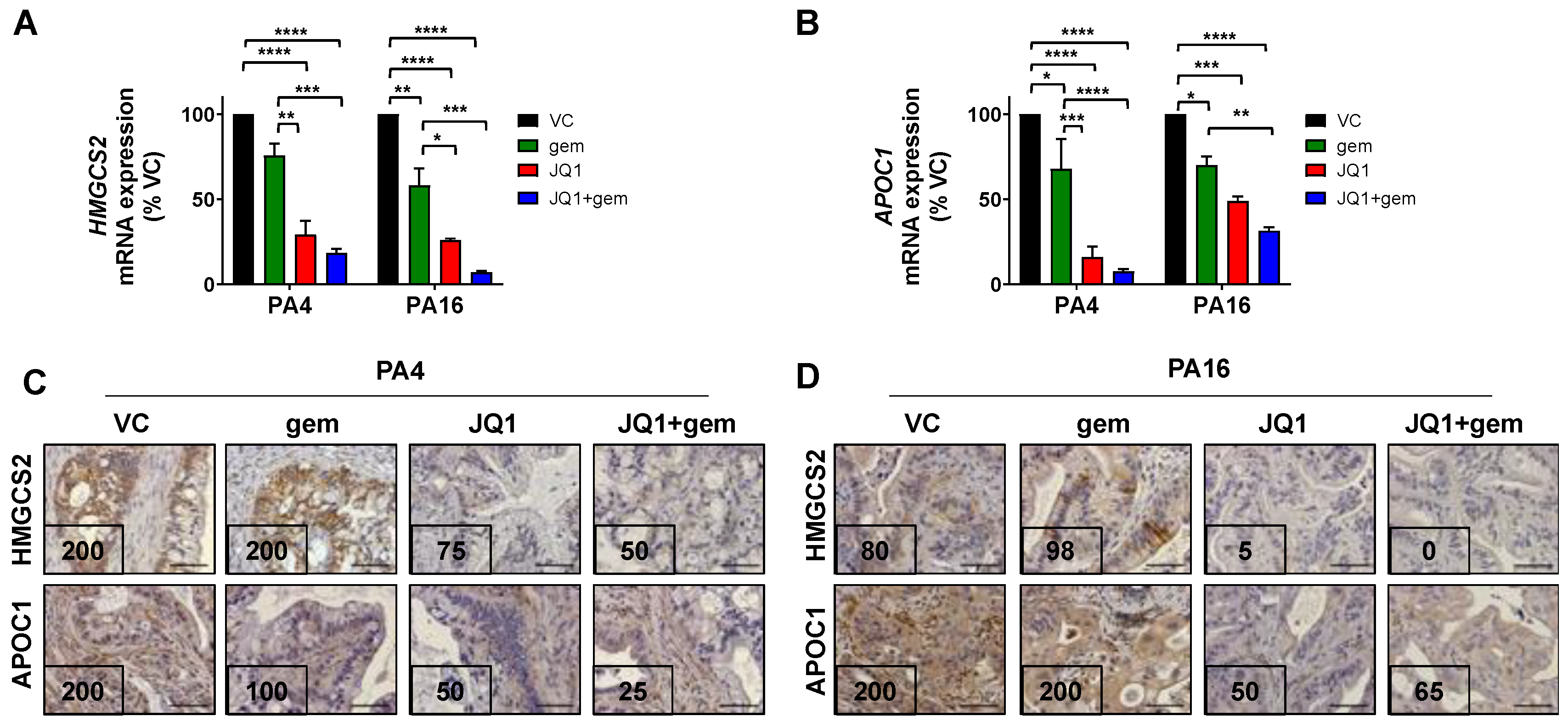
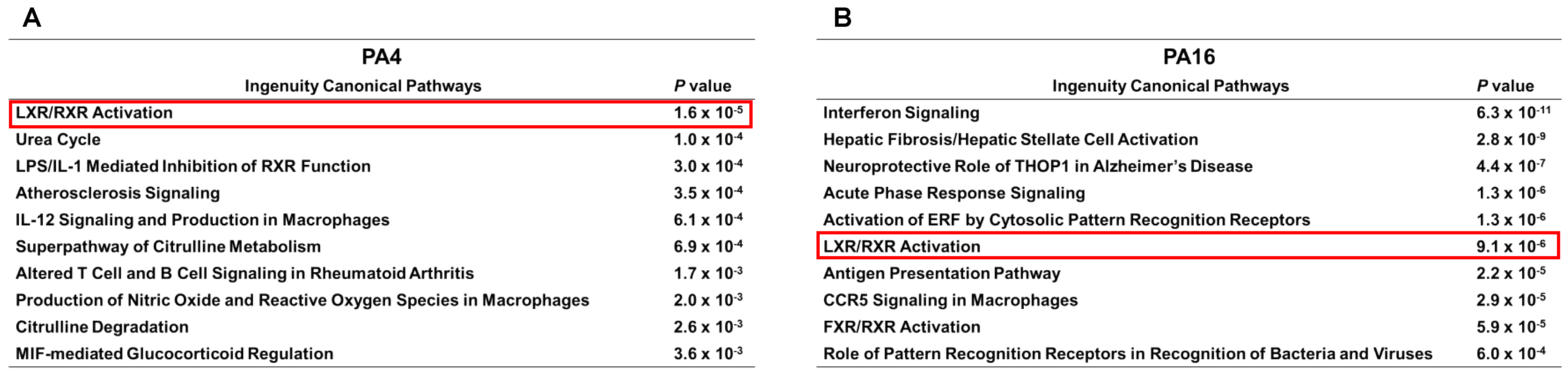
3.5. JQ1 + Gemcitabine Inhibits the Expression of Gene Products of the Lipid Metabolism and the LXR/RXR Activation Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.D.; Ho, M.; Renouf, D.J.; Lim, H.J.; Gill, S.; Ruan, J.Y.; Cheung, W.Y. Eligibility of Metastatic Pancreatic Cancer Patients for First-Line Palliative Intent nab-Paclitaxel Plus Gemcitabine Versus FOLFIRINOX. Am. J. Clin. Oncol. 2017, 40, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Heinemann, V.; Xu, Y.Z.; Chubb, S.; Sen, A.; Hertel, L.W.; Grindey, G.B.; Plunkett, W. Cellular elimination of 2′,2′-difluorodeoxycytidine 5′-triphosphate: A mechanism of self-potentiation. Cancer Res. 1992, 52, 533–539. [Google Scholar] [PubMed]
- Hertel, L.W.; Boder, G.B.; Kroin, J.S.; Rinzel, S.M.; Poore, G.A.; Todd, G.C.; Grindey, G.B. Evaluation of the antitumor activity of gemcitabine (2′,2′-difluoro-2′-deoxycytidine). Cancer Res. 1990, 50, 4417–4422. [Google Scholar] [PubMed]
- Huang, P.; Chubb, S.; Hertel, L.W.; Grindey, G.B.; Plunkett, W. Action of 2′,2′-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991, 51, 6110–6117. [Google Scholar]
- Burris, H.A., III; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Cripps, M.C.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P.; et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. 1997, 15, 2403–2413. [Google Scholar] [CrossRef]
- Parsels, L.A.; Morgan, M.A.; Tanska, D.M.; Parsels, J.D.; Palmer, B.D.; Booth, R.J.; Denny, W.A.; Canman, C.E.; Kraker, A.J.; Lawrence, T.S.; et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol. Cancer Ther. 2009, 8, 45–54. [Google Scholar] [CrossRef]
- Azzariti, A.; Bocci, G.; Porcelli, L.; Fioravanti, A.; Sini, P.; Simone, G.M.; Quatrale, A.E.; Chiarappa, P.; Mangia, A.; Sebastian, S.; et al. Aurora B kinase inhibitor AZD1152: Determinants of action and ability to enhance chemotherapeutics effectiveness in pancreatic and colon cancer. Br. J. Cancer 2011, 104, 769–780. [Google Scholar] [CrossRef]
- Cuneo, K.C.; Morgan, M.A.; Sahai, V.; Schipper, M.J.; Parsels, L.A.; Parsels, J.D.; Devasia, T.; Al-Hawaray, M.; Cho, C.S.; Nathan, H.; et al. Dose Escalation Trial of the Wee1 Inhibitor Adavosertib (AZD1775) in Combination With Gemcitabine and Radiation for Patients With Locally Advanced Pancreatic Cancer. J. Clin. Oncol. 2019, 37, 2643–2650. [Google Scholar] [CrossRef]
- Kumarasamy, V.; Ruiz, A.; Nambiar, R.; Witkiewicz, A.K.; Knudsen, E.S. Chemotherapy impacts on the cellular response to CDK4/6 inhibition: Distinct mechanisms of interaction and efficacy in models of pancreatic cancer. Oncogene 2020, 39, 1831–1845. [Google Scholar] [CrossRef]
- Li, J.; Wang, R.; Schweickert, P.G.; Karki, A.; Yang, Y.; Kong, Y.; Ahmad, N.; Konieczny, S.F.; Liu, X. Plk1 inhibition enhances the efficacy of gemcitabine in human pancreatic cancer. Cell Cycle 2016, 15, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Wallez, Y.; Dunlop, C.R.; Johnson, T.I.; Koh, S.B.; Fornari, C.; Yates, J.W.T.; Bernaldo de Quiros Fernandez, S.; Lau, A.; Richards, F.M.; Jodrell, D.I. The ATR Inhibitor AZD6738 Synergizes with Gemcitabine In Vitro and In Vivo to Induce Pancreatic Ductal Adenocarcinoma Regression. Mol. Cancer Ther. 2018, 17, 1670–1682. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.L.; Garcia, P.L.; Yoon, K.J. Developing effective combination therapy for pancreatic cancer: An overview. Pharmacol. Res. 2020, 155, 104740. [Google Scholar] [CrossRef] [PubMed]
- Garcia, P.L.; Miller, A.L.; Kreitzburg, K.M.; Council, L.N.; Gamblin, T.L.; Christein, J.D.; Heslin, M.J.; Arnoletti, J.P.; Richardson, J.H.; Chen, D.; et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene 2016, 35, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.L.; Fehling, S.C.; Garcia, P.L.; Gamblin, T.L.; Council, L.N.; van Waardenburg, R.; Yang, E.S.; Bradner, J.E.; Yoon, K.J. The BET inhibitor JQ1 attenuates double-strand break repair and sensitizes models of pancreatic ductal adenocarcinoma to PARP inhibitors. EBioMedicine 2019, 44, 419–430. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Z.; Wang, Z.; Wang, S.; Chen, Z.; Li, Z.; Zhang, M.; Zou, J.; Dong, B.; Gao, J.; et al. Targeting c-Myc: JQ1 as a promising option for c-Myc-amplified esophageal squamous cell carcinoma. Cancer Lett. 2018, 419, 64–74. [Google Scholar] [CrossRef]
- Fehling, S.C.; Miller, A.L.; Garcia, P.L.; Vance, R.B.; Yoon, K.J. The combination of BET and PARP inhibitors is synergistic in models of cholangiocarcinoma. Cancer Lett. 2020, 468, 48–58. [Google Scholar] [CrossRef]
- Garcia, P.L.; Council, L.N.; Christein, J.D.; Arnoletti, J.P.; Heslin, M.J.; Gamblin, T.L.; Richardson, J.H.; Bjornsti, M.A.; Yoon, K.J. Development and histopathological characterization of tumorgraft models of pancreatic ductal adenocarcinoma. PLoS ONE 2013, 8, e78183. [Google Scholar] [CrossRef] [PubMed]
- Kreitzburg, K.M.; Fehling, S.C.; Landen, C.N.; Gamblin, T.L.; Vance, R.B.; Arend, R.C.; Katre, A.A.; Oliver, P.G.; van Waardenburg, R.; Alvarez, R.D.; et al. FTY720 enhances the anti-tumor activity of carboplatin and tamoxifen in a patient-derived xenograft model of ovarian cancer. Cancer Lett. 2018, 436, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Dillies, M.A.; Rau, A.; Aubert, J.; Hennequet-Antier, C.; Jeanmougin, M.; Servant, N.; Keime, C.; Marot, G.; Castel, D.; Estelle, J.; et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013, 14, 671–683. [Google Scholar] [CrossRef]
- Miller, A.L.; Garcia, P.L.; Gamblin, T.L.; Vance, R.B.; Yoon, K.J. Development of gemcitabine-resistant patient-derived xenograft models of pancreatic ductal adenocarcinoma. Cancer Drug Resist. 2020, 3, 572–585. [Google Scholar] [CrossRef]
- Mazur, P.K.; Herner, A.; Mello, S.S.; Wirth, M.; Hausmann, S.; Sanchez-Rivera, F.J.; Lofgren, S.M.; Kuschma, T.; Hahn, S.A.; Vangala, D.; et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat. Med. 2015, 21, 1163–1171. [Google Scholar] [CrossRef]
- Xie, F.; Huang, M.; Lin, X.; Liu, C.; Liu, Z.; Meng, F.; Wang, C.; Huang, Q. The BET inhibitor I-BET762 inhibits pancreatic ductal adenocarcinoma cell proliferation and enhances the therapeutic effect of gemcitabine. Sci. Rep. 2018, 8, 8102. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Fei, Q.; Xiong, P.; Yang, J.; Zhang, Z.; Lin, X.; Pan, M.; Lu, F.; Huang, H. Synergistic inhibition of pancreatic cancer with anti-PD-L1 and c-Myc inhibitor JQ1. Oncoimmunology 2019, 8, e1581529. [Google Scholar] [CrossRef]
- Xu, C.; Wang, X.; Zhou, Y.; Chen, F.X.; Wang, H.; Li, K.; Fan, H.; Tang, X.; Jiang, G.; Zhang, J. Synergy between arsenic trioxide and JQ1 on autophagy in pancreatic cancer. Oncogene 2019, 38, 7249–7265. [Google Scholar] [CrossRef]
- Kaoutari, A.E.; Fraunhoffer, N.A.; Hoare, O.; Teyssedou, C.; Soubeyran, P.; Gayet, O.; Roques, J.; Lomberk, G.; Urrutia, R.; Dusetti, N.; et al. Metabolomic profiling of pancreatic adenocarcinoma reveals key features driving clinical outcome and drug resistance. EBioMedicine 2021, 66, 103332. [Google Scholar] [CrossRef]
- Kirby, M.K.; Ramaker, R.C.; Gertz, J.; Davis, N.S.; Johnston, B.E.; Oliver, P.G.; Sexton, K.C.; Greeno, E.W.; Christein, J.D.; Heslin, M.J.; et al. RNA sequencing of pancreatic adenocarcinoma tumors yields novel expression patterns associated with long-term survival and reveals a role for ANGPTL4. Mol. Oncol. 2016, 10, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- Tadros, S.; Shukla, S.K.; King, R.J.; Gunda, V.; Vernucci, E.; Abrego, J.; Chaika, N.V.; Yu, F.; Lazenby, A.J.; Berim, L.; et al. De Novo Lipid Synthesis Facilitates Gemcitabine Resistance through Endoplasmic Reticulum Stress in Pancreatic Cancer. Cancer Res. 2017, 77, 5503–5517. [Google Scholar] [CrossRef]
- Hegardt, F.G. Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: A control enzyme in ketogenesis. Biochem. J. 1999, 338 Pt 3, 569–582. [Google Scholar] [CrossRef]
- Puchalska, P.; Crawford, P.A. Multi-dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef]
- Neuwirt, H.; Bouchal, J.; Kharaishvili, G.; Ploner, C.; Johrer, K.; Pitterl, F.; Weber, A.; Klocker, H.; Eder, I.E. Cancer-associated fibroblasts promote prostate tumor growth and progression through upregulation of cholesterol and steroid biosynthesis. Cell Commun. Signal. 2020, 18, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.W.; Chou, C.T.; Chang, C.C.; Li, Y.J.; Chen, S.T.; Lin, I.C.; Kok, S.H.; Cheng, S.J.; Lee, J.J.; Wu, T.S.; et al. HMGCS2 enhances invasion and metastasis via direct interaction with PPARalpha to activate Src signaling in colorectal cancer and oral cancer. Oncotarget 2017, 8, 22460–22476. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Liu, C.L.; Chiu, W.C.; Twu, Y.C.; Liao, Y.J. HMGCS2 Mediates Ketone Production and Regulates the Proliferation and Metastasis of Hepatocellular Carcinoma. Cancers 2019, 11, 1876. [Google Scholar] [CrossRef] [PubMed]
- Fuior, E.V.; Gafencu, A.V. Apolipoprotein C1: Its Pleiotropic Effects in Lipid Metabolism and Beyond. Int. J. Mol. Sci. 2019, 20, 5939. [Google Scholar] [CrossRef]
- Jong, M.C.; Dahlmans, V.E.; van Gorp, P.J.; van Dijk, K.W.; Breuer, M.L.; Hofker, M.H.; Havekes, L.M. In the absence of the low density lipoprotein receptor, human apolipoprotein C1 overexpression in transgenic mice inhibits the hepatic uptake of very low density lipoproteins via a receptor-associated protein-sensitive pathway. J. Clin. Investig. 1996, 98, 2259–2267. [Google Scholar] [CrossRef][Green Version]
- Cui, Y.; Miao, C.; Hou, C.; Wang, Z.; Liu, B. Apolipoprotein C1 (APOC1): A Novel Diagnostic and Prognostic Biomarker for Clear Cell Renal Cell Carcinoma. Front. Oncol. 2020, 10, 1436. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Wu, L.W.; Zeng, L.H.; Zhang, Z.Y.; Wang, W.; Zhang, C.; Lin, N.M. ApoC1 promotes the metastasis of clear cell renal cell carcinoma via activation of STAT3. Oncogene 2020, 39, 6203–6217. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Chen, Z.; Yang, L.; Xiong, W.; Yang, H.; Xu, K.; Zhai, E.; Ding, L.; He, Y.; Song, X. Apolipoprotein C1 (APOC1) promotes tumor progression via MAPK signaling pathways in colorectal cancer. Cancer Manag. Res. 2019, 11, 4917–4930. [Google Scholar] [CrossRef] [PubMed]
- Calkin, A.C.; Tontonoz, P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 2012, 13, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Candelaria, N.R.; Addanki, S.; Zheng, J.; Nguyen-Vu, T.; Karaboga, H.; Dey, P.; Gabbi, C.; Vedin, L.L.; Liu, K.; Wu, W.; et al. Antiproliferative effects and mechanisms of liver X receptor ligands in pancreatic ductal adenocarcinoma cells. PLoS ONE 2014, 9, e106289. [Google Scholar] [CrossRef] [PubMed]
- Karaboga, H.; Huang, W.; Srivastava, S.; Widmann, S.; Addanki, S.; Gamage, K.T.; Mazhar, Z.; Ebalunode, J.O.; Briggs, J.M.; Gustafsson, J.A.; et al. Screening of Focused Compound Library Targeting Liver X Receptors in Pancreatic Cancer Identified Ligands with Inverse Agonist and Degrader Activity. ACS Chem. Biol. 2020, 15, 2916–2928. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, K.J.; Valfort, A.C.; Steinauer, N.; Chatterjee, A.; Abuirqeba, S.; Majidi, S.; Sengupta, M.; Di Paolo, R.J.; Shornick, L.P.; Zhang, J.; et al. LXR-inverse agonism stimulates immune-mediated tumor destruction by enhancing CD8 T-cell activity in triple negative breast cancer. Sci. Rep. 2019, 9, 19530. [Google Scholar] [CrossRef]
- Flaveny, C.A.; Griffett, K.; El-Gendy Bel, D.; Kazantzis, M.; Sengupta, M.; Amelio, A.L.; Chatterjee, A.; Walker, J.; Solt, L.A.; Kamenecka, T.M.; et al. Broad Anti-tumor Activity of a Small Molecule that Selectively Targets the Warburg Effect and Lipogenesis. Cancer Cell 2015, 28, 42–56. [Google Scholar] [CrossRef]
- Wu, G.; Wang, Q.; Xu, Y.; Li, J.; Zhang, H.; Qi, G.; Xia, Q. Targeting the transcription factor receptor LXR to treat clear cell renal cell carcinoma: Agonist or inverse agonist? Cell Death Dis. 2019, 10, 416. [Google Scholar] [CrossRef]
- Wan, W.; Hou, Y.; Wang, K.; Cheng, Y.; Pu, X.; Ye, X. The LXR-623-induced long non-coding RNA LINC01125 suppresses the proliferation of breast cancer cells via PTEN/AKT/p53 signaling pathway. Cell Death Dis. 2019, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, S.A.; Websdale, A.; Cioccoloni, G.; Røberg-Larsen, H.; Lianto, P.; Kim, B.; Rose, A.; Soteriou, C.; Pramanik, A.; Wastall, L.M.; et al. Liver x receptor alpha drives chemoresistance in response to side-chain hydroxycholesterols in triple negative breast cancer. Oncogene 2021, 40, 2872–2883. [Google Scholar] [CrossRef]
- Lo Sasso, G.; Bovenga, F.; Murzilli, S.; Salvatore, L.; Di Tullio, G.; Martelli, N.; D’Orazio, A.; Rainaldi, S.; Vacca, M.; Mangia, A.; et al. Liver X receptors inhibit proliferation of human colorectal cancer cells and growth of intestinal tumors in mice. Gastroenterology 2013, 144, 1497–1507.e13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ma, X.; Chen, Y.; Zhang, L.; Jiang, M.; Li, X.; Xiang, R.; Miao, R.; Hajjar, D.P.; Duan, Y.; et al. Identification of interferon-gamma as a new molecular target of liver X receptor. Biochem. J. 2014, 459, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Pencheva, N.; Buss, C.G.; Posada, J.; Merghoub, T.; Tavazoie, S.F. Broad-spectrum therapeutic suppression of metastatic melanoma through nuclear hormone receptor activation. Cell 2014, 156, 986–1001. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, A.L.; Garcia, P.L.; Fehling, S.C.; Gamblin, T.L.; Vance, R.B.; Council, L.N.; Chen, D.; Yang, E.S.; van Waardenburg, R.C.A.M.; Yoon, K.J. The BET Inhibitor JQ1 Augments the Antitumor Efficacy of Gemcitabine in Preclinical Models of Pancreatic Cancer. Cancers 2021, 13, 3470. https://doi.org/10.3390/cancers13143470
Miller AL, Garcia PL, Fehling SC, Gamblin TL, Vance RB, Council LN, Chen D, Yang ES, van Waardenburg RCAM, Yoon KJ. The BET Inhibitor JQ1 Augments the Antitumor Efficacy of Gemcitabine in Preclinical Models of Pancreatic Cancer. Cancers. 2021; 13(14):3470. https://doi.org/10.3390/cancers13143470
Chicago/Turabian StyleMiller, Aubrey L., Patrick L. Garcia, Samuel C. Fehling, Tracy L. Gamblin, Rebecca B. Vance, Leona N. Council, Dongquan Chen, Eddy S. Yang, Robert C. A. M. van Waardenburg, and Karina J. Yoon. 2021. "The BET Inhibitor JQ1 Augments the Antitumor Efficacy of Gemcitabine in Preclinical Models of Pancreatic Cancer" Cancers 13, no. 14: 3470. https://doi.org/10.3390/cancers13143470
APA StyleMiller, A. L., Garcia, P. L., Fehling, S. C., Gamblin, T. L., Vance, R. B., Council, L. N., Chen, D., Yang, E. S., van Waardenburg, R. C. A. M., & Yoon, K. J. (2021). The BET Inhibitor JQ1 Augments the Antitumor Efficacy of Gemcitabine in Preclinical Models of Pancreatic Cancer. Cancers, 13(14), 3470. https://doi.org/10.3390/cancers13143470





