Evaluation of Integrated HPV DNA as Individualized Biomarkers for the Detection of Recurrent CIN2/3 during Post-Treatment Surveillance
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Identification of Viral-Cellular Junctions (vcj) and Development of the Individualized vcj-PCR
2.3. Index Tests and Reference Procedures
2.4. Study Endpoints
2.5. Statistical Analysis
3. Results
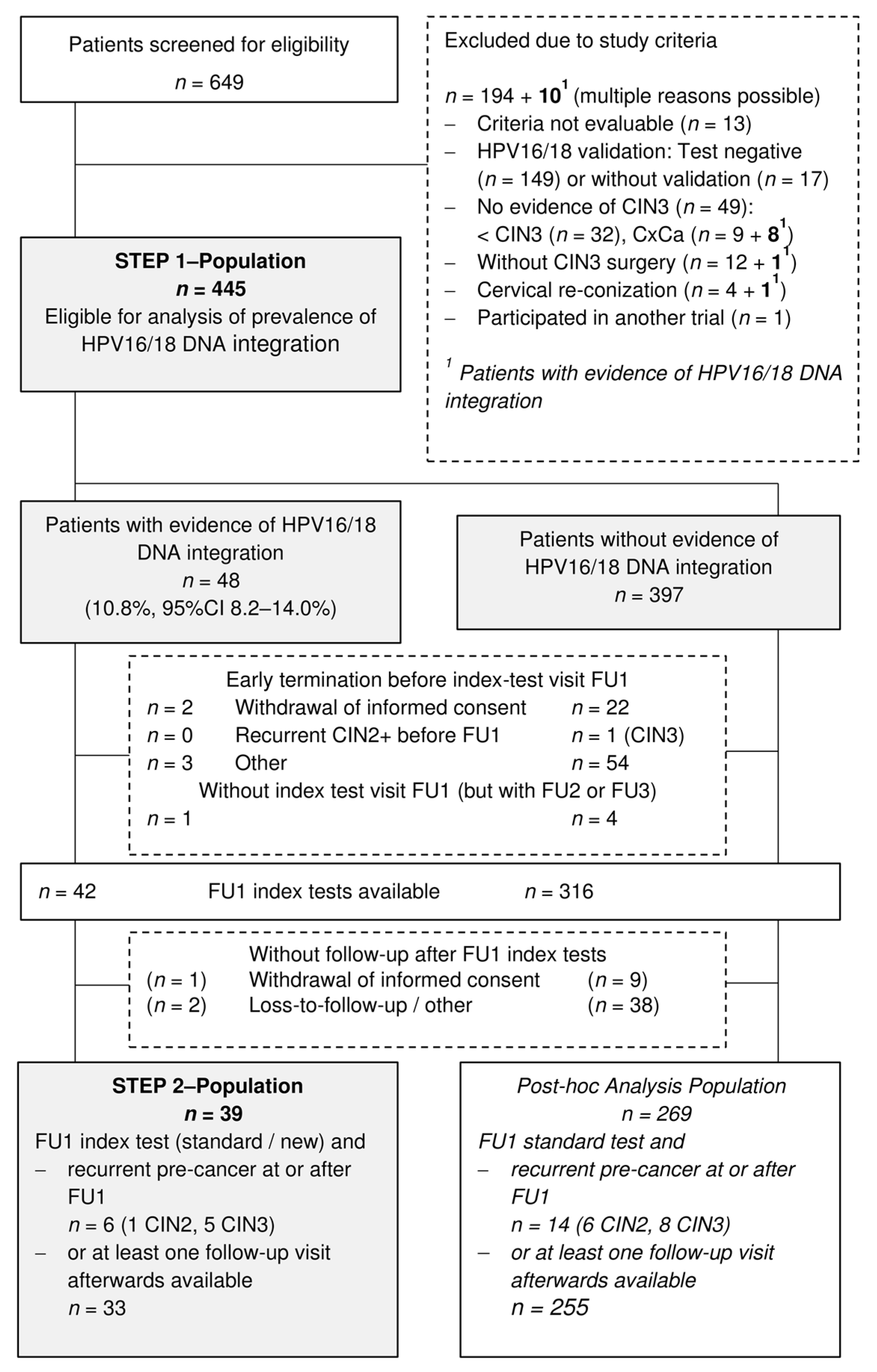
3.1. Recruitment and Patients Characteristics
3.2. Performance of Index Tests in Patients with Evidence of HPV16/18 DNA Integration
3.3. Performance of Standard Tests in Patients without Evidence of HPV16/18 DNA Integration (Post-Hoc Analyses)
4. Discussion
4.1. HPV-DNA Integration in CIN3
4.2. Predictive Accuracy of vcj-PCR/Cytology Co-Testing Compared to the Standard Co-Testing
4.3. Post Hoc Analyses of Patients without HPV16/18 DNA Integration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Sample Size Calculation
| Predictive Tests | Patients with HPV16/18 DNA Integration (n = 39) | ||||||
|---|---|---|---|---|---|---|---|
| with Recurrent Pre-Cancer at or after Visit | without Signs of Recurrence at or after Visit | without Visit | |||||
| First Follow-up | n = 6 (4 before second follow-up) | n = 33 | n = 0 | ||||
| Test results 1, n | Positive | Negative | Missing | Positive | Negative | Missing | |
| Cytology | 4 | 2 | 4 | 28 | 1 | ||
| hr-HPV | 5 | 1 | 3 | 30 | |||
| vcj-PCR | 3 | 3 | 0 | 33 | |||
| Cytology + hr-HPV (standard) | 6 | 0 | 6 | 27 | |||
| Cytology + vcj-PCR (new) | 5 | 1 | 4 | 29 | |||
| Second follow-up | n = 2 (2 before third follow-up) | n = 33 | n = 0 | ||||
| Test results 1, n | Positive | Negative | Missing | Positive | Negative | Missing | |
| Cytology | 1 | 0 | 1 | 2 | 27 | 4 | |
| hr-HPV | 2 | 0 | 1 | 31 | 1 | ||
| vcj-PCR | 1 | 1 | 0 | 32 | 1 | ||
| Cytology + hr-HPV (standard) | 2 | 0 | 3 | 29 | 1 | ||
| Cytology + vcj-PCR (new) | 2 | 0 | 2 | 30 | 1 | ||
| Third follow-up | n = 0 | n = 28 | n = 5 | ||||
| Test results 1, n | Positive | Negative | Missing | Positive | Negative | Missing | |
| Cytology | 1 | 26 | 1 | 5 | |||
| hr-HPV | 0 | 27 | 1 | 5 | |||
| vcj-PCR | 0 | 27 | 1 | 5 | |||
| Cytology + hr-HPV (standard) | 1 | 27 | 0 | 5 | |||
| Cytology + vcj-PCR (new) | 1 | 27 | 0 | 5 | |||
| Predictive Tests | Patients without HPV16/18 DNA Integration (n = 269) | ||||||
|---|---|---|---|---|---|---|---|
| with Recurrent Pre-Cancer at or after Visit | without Signs of Recurrence at or after Visit | without Visit | |||||
| First Follow-up | n = 14 (7 before second follow-up) | n = 255 | n = 0 | ||||
| Test results 1, n | Positive | Negative | Missing | Positive | Negative | Missing | |
| Cytology | 5 | 8 | 1 | 16 | 226 | 13 | |
| hr-HPV | 9 | 5 | 28 | 227 | |||
| Cytology + hr-HPV (standard) | 10 | 4 | 40 | 215 | |||
| Second follow-up | n = 7 (4 before third follow-up) | n = 255 | n = 0 | ||||
| Test results 1, n | Positive | Negative | Missing | Positive | Negative | Missing | |
| Cytology | 2 | 4 | 1 | 12 | 206 | 37 | |
| hr-HPV | 5 | 2 | 24 | 207 | 24 | ||
| Cytology + hr-HPV (standard) | 6 | 1 | 35 | 196 | 24 | ||
| Third follow-up | n = 3 (3 at third follow-up) | n = 232 | n = 23 | ||||
| Test results 1, n | Positive | Negative | Missing | Positive | Negative | Missing | |
| Cytology | 2 | 1 | 11 | 215 | 6 | 23 | |
| hr-HPV | 2 | 1 | 25 | 207 | 23 | ||
| Cytology + hr-HPV (standard) | 3 | 0 | 34 | 198 | 23 | ||
References
- Arbyn, M.; Sasieni, P.; Meijer, C.J.; Clavel, C.; Koliopoulos, G.; Dillner, J. Chapter 9: Clinical applications of hpv testing: A summary of meta-analyses. Vaccine 2006, 24 (Suppl. 3), 78–89. [Google Scholar] [CrossRef]
- Lili, E.; Chatzistamatiou, K.; Kalpaktsidou-Vakiani, A.; Moysiadis, T.; Agorastos, T. Low recurrence rate of high-grade cervical intraepithelial neoplasia after successful excision and routine colposcopy during follow-up. Medicine 2018, 97, e9719. [Google Scholar] [CrossRef]
- Ghaem-Maghami, S.; Sagi, S.; Majeed, G.; Soutter, W.P. Incomplete excision of cervical intraepithelial neoplasia and risk of treatment failure: A meta-analysis. Lancet Oncol. 2007, 8, 985–993. [Google Scholar] [CrossRef]
- Bjornerem, M.S.; Sorbye, S.W.; Skjeldestad, F.E. Recurrent disease after treatment for cervical intraepithelial neoplasia-the importance of a flawless definition of residual disease and length of follow-up. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 248, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Friebe, K.; Klapdor, R.; Hillemanns, P.; Jentschke, M. The value of partial hpv genotyping after conization of cervical dysplasias. Geburtshilfe Frauenheilkd. 2017, 77, 887–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruno, M.T.; Cassaro, N.; Garofalo, S.; Boemi, S. Hpv16 persistent infection and recurrent disease after leep. Virol. J. 2019, 16, 148. [Google Scholar] [CrossRef]
- Codde, E.; Munro, A.; Stewart, C.; Spilsbury, K.; Bowen, S.; Codde, J.; Steel, N.; Leung, Y.; Tan, J.; Salfinger, S.G.; et al. Risk of persistent or recurrent cervical neoplasia in patients with ‘pure’ adenocarcinoma-in-situ (ais) or mixed ais and high-grade cervical squamous neoplasia (cervical intra-epithelial neoplasia grades 2 and 3 (cin 2/3)): A population-based study. BJOG Int. J. Obstet. Gynaecol. 2018, 125, 74–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, W.D.; Oh, M.J.; Kim, S.M.; Nam, J.H.; Park, C.S.; Choi, H.S. Significance of human papillomavirus genotyping with high-grade cervical intraepithelial neoplasia treated by a loop electrosurgical excision procedure. Am. J. Obstet. Gynecol. 2010, 203, 72.e1–72.e6. [Google Scholar] [CrossRef] [PubMed]
- Kocken, M.; Uijterwaal, M.H.; de Vries, A.L.; Berkhof, J.; Ket, J.C.; Helmerhorst, T.J.; Meijer, C.J. High-risk human papillomavirus testing versus cytology in predicting post-treatment disease in women treated for high-grade cervical disease: A systematic review and meta-analysis. Gynecol. Oncol. 2012, 125, 500–507. [Google Scholar] [CrossRef]
- Massad, L.S.; Einstein, M.H.; Huh, W.K.; Katki, H.A.; Kinney, W.K.; Schiffman, M.; Solomon, D.; Wentzensen, N.; Lawson, H.W. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. Obstet. Gynecol. 2013, 121, 829–846. [Google Scholar] [CrossRef] [Green Version]
- Munro, A.; Codde, J.; Semmens, J.; Leung, Y.; Spilsbury, K.; Williams, V.; Steel, N.; Cohen, P.; Pavicic, H.; Westoby, V.; et al. Utilisation of co-testing (human papillomavirus DNA testing and cervical cytology) after treatment of cin: A survey of gps’ awareness and knowledge. Aust. Fam. Physician 2015, 44, 64–68. [Google Scholar] [PubMed]
- S3-Leitlinie. Prävention des Zervixkarzinoms. AWMF Registernummer: 015/027OL. Available online: http://www.leitlinienprogramm-onkologie.de/leitlinien/zervixkarzinom-praevention/ (accessed on 2 April 2021).
- Uijterwaal, M.H.; van Zummeren, M.; Kocken, M.; Luttmer, R.; Berkhof, J.; Witte, B.I.; van Baal, W.M.; Graziosi, G.C.M.; Verheijen, R.H.M.; Helmerhorst, T.J.M.; et al. Performance of cadm1/mal-methylation analysis for monitoring of women treated for high-grade cin. Gynecol. Oncol. 2016, 143, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Gong, J.; Xu, H.; Zhang, D.; Xia, N.; Li, H.; Song, K.; Lv, T.; Chen, Y.; Diao, Y.; et al. Good performance of p16/ki-67 dual-stain cytology for detection and post-treatment surveillance of high-grade cin/vain in a prospective, cross-sectional study. Diagn. Cytopathol. 2020, 48, 635–644. [Google Scholar] [CrossRef]
- Polman, N.J.; Uijterwaal, M.H.; Witte, B.I.; Berkhof, J.; van Kemenade, F.J.; Spruijt, J.W.; van Baal, W.M.; Graziosi, P.G.; van Dijken, D.K.; Verheijen, R.H.; et al. Good performance of p16/ki-67 dual-stained cytology for surveillance of women treated for high-grade cin. Int. J. Cancer 2017, 140, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Durst, M.; Kleinheinz, A.; Hotz, M.; Gissmann, L. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumours. J. Gen. Virol. 1985, 66 Pt 7, 1515–1522. [Google Scholar] [CrossRef]
- Luft, F.; Klaes, R.; Nees, M.; Durst, M.; Heilmann, V.; Melsheimer, P.; von Knebel Doeberitz, M. Detection of integrated papillomavirus sequences by ligation-mediated pcr (dips-pcr) and molecular characterization in cervical cancer cells. Int. J. Cancer 2001, 92, 9–17. [Google Scholar] [CrossRef]
- Hu, Z.; Zhu, D.; Wang, W.; Li, W.; Jia, W.; Zeng, X.; Ding, W.; Yu, L.; Wang, X.; Wang, L.; et al. Genome-wide profiling of hpv integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat. Genet. 2015, 47, 158–163. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of hpv integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, C.; Gao, W.; Wang, L.; Pan, Y.; Gao, Y.; Lu, Z.; Ke, Y. Genome-wide profiling of the human papillomavirus DNA integration in cervical intraepithelial neoplasia and normal cervical epithelium by hpv capture technology. Sci. Rep. 2016, 6, 35427. [Google Scholar] [CrossRef]
- Durst, M.; Croce, C.M.; Gissmann, L.; Schwarz, E.; Huebner, K. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc. Natl. Acad. Sci. USA 1987, 84, 1070–1074. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, M.; Driesch, C.; Jansen, L.; Runnebaum, I.B.; Durst, M. Non-random integration of the hpv genome in cervical cancer. PLoS ONE 2012, 7, e39632. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, M.; Driesch, C.; Beer-Grondke, K.; Jansen, L.; Runnebaum, I.B.; Durst, M. Loss of gene function as a consequence of human papillomavirus DNA integration. Int. J. Cancer 2012, 131, E593–E602. [Google Scholar] [CrossRef]
- Xu, B.; Chotewutmontri, S.; Wolf, S.; Klos, U.; Schmitz, M.; Durst, M.; Schwarz, E. Multiplex identification of human papillomavirus 16 DNA integration sites in cervical carcinomas. PLoS ONE 2013, 8, e66693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraus, I.; Driesch, C.; Vinokurova, S.; Hovig, E.; Schneider, A.; von Knebel Doeberitz, M.; Durst, M. The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res. 2008, 68, 2514–2522. [Google Scholar] [CrossRef] [Green Version]
- Carow, K.; Golitz, M.; Wolf, M.; Hafner, N.; Jansen, L.; Hoyer, H.; Schwarz, E.; Runnebaum, I.B.; Durst, M. Viral-cellular DNA junctions as molecular markers for assessing intra-tumor heterogeneity in cervical cancer and for the detection of circulating tumor DNA. Int. J. Mol. Sci. 2017, 18, 2032. [Google Scholar] [CrossRef] [Green Version]
- Woodman, C.B.; Collins, S.I.; Young, L.S. The natural history of cervical hpv infection: Unresolved issues. Nat. Rev. 2007, 7, 11–22. [Google Scholar] [CrossRef]
- Jacobs, M.V.; Snijders, P.J.; van den Brule, A.J.; Helmerhorst, T.J.; Meijer, C.J.; Walboomers, J.M. A general primer gp5+/gp6(+)-mediated pcr-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 1997, 35, 791–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newcombe, R.G. Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat. Med. 1998, 17, 857–872. [Google Scholar] [CrossRef]
- Alonzo, T.A.; Pepe, M.S.; Moskowitz, C.S. Sample size calculations for comparative studies of medical tests for detecting presence of disease. Stat. Med. 2002, 21, 835–852. [Google Scholar] [CrossRef] [PubMed]
- Dyer, N.; Young, L.; Ott, S. Artifacts in the data of hu et al. Nat. Genet. 2016, 48, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, P.M.; Irwig, L.; Craig, J.; Glasziou, P. Comparative accuracy: Assessing new tests against existing diagnostic pathways. BMJ 2006, 332, 1089–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Patient Characteristics | with Integration (n = 48) | without Integration (n = 397) | Total (n = 445) |
|---|---|---|---|
| Age (years), median (range) | 33 (23–51) | 32 (21–57) | 32 (21–57) |
| Number of life births, n [%] | |||
| None | 24 (50.0) | 193 (48.6) | 217 (48.8) |
| One | 13 (27.1) | 113 (28.5) | 126 (28.3) |
| More than one | 11 (22.9) | 91 (22.9) | 102 (22.9) |
| Any contraception, n [%] | 34 (70.8) | 251 (63.4) | 285 (64.2) |
| Thereof contraceptive pill | 23 (67.6) | 162 (64.5) | 185 (64.9) |
| Thereof condom | 4 (11.8) | 52 (20.7) | 56 (19.6) |
| Pregnant at study entry, n [%] | 0 | 2 (0.5) | 2 (0.4) |
| Menopause, n [%] | 0 | 11 (2.8) | 11 (2.5) |
| Smoking status, n [%] | |||
| Current smoker | 19 (39.6) | 180 (45.3) | 199 (44.7) |
| Ex-smoker | 7 (14.6) | 42 (10.6) | 49 (11.0) |
| Non-smoker | 21 (43.7) | 168 (42.3) | 189 (42.5) |
| Unknown | 1 (2.1) | 7 (1.8) | 8 (1.8) |
| HPV vaccination, n [%] | 5 (12.8) | 46 (13.7) | 51 (13.6) |
| Type of primary surgery, n [%] | |||
| High-frequency (HF) loop | 33 (68.8) | 282 (71.0) | 315 (70.8) |
| Laser conisation | 10 (20.8) | 75 (18.9) | 85 (19.1) |
| HF loop and laser conisation | 5 (10.4) | 40 (10.1) | 45 (10.1) |
| Resection margin, n [%] | |||
| Negative | 30 (62.5) | 266 (67.0) | 296 (66.5) |
| Positive | 8 (16.7) | 52 (13.1) | 60 (13.5) |
| Unknown | 10 (20.8) | 79 (19.9) | 89 (20.0) |
| First Follow-Up 6 Months after Surgery | True Positive Rate (a) | False Positive Rate 95% CI [%] | Test Positive Rate 95% CI [%] | ||||
|---|---|---|---|---|---|---|---|
| With integration (Step 2 analysis population, n = 39) | |||||||
| Cytology | (4/6) | (4/32) | 12.5 | 5.0–28.1 | (8/38) | 21.1 | 11.1–36.3 |
| hr-HPV | (5/6) | (3/33) | 9.1 | 3.1–23.6 | (8/39) | 20.5 | 10.8–35.5 |
| vcj-PCR | (3/6) | (0/33) | 0.0 | 0.0–10.4 | (3/39) | 7.7 | 2.7–20.3 |
| Cytology + vcj-PCR (new) | (5/6) | (4/33) | 12.1 | 4.8–27.3 | (9/39) | 23.1 | 12.6–38.3 |
| Cytology + hrHPV (standard) | (6/6) | (6/33) | 18.2 | 8.6–34.4 | (12/39) | 30.8 | 18.6–46.4 |
| Rate ratio (new/standard) | 0.67 | 0.38–1.17 | 0.75 | 0.54–1.04 | |||
| Without integration (post-hoc analysis population, n = 269) | |||||||
| Cytology | (5/13) | (16/242) | 6.6 | 4.1–10.5 | (21/255) | 8.2 | 5.4–12.3 |
| hr-HPV | (9/14) | (28/255) | 11.0 | 7.7–15.4 | (37/269) | 13.8 | 10.1–18.4 |
| Cytology + hrHPV (standard) | (10/14) | (40/255) | 15.7 | 11.7–20.7 | (50/269) | 18.6 | 14.4–23.7 |
| First Follow-Up 6 Months after Surgery | Positive Predictive Value 95% CI [%] | Negative Predictive Value 95% CI [%] | ||||
|---|---|---|---|---|---|---|
| With integration (Step 2, analysis population, n = 39) | ||||||
| Cytology | (4/8) | (a) | (28/30) | 93.3 | 78.7–98.2 | |
| hr-HPV | (5/8) | (a) | (30/31) | 96.8 | 83.8–99.4 | |
| vcj-PCR | (3/3) | (a) | (33/36) | 91.7 | 78.2–97.1 | |
| Cytology + vcj-PCR (new) | (5/9) | (a) | (29/30) | 96.7 | 83.3–99.4 | |
| Cytology + hr-HPV (standard) | (6/12) | (a) | (27/27) | 100 | 87.5–100 | |
| Without integration (post-hoc analysis population, n = 269) | ||||||
| Cytology | (5/21) | 23.8 | 10.6–45.1 | (226/234) | 96.6 | 93.4–98.3 |
| hr-HPV | (9/37) | 24.3 | 13.4–40.1 | (227/232) | 97.8 | 95.1–99.1 |
| Cytology + hr-HPV (standard) | (10/50) | 20.0 | 11.2–33.0 | (215/219) | 98.2 | 95.4–99.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoyer, H.; Mehlhorn, G.; Scheungraber, C.; Hagemann, I.; Hirchenhain, C.; Woelber, L.; Stolte, C.; Hampl, M.; Scherbring, S.; Denecke, A.; et al. Evaluation of Integrated HPV DNA as Individualized Biomarkers for the Detection of Recurrent CIN2/3 during Post-Treatment Surveillance. Cancers 2021, 13, 3309. https://doi.org/10.3390/cancers13133309
Hoyer H, Mehlhorn G, Scheungraber C, Hagemann I, Hirchenhain C, Woelber L, Stolte C, Hampl M, Scherbring S, Denecke A, et al. Evaluation of Integrated HPV DNA as Individualized Biomarkers for the Detection of Recurrent CIN2/3 during Post-Treatment Surveillance. Cancers. 2021; 13(13):3309. https://doi.org/10.3390/cancers13133309
Chicago/Turabian StyleHoyer, Heike, Grit Mehlhorn, Cornelia Scheungraber, Ingke Hagemann, Christine Hirchenhain, Linn Woelber, Claudia Stolte, Monika Hampl, Sarah Scherbring, Agnieszka Denecke, and et al. 2021. "Evaluation of Integrated HPV DNA as Individualized Biomarkers for the Detection of Recurrent CIN2/3 during Post-Treatment Surveillance" Cancers 13, no. 13: 3309. https://doi.org/10.3390/cancers13133309






