Comprehensive Somatic Copy Number Analysis Using Aqueous Humor Liquid Biopsy for Retinoblastoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Patient Demographics and Clinical Outcomes
3.2. Whole-Genome RB SCNA Analysis
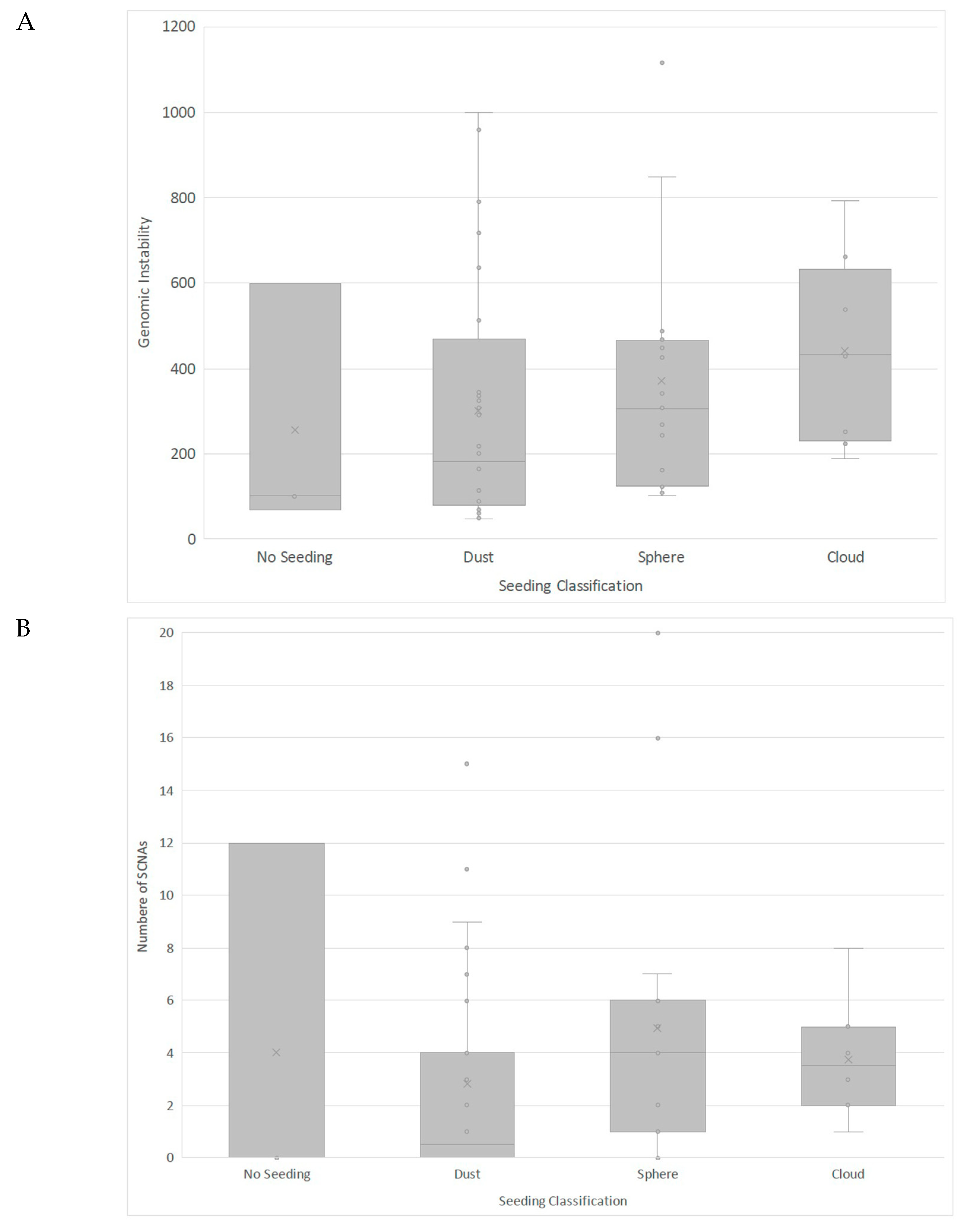
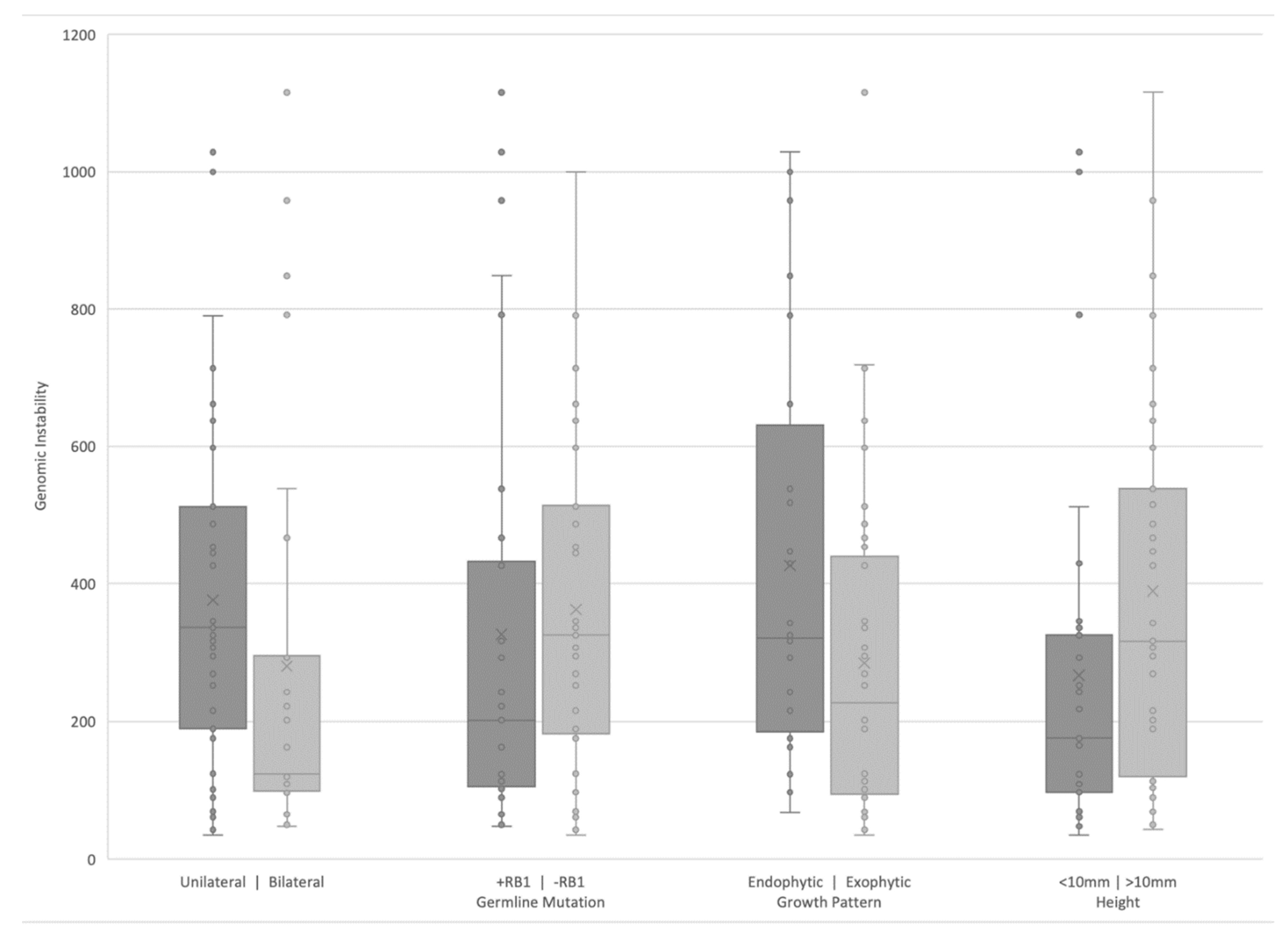
3.3. Clinical Correlates of RB SCNAs
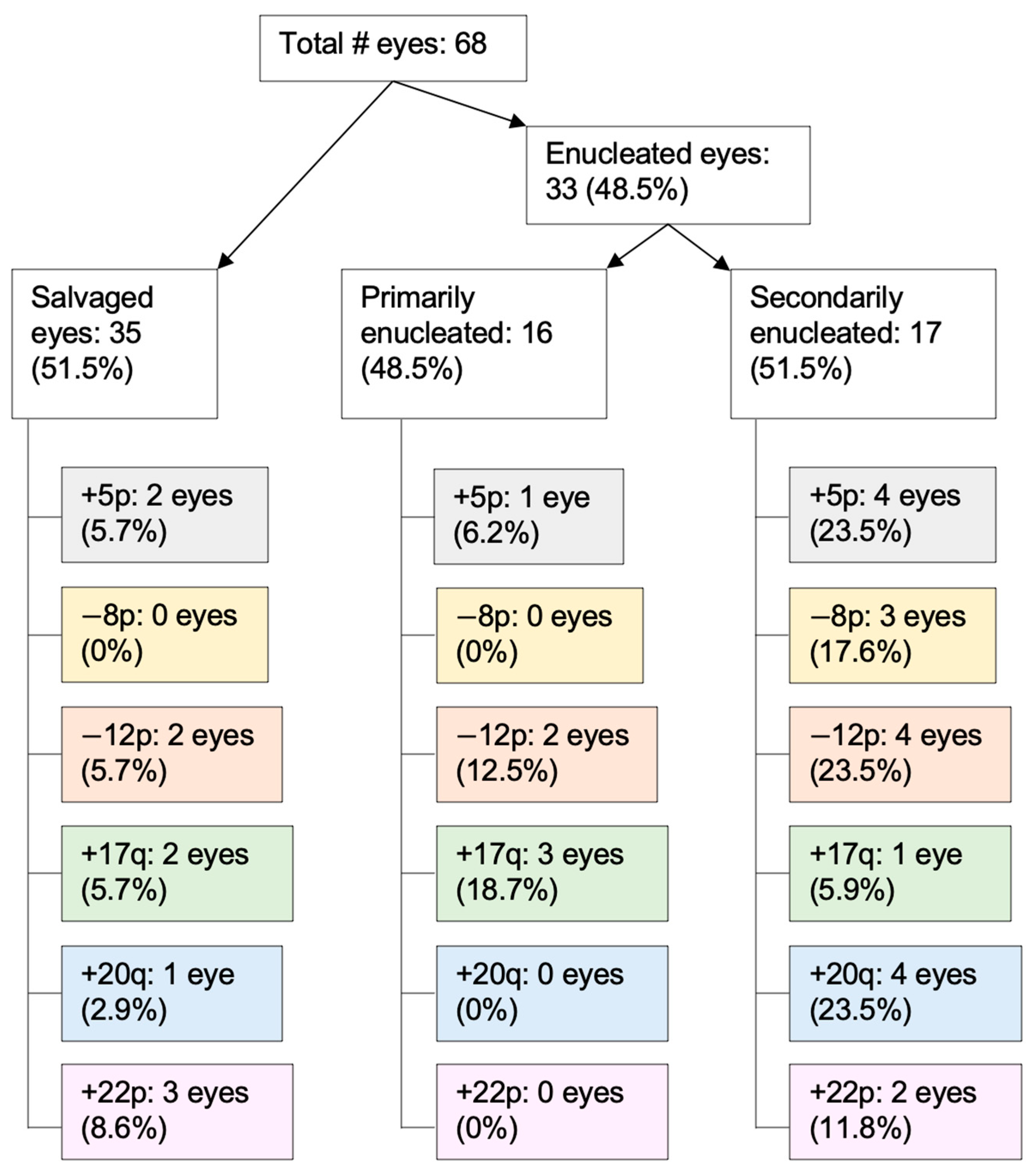
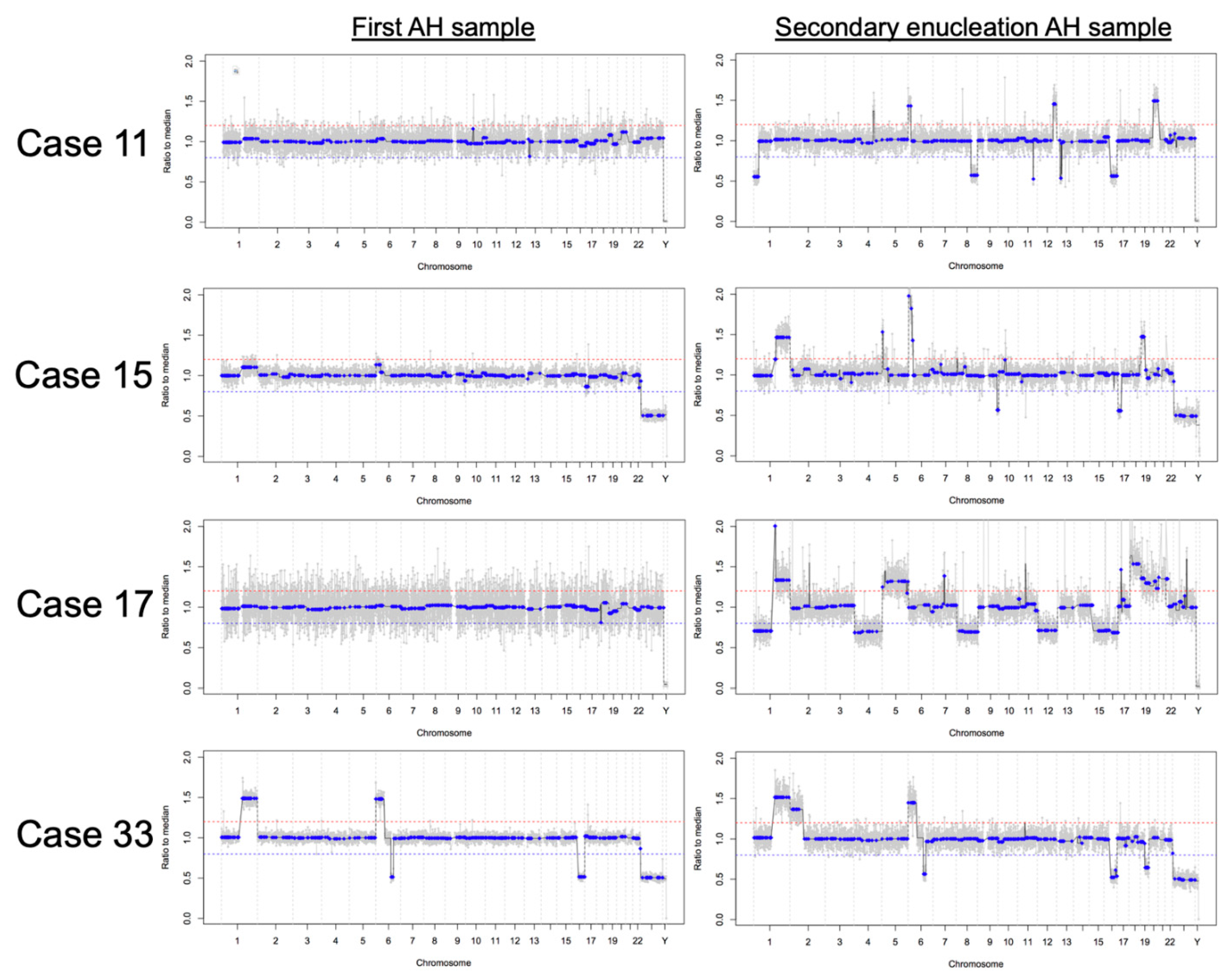
3.4. RB SCNAs in Enucleated Eyes
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parkin, D.M.; Stiller, C.A.; Draper, G.J.; Bieber, C.A. The international incidence of childhood cancer. Int. J. Cancer 1988, 42, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, S.S.; Smith, L.M. Retinoblastoma: Biology, presentation, and current management. Oncology 1989, 3, 45–51. [Google Scholar]
- Corson, T.W.; Gallie, B.L. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosomes Cancer 2007, 46, 617–634. [Google Scholar] [CrossRef]
- Dimaras, H.; Khetan, V.; Halliday, W.; Orlic, M.; Prigoda, N.L.; Piovesan, B.; Marrano, P.; Corson, T.W.; Eagle Jr, R.C.; Squire, J.A. Loss of RB1 induces non-proliferative retinoma: Increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet. 2008, 17, 1363–1372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampieri, K.; Mencarelli, M.A.; Carmela Epistolato, M.; Toti, P.; Lazzi, S.; Bruttini, M.; Francesco, S.D.; Longo, I.; Meloni, I.; Mari, F. Genomic differences between retinoma and retinoblastoma. Acta Oncol. 2008, 47, 1483–1492. [Google Scholar] [CrossRef] [Green Version]
- Sampieri, K.; Amenduni, M.; Papa, F.T.; Katzaki, E.; Mencarelli, M.A.; Marozza, A.; Epistolato, M.C.; Toti, P.; Lazzi, S.; Bruttini, M. Array comparative genomic hybridization in retinoma and retinoblastoma tissues. Cancer Sci. 2009, 100, 465–471. [Google Scholar] [CrossRef]
- Singh, H.P.; Wang, S.; Stachelek, K.; Lee, S.; Reid, M.W.; Thornton, M.E.; Craft, C.M.; Grubbs, B.H.; Cobrinik, D. Developmental stage-specific proliferation and retinoblastoma genesis in RB-deficient human but not mouse cone precursors. Proc. Natl. Acad. Sci. USA 2018, 115, E9391–E9400. [Google Scholar] [CrossRef] [Green Version]
- Gallie, B.L.; Campbell, C.; Devlin, H.; Duckett, A.; Squire, J.A. Developmental basis of retinal-specific induction of cancer by RB mutation. Cancer Res. 1999, 59, 1731s–1735s. [Google Scholar]
- Bowles, E.; Corson, T.W.; Bayani, J.; Squire, J.A.; Wong, N.; Lai, P.B.S.; Gallie, B.L. Profiling genomic copy number changes in retinoblastoma beyond loss of RB1. Genes Chromosomes Cancer 2007, 46, 118–129. [Google Scholar] [CrossRef]
- Kooi, I.E.; Mol, B.M.; Massink, M.P.G.; Ameziane, N.; Meijers-Heijboer, H.; Dommering, C.J.; Van Mil, S.E.; De Vries, Y.; Van Der Hout, A.H.; Kaspers, G.J.L. Somatic genomic alterations in retinoblastoma beyond RB1 are rare and limited to copy number changes. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kooi, I.E.; Mol, B.M.; Massink, M.P.G.; de Jong, M.C.; de Graaf, P.; van der Valk, P.; Meijers-Heijboer, H.; Kaspers, G.J.L.; Moll, A.C.; Te Riele, H. A meta-analysis of retinoblastoma copy numbers refines the list of possible driver genes involved in tumor progression. PLoS ONE 2016, 11, e0153323. [Google Scholar] [CrossRef] [Green Version]
- Grasemann, C.; Gratias, S.; Stephan, H.; Schüler, A.; Schramm, A.; Klein-Hitpass, L.; Rieder, H.; Schneider, S.; Kappes, F.; Eggert, A. Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene 2005, 24, 6441–6449. [Google Scholar] [CrossRef] [Green Version]
- Cobrinik, D. Retinoblastoma progression. EBioMedicine 2015, 2, 623–624. [Google Scholar] [CrossRef] [Green Version]
- Rushlow, D.E.; Mol, B.M.; Kennett, J.Y.; Yee, S.; Pajovic, S.; Thériault, B.L.; Prigoda-Lee, N.L.; Spencer, C.; Dimaras, H.; Corson, T.W. Characterisation of retinoblastomas without RB1 mutations: Genomic, gene expression, and clinical studies. Lancet Oncol. 2013, 14, 327–334. [Google Scholar] [CrossRef] [Green Version]
- Thériault, B.L.; Dimaras, H.; Gallie, B.L.; Corson, T.W. The genomic landscape of retinoblastoma: A review. Clin. Exp. Ophthalmol. 2014, 42, 33–52. [Google Scholar] [CrossRef] [Green Version]
- van der Wal, J.E.; Hermsen, M.A.J.A.; Gille, H.J.P.; Schouten-Van Meeteren, N.Y.N.; Moll, A.C.; Imhof, S.M.; Meijer, G.A.; Baak, J.P.A.; van der Valk, P. Comparative genomic hybridisation divides retinoblastomas into a high and a low level chromosomal instability group. J. Clin. Pathol. 2003, 56, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karcioglu, Z.A.; Gordon, R.A.; Karcioglu, G.L. Tumor seeding in ocular fine needle aspiration biopsy. Ophthalmology 1985, 92, 1763–1767. [Google Scholar] [CrossRef]
- Shields, J.A.; Shields, C.L.; Ehya, H.; Eagle Jr, R.C.; Patrick De Potter, M.D. Fine-needle aspiration biopsy of suspected intraocular tumors: The 1992 Urwick lecture. Ophthalmology 1993, 100, 1677–1684. [Google Scholar] [CrossRef]
- Berry, J.L.; Xu, L.; Murphree, A.L.; Krishnan, S.; Stachelek, K.; Zolfaghari, E.; McGovern, K.; Lee, T.C.; Carlsson, A.; Kuhn, P.; et al. Potential of aqueous humor as a surrogate tumor biopsy for retinoblastoma. JAMA Ophthalmol. 2017, 135, 1221–1230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, J.L.; Xu, L.Y.; Kooi, I.; Murphree, A.L.; Prabakar, R.K.; Reid, M.; Stachelek, K.; Le, B.H.A.; Welter, L.; Reiser, B.J.; et al. Genomic cfDNA analysis of aqueous humor in retinoblastoma predicts eye salvage: The surrogate tumor biopsy for retinoblastoma. Mol. Cancer Res. 2018, 16, 701–1712. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Kim, M.E.; Polski, A.; Prabakar, R.K.; Shen, L.; Peng, C.-C.; Reid, M.W.; Chévez-Barrios, P.; Kim, J.W.; Shah, R.; et al. Establishing the clinical utility of ctDNA analysis for diagnosis, prognosis, and treatment monitoring of retinoblastoma: The aqueous humor liquid biopsy. Cancers 2021, 13, 1282. [Google Scholar] [CrossRef]
- Xu, L.; Polski, A.; Prabakar, R.K.; Reid, M.W.; Chevez-Barrios, P.; Jubran, R.; Kim, J.W.; Kuhn, P.; Cobrinik, D.; Hicks, J.; et al. Chromosome 6p amplification in aqueous humor cell-free DNA is a prognostic biomarker for retinoblastoma ocular survival. Mol. Cancer Res. 2020, 18, 1166–1175. [Google Scholar] [CrossRef]
- Berry, J.L.; Xu, L.; Polski, A.; Jubran, R.; Kuhn, P.; Kim, J.W.; Hicks, J. Aqueous humor is superior to blood as a liquid biopsy for retinoblastoma. Ophthalmology 2020, 127, 552–554. [Google Scholar] [CrossRef]
- Polski, A.; Xu, L.; Prabakar, R.K.; Gai, X.; Kim, J.W.; Shah, R.; Jubran, R.; Kuhn, P.; Cobrinik, D.; Hicks, J.; et al. Variability in retinoblastoma genome stability is driven by age and not heritability. Genes Chromosomes Cancer 2020, 59, 584–590. [Google Scholar] [CrossRef]
- Polski, A.; Xu, L.; Prabakar, R.; Kim, J.W.; Cobrinik, D.; Hicks, J.; Berry, J.L. Longitudinal aqueous humor sampling reflects treatment response in retinoblastoma patients. Investig. Ophthalmol. Vis. Sci. 2020, 61, 1394. [Google Scholar]
- Xu, L.; Shen, L.; Polski, A.; Prabakar, R.K.; Shah, R.; Jubran, R.; Kim, J.W.; Biegel, J.; Kuhn, P.; Cobrinik, D.; et al. Simultaneous identification of clinically relevant RB1 mutations and copy number alterations in aqueous humor of retinoblastoma eyes. Ophthalmic Genet. 2020, 41, 526–532. [Google Scholar] [CrossRef]
- Berry, J.L.; Jubran, R.; Kim, J.W.; Wong, K.; Bababeygy, S.R.; Almarzouki, H.; Lee, T.C.; Murphree, A.L. Long-term outcomes of Group D eyes in bilateral retinoblastoma patients treated with chemoreduction and low-dose IMRT salvage. Pediatric Blood Cancer 2013, 60, 688–693. [Google Scholar] [CrossRef]
- Berry, J.L.; Bechtold, M.; Shah, S.; Zolfaghari, E.; Reid, M.; Jubran, R.; Kim, J.W. Not all seeds are created equal: Seed classification is predictive of outcomes in retinoblastoma. Ophthalmology 2017, 124, 1817–1825. [Google Scholar] [CrossRef]
- Berry, J.L.; Kogachi, K.; Aziz, H.A.; McGovern, K.; Zolfaghari, E.; Murphree, A.L.; Jubran, R.; Kim, J.W. Risk of metastasis and orbital recurrence in advanced retinoblastoma eyes treated with systemic chemoreduction versus primary enucleation. Pediatric Blood Cancer 2017, 64, e26270. [Google Scholar] [CrossRef]
- Linn, A.M. Intraocular retinoblastoma: The case for a new group classification. Ophthalmol. Clin. N. Am. 2005, 18, 41–53. [Google Scholar]
- Baslan, T.; Kendall, J.; Rodgers, L.; Cox, H.; Riggs, M.; Stepansky, A.; Troge, J.; Ravi, K.; Esposito, D.; Lakshmi, B. Genome-wide copy number analysis of single cells. Nat. Protoc. 2012, 7, 1024–1041. [Google Scholar] [CrossRef] [PubMed]
- Baslan, T.; Kendall, J.; Rodgers, L.; Cox, H.; Riggs, M.; Stepansky, A.; Troge, J.; Ravi, K.; Esposito, D.; Lakshmi, B. Erratum: Genome-wide copy number analysis of single cells. Nat. Protoc. 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Zielinski, B.; Gratias, S.; Toedt, G.; Mendrzyk, F.; Stange, D.E.; Radlwimmer, B.; Lohmann, D.R.; Lichter, P. Detection of chromosomal imbalances in retinoblastoma by matrix-based comparative genomic hybridization. Genes Chromosomes Cancer 2005, 43, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Gröbner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhsng, C.Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Shay, J.W.; Wright, W.E. Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 2019, 20, 299–309. [Google Scholar] [CrossRef]
- Seluanov, A.; Gladyshev, V.N.; Vijg, J.; Gorbunova, V. Mechanisms of cancer resistance in long-lived mammals. Nat. Rev. Cancer 2018, 18, 433–441. [Google Scholar] [CrossRef]
- Francis, J.H.; Richards, A.L.; Mandelker, D.L.; Berger, M.F.; Walsh, M.F.; Dunkel, I.J.; Donoghue, M.T.A.; Abramson, D.H. Molecular changes in retinoblastoma beyond RB1: Findings from next-generation sequencing. Cancers 2021, 13, 149. [Google Scholar] [CrossRef]
- Mlakar, V.; Berginc, G.; Volavšek, M.; Štor, Z.; Rems, M.; Glavač, D. Presence of activating KRAS mutations correlates significantly with expression of tumour suppressor genes DCN and TPM1 in colorectal cancer. BMC Cancer 2009, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Pan, H.; Gu, L.; Liu, B.; Li, Y.; Wang, Y.; Bai, X.; Li, L.; Wang, B.; Peng, Q.; Yao, Z. Tropomyosin-1 acts as a potential tumor suppressor in human oral squamous cell carcinoma. PLoS ONE 2017, 12, e0168900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Tang, C.; Yang, C.; Zheng, Q.; Hou, Y. Tropomyosin-1 functions as a tumor suppressor with respect to cell proliferation, angiogenesis and metastasis in renal cell carcinoma. J. Cancer 2019, 10, 2220. [Google Scholar] [CrossRef]
- Hu, J.; Ho, A.L.; Yuan, L.; Hu, B.; Hua, S.; Hwang, S.S.; Zhang, J.; Hu, T.; Zheng, H.; Gan, B. Neutralization Termin. Differ. gliomagenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 14520–14527. [Google Scholar] [CrossRef] [Green Version]
- Bharadwaj, S.; Prasad, G.L. Tropomyosin-1, a novel suppressor of cellular transformation is downregulated by promoter methylation in cancer cells. Cancer Lett. 2002, 183, 205–213. [Google Scholar] [CrossRef]
- Collins, C.; Rommens, J.M.; Kowbel, D.; Godfrey, T.; Tanner, M.; Hwang, S.I.; Polikoff, D.; Nonet, G.; Cochran, J.; Myambo, K.; et al. Positional cloning of ZNF217 and NABC1: Genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc. Natl. Acad. Sci. USA 1998, 95, 8703–8708. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Linn, J.F.; Wu, G.; Anzick, S.L.; Eisenberger, C.F.; Halachmi, S.; Cohen, Y.; Fomenkov, A.; Hoque, M.O.; Okami, K.; et al. CDC91L1 (PIG-U) is a newly discovered oncogene in human bladder cancer. Nat. Med. 2004, 10, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Cuthill, S.; Agarwal, P.; Sarkar, S.; Savelieva, E.; Reznikoff, C.A. Dominant genetic alterations in immortalization: Role for 20q gain. Genes Chromosomes Cancer 1999, 26, 304–311. [Google Scholar] [CrossRef]
- Chantada, G.L.; Gonzalez, A.; Fandino, A.; de Davila, M.T.; Demirdjian, G.; Scopinaro, M.; Abramson, D. Some clinical findings at presentation can predict high-risk pathology features in unilateral retinoblastoma. J. Pediatric Hematol. Oncol. 2009, 31, 325–329. [Google Scholar] [CrossRef]
- Chawla, B.; Sharma, S.; Sen, S.; Azad, R.; Bajaj, M.S.; Kashyap, S.; Pushker, N.; Ghose, S. Correlation between clinical features, magnetic resonance imaging, and histopathologic findings in retinoblastoma: A prospective study. Ophthalmology 2012, 119, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Kaliki, S.; Srinivasan, V.; Gupta, A.; Mishra, D.K.; Naik, M.N. Clinical features predictive of high-risk retinoblastoma in 403 Asian Indian patients: A case-control study. Ophthalmology 2015, 122, 1165–1172. [Google Scholar] [CrossRef]
- Francis, J.H.; Abramson, D.H.; Gaillard, M.-C.; Marr, B.P.; Beck-Popovic, M.; Munier, F.L. The classification of vitreous seeds in retinoblastoma and response to intravitreal melphalan. Ophthalmology 2015, 122, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.A.; Yoon, Y.H.; Kim, J.G.; Lee, J.Y. Clinical Features and Long-term Prognosis of Retinoblastoma according to Age at Diagnosis. Korean J Ophthalmol 2020, 34, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Mol, B.M.; Massink, M.P.G.; van der Hout, A.H.; Dommering, C.J.; Zaman, J.M.A.; Bosscha, M.I.; Kors, W.A.; Meijers-Heijboer, H.E.; Kaspers, G.J.L.; Riele, H.t. High resolution SNP array profiling identifies variability in retinoblastoma genome stability. Genes Chromosomes Cancer 2014, 53, 1–14. [Google Scholar] [CrossRef]
- Von Lindern, M.; Fornerod, M.; Soekarman, N.; Van Baal, S.; Jaegle, M.; Hagemeijer, A.; Bootsma, D.; Grosveld, G. Translocation t (6; 9) in acute non-lymphocytic leukaemia results in the formation of a DEK-CAN fusion gene. Bailliere’s Clin. Haematol. 1992, 5, 857–879. [Google Scholar] [CrossRef]
- Carro, M.S.; Spiga, F.M.; Quarto, M.; Ninni, V.D.; Volorio, S.; Alcalay, M.; Müller, H. DEK expression is controlled by E2F and deregulated in diverse tumor type. Cell Cycle 2006, 5, 1202–1207. [Google Scholar] [CrossRef] [Green Version]
- Adams, M.R.; Sears, R.; Nuckolls, F.; Leone, G.; Nevins, J.R. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol. Cell. Biol. 2000, 20, 3633–3639. [Google Scholar] [CrossRef]
- Amram, A.L.; Rico, G.; Kim, J.W.; Chintagumpala, M.; Herzog, C.E.; Gombos, D.S.; Chévez-Barrios, P. Vitreous seeds in retinoblastoma: Clinicopathologic classification and correlation. Ophthalmology 2017, 124, 1540–1547. [Google Scholar] [CrossRef]
- Gustmann, S.; Klein-Hitpass, L.; Stephan, H.; Weber, S.; Bornfeld, N.; Kaulisch, M.; Lohmann, D.R.; Dunker, N. Loss at chromosome arm 16q in retinoblastoma: Confirmation of the association with diffuse vitreous seeding and refinement of the recurrently deleted region. Genes Chromosomes Cancer 2011, 50, 327–337. [Google Scholar] [CrossRef]
- Marchong, M.N.; Chen, D.; Corson, T.W.; Lee, C.; Harmandayan, M.; Bowles, E.; Chen, N.; Gallie, B.L. Minimal 16q genomic loss implicates cadherin-11 in retinoblastoma. Mol. Cancer Res. 2004, 2, 495–503. [Google Scholar] [PubMed]
- Nawaiseh, I.; Al-Hussaini, M.; Alhamwi, A.; Meyar, M.; Sultan, I.; Alrawashdeh, K.; Jaradat, I.; Yousef, Y.A. The impact of growth patterns of retinoblastoma (endophytic, exophytic, and mixed patterns). Turk. Patoloji. Derg. 2015, 31, 45–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Kim, J.H.; Yu, Y.S.; Kim, D.H.; Kim, Y.K.; Kim, K.-W. Comparative genomic hybridization analysis of newly established retinoblastoma cell lines of adherent growth compared with Y79 of nonadherent growth. J. Pediatric Hematol. Oncol. 2008, 30, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Klingelhutz, A.J.; Qian, Q.; Phillips, S.L.; Gourronc, F.A.; Darbro, B.W.; Patil, S.R. Amplification of the chromosome 20q region is associated with expression of HPV-16 E7 in human airway and anogenital epithelial cells. Virology 2005, 340, 237–244. [Google Scholar] [CrossRef] [Green Version]
- Afshar, A.R.; Pekmezci, M.; Bloomer, M.M.; Cadenas, N.J.; Stevers, M.; Banerjee, A.; Roy, R.; Olshen, A.B.; Van Ziffle, J.; Onodera, C.; et al. Next generation sequencing of retinoblastoma identifies pathogenic alterations beyond RB1 inactivation that correlate with aggressive histopathologic features. Ophthalmology 2020, 127, 804–813. [Google Scholar] [CrossRef]
- Fabian, I.D.; Abdallah, E.; Abdullahi, S.U.; Abdulqader, R.A.; Boubacar, S.A.; Ademola-Popoola, D.S.; Adio, A.; Afshar, A.R.; Aggarwal, P.; Aghaji, A.E. Global retinoblastoma presentation and analysis by national income level. JAMA Oncol. 2020, 6, 685–695. [Google Scholar]
- Schwermer, M.; Hiber, M.; Dreesmann, S.; Rieb, A.; Theißen, J.; Herold, T.; Schramm, A.; Temming, P.; Steenpass, L. Comprehensive characterization of RB1 mutant and MYCN amplified retinoblastoma cell lines. Exp. Cell. Res. 2019, 375, 92–99. [Google Scholar] [CrossRef]
- Davies, H.R.; Broad, K.D.; Onadim, Z.; Price, E.A.; Zou, X.; Sheriff, I.; Karaa, E.K.; Scheimberg, I.; Reddy, M.A.; Sagoo, M.S. Whole-genome sequencing of retinoblastoma reveals the diversity of rearrangements disrupting RB1 and uncovers a treatment-related mutational signature. Cancers 2021, 13, 754. [Google Scholar] [CrossRef]
- Lillington, D.M.; Kingston, J.E.; Coen, P.G.; Price, E.; Hungerford, J.; Domizio, P.; Young, B.D.; Onadim, Z. Comparative genomic hybridization of 49 primary retinoblastoma tumors identifies chromosomal regions associated with histopathology, progression, and patient outcome. Genes Chromosomes Cancer 2003, 36, 121–128. [Google Scholar] [CrossRef] [PubMed]

| Case | Laterality | Age at Diagnosis (months) | Germline RB1 Mutation | Timing of Enucleation | Other SCNAs | Somatic RB1 Mutation in AH | Somatic RB1 Mutation inTumor | Distinct Histologic Features for MYCN Amplified Tumors * |
|---|---|---|---|---|---|---|---|---|
| 3 | Unilateral | 38 | Negative | Primary | 1q and 6p gain, 16p and 16q loss | None | None | None |
| 10 | Bilateral | 2 | Positive | Secondary for persistent seeding | None | None | None | None |
| 31 | Unilateral | 9 | Negative | Primary | None | None | None | None |
| 48 | Unilateral | 18 | Negative | Primary | 1q gain | None | None | None |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.E.; Polski, A.; Xu, L.; Prabakar, R.K.; Peng, C.-C.; Reid, M.W.; Shah, R.; Kuhn, P.; Cobrinik, D.; Hicks, J.; et al. Comprehensive Somatic Copy Number Analysis Using Aqueous Humor Liquid Biopsy for Retinoblastoma. Cancers 2021, 13, 3340. https://doi.org/10.3390/cancers13133340
Kim ME, Polski A, Xu L, Prabakar RK, Peng C-C, Reid MW, Shah R, Kuhn P, Cobrinik D, Hicks J, et al. Comprehensive Somatic Copy Number Analysis Using Aqueous Humor Liquid Biopsy for Retinoblastoma. Cancers. 2021; 13(13):3340. https://doi.org/10.3390/cancers13133340
Chicago/Turabian StyleKim, Mary E., Ashley Polski, Liya Xu, Rishvanth K. Prabakar, Chen-Ching Peng, Mark W. Reid, Rachana Shah, Peter Kuhn, David Cobrinik, James Hicks, and et al. 2021. "Comprehensive Somatic Copy Number Analysis Using Aqueous Humor Liquid Biopsy for Retinoblastoma" Cancers 13, no. 13: 3340. https://doi.org/10.3390/cancers13133340








