Predicting Postoperative Complications in Cancer Patients: A Survey Bridging Classical and Machine Learning Contributions to Postsurgical Risk Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Background
2.1. Classic Prognostication: Calculators, Scores and Indexes
2.2. Machine Learning Models
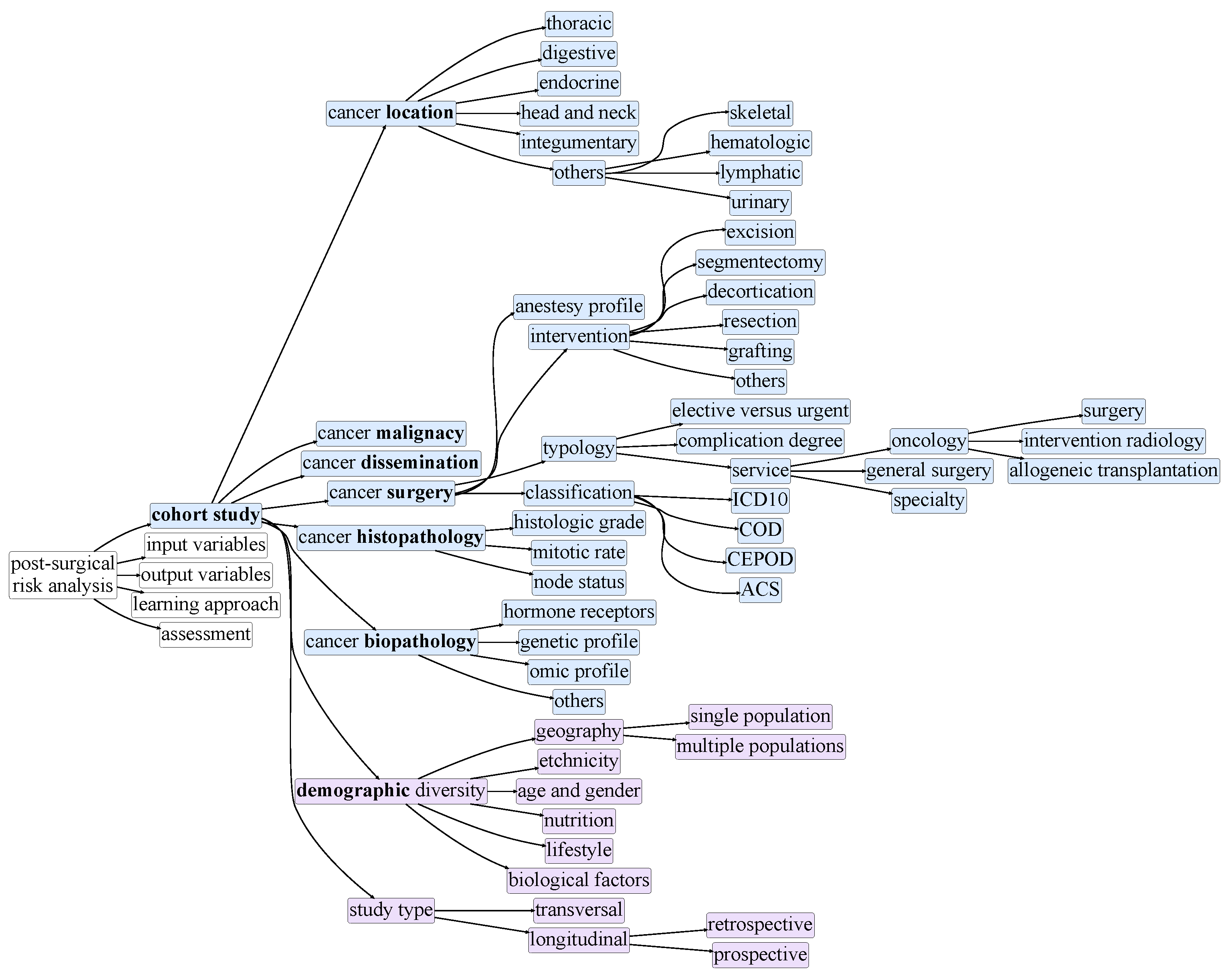
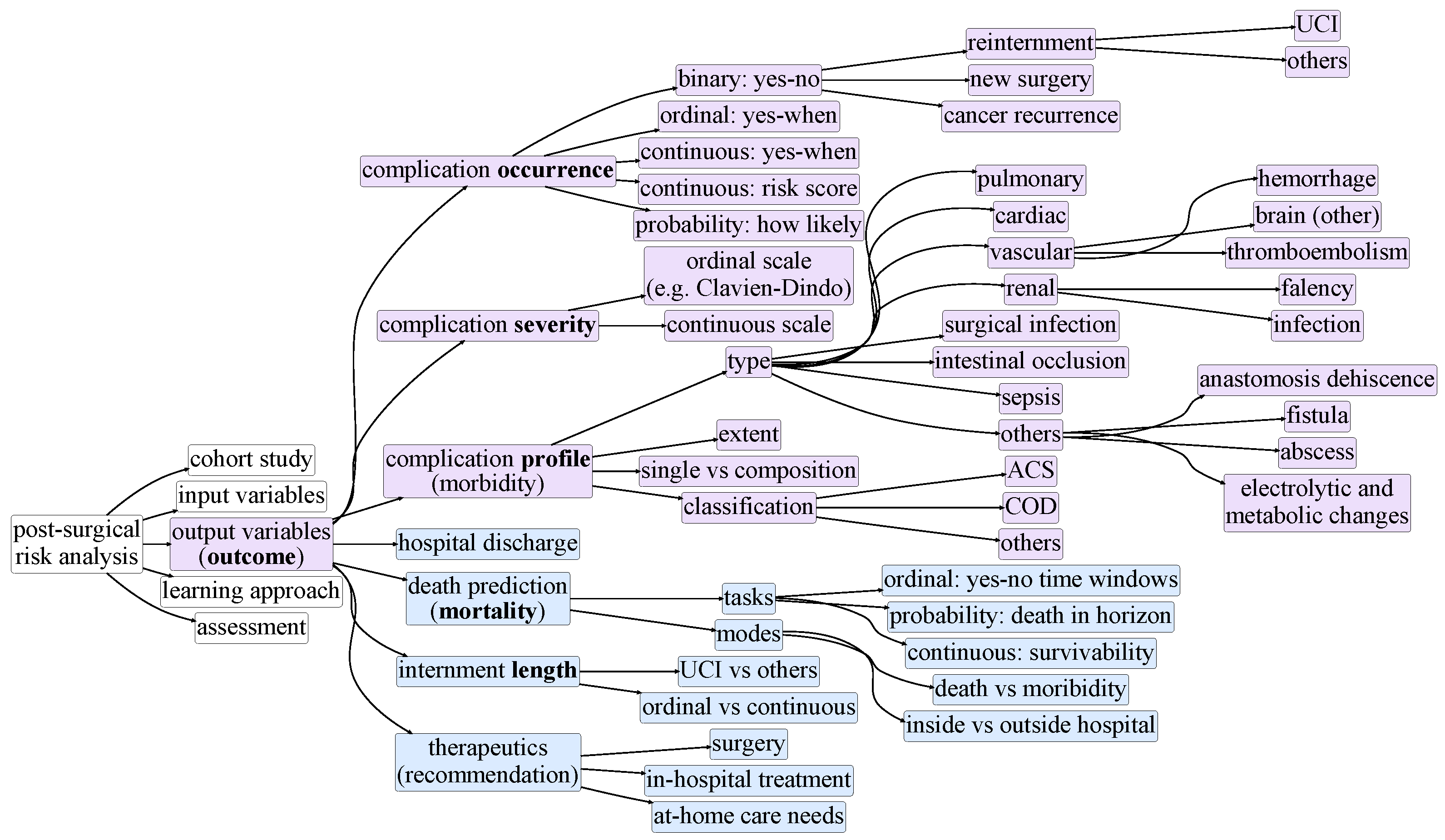
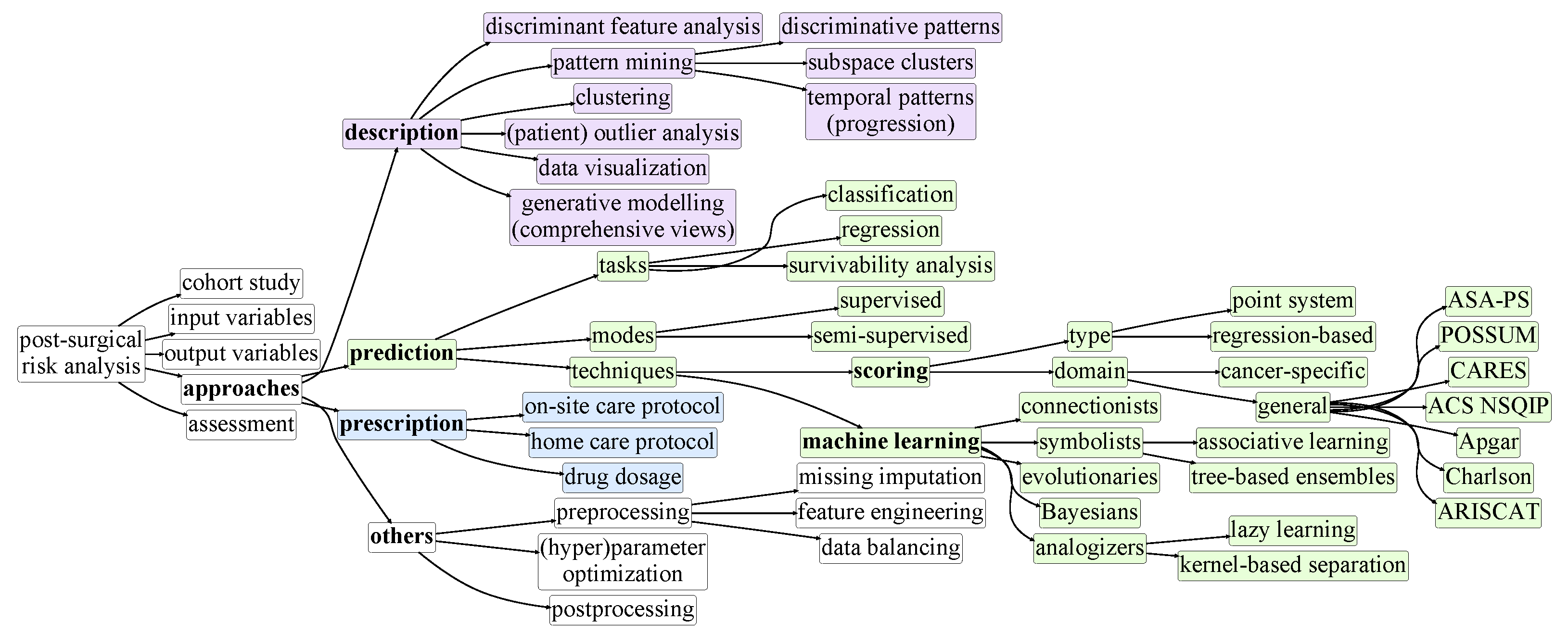
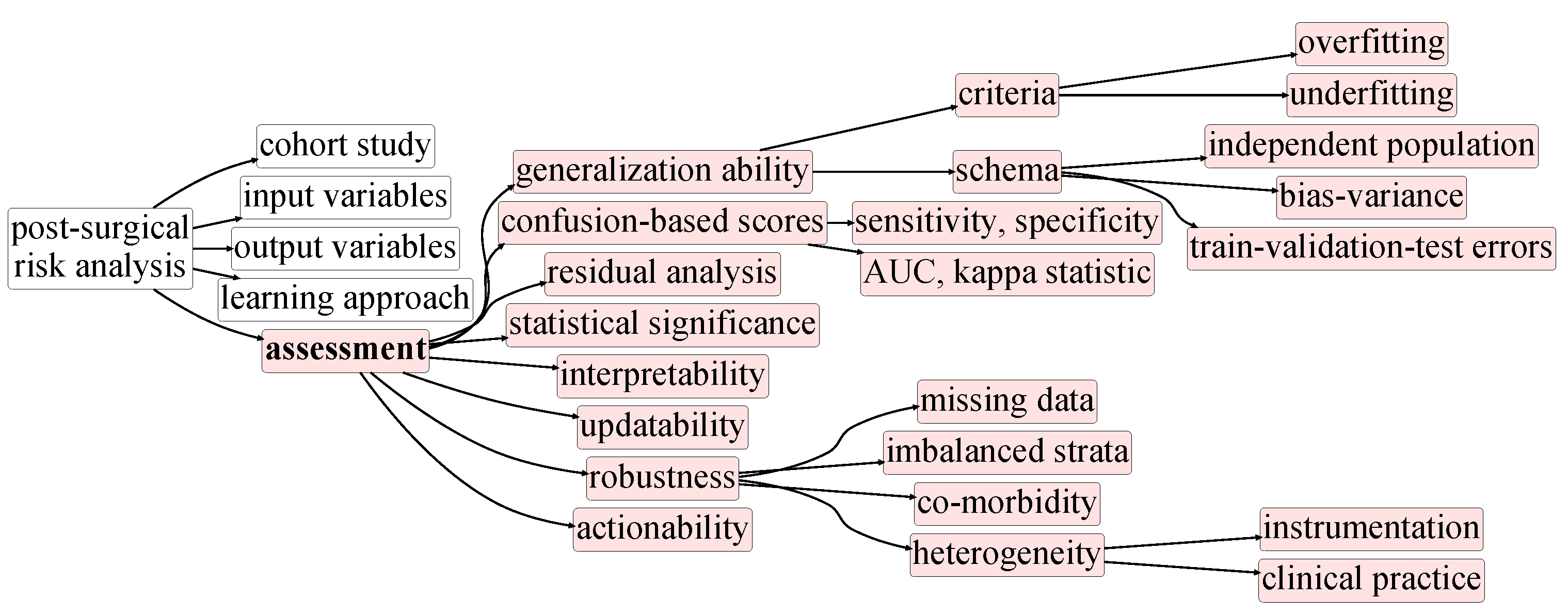
3. Taxonomy of Postsurgical Risk Analysis
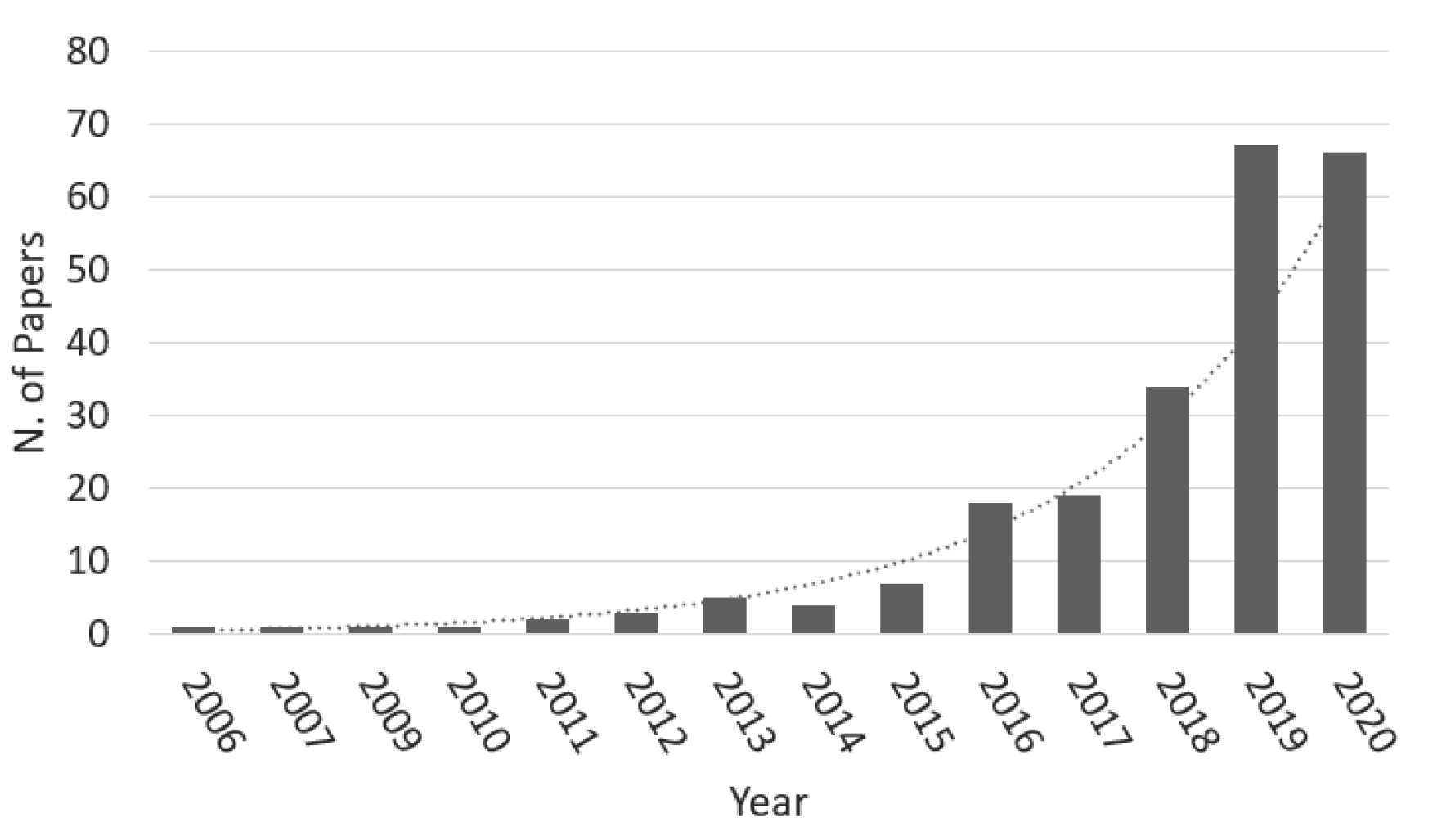
4. Postoperative Prognostics: A Literature Review
4.1. Traditional Risk Scores Studies
4.2. Machine Learning Studies
4.3. Preprocessing
4.4. Prognostic Accuracy
5. Discussion
- Capture non-linear relationships within data;
- Translate risk scores into well-defined clinical decisions;
- Properly deal with the high-dimensional nature of clinical data;
- Identify local dependencies between variables;
- Be incrementally updated in the presence of new data;
- Tolerate arbitrarily-high levels of missing data;
- Explore the inherent temporal nature of clinical data.
5.1. On the Interpretability of Predictive Models
5.2. A final note on quantitative assessments
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASA-PS | American Society of Anesthesiologists for the classification of Physical Status |
| ACS | American College of Surgeons |
| ANFIS | Adaptive Neuro-Fuzzy Inference System |
| ANN | Artificial Neural Network |
| CARES | Combined Assessment of Risk Encountered in Surgery |
| AUC | Area Under the Curve |
| DT | Decision Tree |
| ELM | Extreme Learning Machines |
| FP | False Positive |
| FN | False Negative |
| HL | Hosmer–Lemeshow Test |
| IPO | Instituto Português de Oncologia |
| GAM | Generalized Additive Model |
| GB | Gradient Boosting |
| kNN | k-Nearest Neighbors |
| LR | Logistic Regression |
| MAE | Mean Absolute Error |
| ML | Machine Learning |
| MLP | Multi-Layer Perceptron |
| NB | Naive Bayes |
| NN | Neural Network |
| NSQIP | National Surgical Quality Improvement Program |
| POSSUM | Physiological and Operative Severity Score for enumeration of Mortality and Morbidity |
| PS | Point System |
| TP | True Positive |
| TN | True Negative |
| RAE | Relative Absolute Error |
| RELM | Regularized Extreme Learning Machines |
| RF | Random Forest |
| ROC | Receiver Operating Curve |
| RMSE | Root Mean Squared Error |
| RRSE | Root Relative Squared Error |
| SVM | Support Vector Machine |
| WI | Weighted Index |
| XGBoost | Extreme Gradient Boosting |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Nowakowski, M.; Pisarska, M.; Rubinkiewicz, M.; Torbicz, G.; Gajewska, N.; Mizera, M.; Major, P.; Potocki, P.; Radkowiak, D.; Pedziwiatr, M. Postoperative complications are associated with worse survival after laparoscopic surgery for non-metastatic colorectal cancer–interim analysis of 3-year overall survival. Videosurgery Other Miniinvasive Tech. 2018, 13, 326. [Google Scholar] [CrossRef] [PubMed]
- Tekkis, P.P.; Poloniecki, J.D.; Thompson, M.R.; Stamatakis, J.D. Operative mortality in colorectal cancer: Prospective national study. BMJ 2003, 327, 1196–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, W.L.; Choi, H.K.; Lee, Y.M.; Ho, J.W. The impact of postoperative complications on long-term outcomes following curative resection for colorectal cancer. Ann. Surg. Oncol. 2007, 14, 2559–2566. [Google Scholar] [CrossRef]
- Wijeysundera, D.N. Predicting outcomes: Is there utility in risk scores? Can. J. Anesth. Can. D’anesthésie 2016, 63, 148–158. [Google Scholar] [CrossRef]
- Cruz, J.A.; Wishart, D.S. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. 2006, 2, 117693510600200030. [Google Scholar] [CrossRef]
- Yu, Y.; Li, M.; Liu, L.; Li, Y.; Wang, J. Clinical big data and deep learning: Applications, challenges, and future outlooks. Big Data Min. Anal. 2019, 2, 288–305. [Google Scholar] [CrossRef] [Green Version]
- Maclin, P.S.; Dempsey, J.; Brooks, J.; Rand, J. Using neural networks to diagnose cancer. J. Med. Syst. 1991, 15, 11–19. [Google Scholar] [CrossRef]
- Naguib, R.N.; Sherbet, G.V. Artificial Neural Networks in Cancer Diagnosis, Prognosis, and Patient Management; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Saklad, M. Grading of patients for surgical procedures. Anesthesiol. J. Am. Soc. Anesthesiol. 1941, 2, 281–284. [Google Scholar] [CrossRef]
- Hackett, N.J.; De Oliveira, G.S.; Jain, U.K.; Kim, J.Y. ASA class is a reliable independent predictor of medical complications and mortality following surgery. Int. J. Surg. 2015, 18, 184–190. [Google Scholar] [CrossRef]
- Aronson, W.; McAuliffe, M.S.; Miller, K. Variability in the American Society of Anesthesiologists physical status classification scale. AANA J. 2003, 71, 265–276. [Google Scholar]
- Copeland, G.; Jones, D.; Walters, M. POSSUM: A scoring system for surgical audit. Br. J. Surg. 1991, 78, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Bilimoria, K.Y.; Liu, Y.; Paruch, J.L.; Zhou, L.; Kmiecik, T.E.; Ko, C.Y.; Cohen, M.E. Development and evaluation of the universal ACS NSQIP surgical risk calculator: A decision aid and informed consent tool for patients and surgeons. J. Am. Coll. Surg. 2013, 217, 833–842. [Google Scholar] [CrossRef] [Green Version]
- Zupan, B.; DemšAr, J.; Kattan, M.W.; Beck, J.R.; Bratko, I. Machine learning for survival analysis: A case study on recurrence of prostate cancer. Artif. Intell. Med. 2000, 20, 59–75. [Google Scholar] [CrossRef] [Green Version]
- Takada, M.; Sugimoto, M.; Masuda, N.; Iwata, H.; Kuroi, K.; Yamashiro, H.; Ohno, S.; Ishiguro, H.; Inamoto, T.; Toi, M. Prediction of postoperative disease-free survival and brain metastasis for HER2-positive breast cancer patients treated with neoadjuvant chemotherapy plus trastuzumab using a machine learning algorithm. Breast Cancer Res. Treat. 2018, 172, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.A.; Collins, G.S. The rise of big clinical databases. Br. J. Surg. 2015, 102, e93–e101. [Google Scholar] [CrossRef]
- Weston, A.D.; Hood, L. Systems biology, proteomics, and the future of health care: Toward predictive, preventative, and personalized medicine. J. Proteome Res. 2004, 3, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidey-Gibbons, J.A.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Patrick, E.A.; Fischer, F.P., III. A generalized k-nearest neighbor rule. Inf. Control 1970, 16, 128–152. [Google Scholar] [CrossRef] [Green Version]
- Langley, P.; Iba, W.; Thompson, K. An analysis of Bayesian classifiers. In AAAI; AAAI Press: San Jose, CA, USA, 1992; Volume 90, pp. 223–228. [Google Scholar]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef] [Green Version]
- Shaik, A.B.; Srinivasan, S. A brief survey on random forest ensembles in classification model. In International Conference on Innovative Computing and Communications; Springer: Singapore, 2019; pp. 253–260. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Dindo, D.; Demartines, N.; Clavien, P.A. Classification of surgical complications: A new proposal with evaluation in a cohort of 6336 patients and results of a survey. Ann. Surg. 2004, 240, 205. [Google Scholar] [CrossRef]
- Alexandre, L.; Costa, R.S.; Santos, L.L.; Henriques, R. Mining pre-surgical patterns able to discriminate post-surgical outcomes in the oncological domain. IEEE J. Biomed. Health Inform. 2021. [Google Scholar] [CrossRef] [PubMed]
- Knaus, W.A.; Draper, E.A.; Wagner, D.P.; Zimmerman, J.E. APACHE II: A severity of disease classification system. Crit. Care Med. 1985, 13, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Charlson, M.E.; Pompei, P.; Ales, K.L.; MacKenzie, C.R. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J. Chronic Dis. 1987, 40, 373–383. [Google Scholar] [CrossRef]
- Marcantonio, E.R.; Goldman, L.; Mangione, C.M.; Ludwig, L.E.; Muraca, B.; Haslauer, C.M.; Donaldson, M.C.; Whittemore, A.D.; Sugarbaker, D.J.; Poss, R.; et al. A clinical prediction rule for delirium after elective noncardiac surgery. JAMA 1994, 271, 134–139. [Google Scholar] [CrossRef]
- Whiteley, M.; Prytherch, D.; Higgins, B.; Weaver, P.; Prout, W. An evaluation of the POSSUM surgical scoring system. Br. J. Surg. 1996, 83, 812–815. [Google Scholar] [CrossRef]
- Roques, F.; Nashef, S.; Michel, P.; Gauducheau, E.; De Vincentiis, C.; Baudet, E.; Cortina, J.; David, M.; Faichney, A.; Gavrielle, F.; et al. Risk factors and outcome in European cardiac surgery: Analysis of the EuroSCORE multinational database of 19030 patients. Eur. J. Cardio-Thorac. Surg. 1999, 15, 816–823. [Google Scholar] [CrossRef] [Green Version]
- Dupuis, J.Y.; Wang, F.; Nathan, H.; Lam, M.; Grimes, S.; Bourke, M. The Cardiac Anesthesia Risk Evaluation ScoreA Clinically Useful Predictor of Mortality and Morbidity after Cardiac Surgery. Anesthesiol. J. Am. Soc. Anesthesiol. 2001, 94, 194–204. [Google Scholar]
- Arozullah, A.M.; Khuri, S.F.; Henderson, W.G.; Daley, J. Development and validation of a multifactorial risk index for predicting postoperative pneumonia after major noncardiac surgery. Ann. Intern. Med. 2001, 135, 847–857. [Google Scholar] [CrossRef]
- Sutton, R.; Bann, S.; Brooks, M.; Sarin, S. The Surgical Risk Scale as an improved tool for risk-adjusted analysis in comparative surgical audit. Br. J. Surg. 2002, 89, 763–768. [Google Scholar] [CrossRef]
- Donati, A.; Ruzzi, M.; Adrario, E.; Pelaia, P.; Coluzzi, F.; Gabbanelli, V.; Pietropaoli, P. A new and feasible model for predicting operative risk. Br. J. Anaesth. 2004, 93, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Gawande, A.A.; Kwaan, M.R.; Regenbogen, S.E.; Lipsitz, S.A.; Zinner, M.J. An Apgar score for surgery. J. Am. Coll. Surg. 2007, 204, 201–208. [Google Scholar] [CrossRef]
- Canet, J.; Gallart, L.; Gomar, C.; Paluzie, G.; Valles, J.; Castillo, J.; Sabate, S.; Mazo, V.; Briones, Z.; Sanchis, J. Prediction of postoperative pulmonary complications in a population-based surgical cohort. Anesthesiol. J. Am. Soc. Anesthesiol. 2010, 113, 1338–1350. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Gupta, H.; Sundaram, A.; Kaushik, M.; Fang, X.; Miller, W.J.; Esterbrooks, D.J.; Hunter, C.B.; Pipinos, I.I.; Johanning, J.M.; et al. Development and validation of a risk calculator for prediction of cardiac risk after surgery. Circulation 2011, 124, 381–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaid, S.; Bell, T.; Grim, R.; Ahuja, V. Predicting risk of death in general surgery patients on the basis of preoperative variables using American College of Surgeons National Surgical Quality Improvement Program data. Perm. J. 2012, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- L’heureux, A.; Grolinger, K.; Elyamany, H.F.; Capretz, M.A. Machine learning with big data: Challenges and approaches. IEEE Access 2017, 5, 7776–7797. [Google Scholar] [CrossRef]
- Simester, D.; Timoshenko, A.; Zoumpoulis, S.I. Targeting prospective customers: Robustness of machine-learning methods to typical data challenges. Manag. Sci. 2020, 66, 2495–2522. [Google Scholar] [CrossRef]
- Khan, M.U.; Choi, J.P.; Shin, H.; Kim, M. Predicting breast cancer survivability using fuzzy decision trees for personalized healthcare. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; pp. 5148–5151. [Google Scholar]
- Chang, S.W.; Abdul-Kareem, S.; Merican, A.F.; Zain, R.B. Oral cancer prognosis based on clinicopathologic and genomic markers using a hybrid of feature selection and machine learning methods. BMC Bioinform. 2013, 14, 170. [Google Scholar] [CrossRef] [Green Version]
- Zięba, M.; Tomczak, J.M.; Lubicz, M.; Świątek, J. Boosted SVM for extracting rules from imbalanced data in application to prediction of the post-operative life expectancy in the lung cancer patients. Appl. Soft Comput. 2014, 14, 99–108. [Google Scholar] [CrossRef]
- Danjuma, K.J. Performance evaluation of machine learning algorithms in post-operative life expectancy in the lung cancer patients. arXiv 2015, arXiv:1504.04646. [Google Scholar]
- Parmar, C.; Grossmann, P.; Rietveld, D.; Rietbergen, M.M.; Lambin, P.; Aerts, H.J. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front. Oncol. 2015, 5, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Lam, K.M.; Deng, Z.; Choi, K.S. Prediction of mortality after radical cystectomy for bladder cancer by machine learning techniques. Comput. Biol. Med. 2015, 63, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Thottakkara, P.; Ozrazgat-Baslanti, T.; Hupf, B.B.; Rashidi, P.; Pardalos, P.; Momcilovic, P.; Bihorac, A. Application of machine learning techniques to high-dimensional clinical data to forecast postoperative complications. PLoS ONE 2016, 11, e0155705. [Google Scholar] [CrossRef] [Green Version]
- Soguero-Ruiz, C.; Hindberg, K.; Mora-Jiménez, I.; Rojo-Álvarez, J.L.; Skrøvseth, S.O.; Godtliebsen, F.; Mortensen, K.; Revhaug, A.; Lindsetmo, R.O.; Augestad, K.M.; et al. Predicting colorectal surgical complications using heterogeneous clinical data and kernel methods. J. Biomed. Inform. 2016, 61, 87–96. [Google Scholar] [CrossRef]
- Kim, D.W.; Lee, S.; Kwon, S.; Nam, W.; Cha, I.H.; Kim, H.J. Deep learning-based survival prediction of oral cancer patients. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Parikh, R.B.; Manz, C.; Chivers, C.; Regli, S.H.; Braun, J.; Draugelis, M.E.; Schuchter, L.M.; Shulman, L.N.; Navathe, A.S.; Patel, M.S.; et al. Machine Learning Approaches to Predict 6-Month Mortality among Patients with Cancer. JAMA Netw. Open 2019, 2, e1915997. [Google Scholar] [CrossRef] [Green Version]
- Bellman, R.E. Adaptive Control Processes: A Guided Tour; Princeton University Press: Princeton, NJ, USA, 1961. [Google Scholar]
- Ibrahim, J.G.; Chu, H.; Chen, M.H. Missing data in clinical studies: Issues and methods. J. Clin. Oncol. 2012, 30, 3297. [Google Scholar] [CrossRef]
- Van Stiphout, R.; Postma, E.; Valentini, V.; Lambin, P. The contribution of machine learning to predicting cancer outcome. Artif. Intell. 2010, 350, 400. [Google Scholar]
- Jiang, C.; Yang, Z. CKNNI: An improved knn-based missing value handling technique. In Advanced Intelligent Computing Theories and Applications, Proceedings of the International Conference on Intelligent Computing, Fuzhou, China, 20–23 August 2015; Springer International Publishing: Berlin, Germany, 2015; pp. 441–452. [Google Scholar]
- Wilk, S.; Stefanowski, J.; Wojciechowski, S.; Farion, K.J.; Michalowski, W. Application of preprocessing methods to imbalanced clinical data: An experimental study. In Conference of Information Technologies in Biomedicine; Springer International Publishing: Berlin, Germany, 2016; pp. 503–515. [Google Scholar]
- He, H.; Garcia, E.A. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Klecka, W.R.; Iversen, G.R.; Klecka, W.R. Discriminant Analysis; SAGE Publications: New York, NY, USA, 1980; Volume 19. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Kuncheva, L.I.; Rodríguez, J.J. On feature selection protocols for very low-sample-size data. Pattern Recognit. 2018, 81, 660–673. [Google Scholar] [CrossRef] [Green Version]
- Hossin, M.; Sulaiman, M. A review on evaluation metrics for data classification evaluations. Int. J. Data Min. Knowl. Manag. Process 2015, 5, 1. [Google Scholar]
- Pearson, K.X. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1900, 50, 157–175. [Google Scholar] [CrossRef] [Green Version]
- Hosmer, D.W., Jr.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression; John Wiley & Sons: Hoboken, NJ, USA, 2013; Volume 398. [Google Scholar]
- Patricio, A.; Costa, R.S.; Henriques, R. Predictability of COVID-19 Hospitalizations, Intensive Care Unit Admissions, and Respiratory Assistance in Portugal: Longitudinal Cohort Study. J. Med. Internet Res. 2020, 23, e26075. [Google Scholar] [CrossRef] [PubMed]
- Garofallo, S.B.; Machado, D.P.; Rodrigues, C.G.; Bordim, O., Jr.; Kalil, R.A.; Portal, V.L. Applicability of two international risk scores in cardiac surgery in a reference center in Brazil. Arq. Bras. Cardiol. 2014, 102, 539–548. [Google Scholar] [CrossRef]
- Chin, C.T.; Chua, T.; LIM, S. Risk assessment models in acute coronary syndromes and their applicability in Singapore. Ann. Acad. Med. Singap. 2010, 39, 216–220. [Google Scholar]
- Formiga, F.; Masip, J.; Chivite, D.; Corbella, X. Applicability of the heart failure Readmission Risk score: A first European study. Int. J. Cardiol. 2017, 236, 304–309. [Google Scholar] [CrossRef]
- Goh, L.G.; Dhaliwal, S.S.; Welborn, T.A.; Thompson, P.L.; Maycock, B.R.; Kerr, D.A.; Lee, A.H.; Bertolatti, D.; Clark, K.M.; Naheed, R.; et al. Cardiovascular disease risk score prediction models for women and its applicability to Asians. Int. J. Women’s Health 2014, 6, 259. [Google Scholar]
- Sutton, C.; McCallum, A. An introduction to conditional random fields. Found. Trends Mach. Learn. 2012, 4, 267–373. [Google Scholar] [CrossRef]
- Greenland, S.; Schwartzbaum, J.A.; Finkle, W.D. Problems due to small samples and sparse data in conditional logistic regression analysis. Am. J. Epidemiol. 2000, 151, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Domingos, P. The Master Algorithm: How the Quest for the Ultimate Learning Machine Will Remake Our World; Basic Books, Inc.: New York, NY, USA, 2018. [Google Scholar]
- Millerand, G. Enhancing Decision Tree Accuracy and Compactness with Improved Categorical Split and Sampling Techniques. Master’s Thesis, KTH, School of Electrical Engineering and Computer Science, Stockholm, Sweden, 2020. TRITA-EECS-EX 254. [Google Scholar]
- Katz, G.; Shabtai, A.; Rokach, L.; Ofek, N. ConfDtree: A statistical method for improving decision trees. J. Comput. Sci. Technol. 2014, 29, 392–407. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased boosting with categorical features. In Proceedings of the 32nd Conference on Neural Information Processing Systems (NeurIPS 2018), Montreal, QC, Canada, 3–8 December 2018; pp. 6638–6648. [Google Scholar]
- Auret, L.; Aldrich, C. Interpretation of nonlinear relationships between process variables by use of random forests. Miner. Eng. 2012, 35, 27–42. [Google Scholar] [CrossRef]
- Henriques, R.; Madeira, S.C. FleBiC: Learning classifiers from high-dimensional biomedical data using discriminative biclusters with non-constant patterns. Pattern Recognit. 2021, 115, 107900. [Google Scholar] [CrossRef]
- Ghahramani, Z. An introduction to hidden Markov models and Bayesian networks. In Hidden Markov Models: Applications in Computer Vision; World Scientific: Singapore, 2001; pp. 9–41. [Google Scholar]
- Jadhav, S.D.; Channe, H. Comparative study of K-NN, naive Bayes and decision tree classification techniques. Int. J. Sci. Res. 2016, 5, 1842–1845. [Google Scholar]
- Kalantar, B.; Pradhan, B.; Naghibi, S.A.; Motevalli, A.; Mansor, S. Assessment of the effects of training data selection on the landslide susceptibility mapping: A comparison between support vector machine (SVM), logistic regression (LR) and artificial neural networks (ANN). Geomat. Nat. Hazards Risk 2018, 9, 49–69. [Google Scholar] [CrossRef]
- Zhang, Q.s.; Zhu, S.C. Visual interpretability for deep learning: A survey. Front. Inf. Technol. Electron. Eng. 2018, 19, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Kendall, A.; Gal, Y. What uncertainties do we need in bayesian deep learning for computer vision? In Advances in Neural Information Processing Systems; EUA: Long Beach, CA, USA, 2017; pp. 5574–5584. [Google Scholar]
- Calhas, D.; Romero, E.; Henriques, R. On the use of Pairwise Distance Learning for Brain Signal Classification with Limited Observations. arXiv 2019, arXiv:1906.02076. [Google Scholar]
- Gilpin, L.H.; Bau, D.; Yuan, B.Z.; Bajwa, A.; Specter, M.; Kagal, L. Explaining explanations: An overview of interpretability of machine learning. In Proceedings of the 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy, 1–3 October 2018; pp. 80–89. [Google Scholar]
- Carvalho, D.V.; Pereira, E.M.; Cardoso, J.S. Machine learning interpretability: A survey on methods and metrics. Electronics 2019, 8, 832. [Google Scholar] [CrossRef] [Green Version]
- Burkart, N.; Huber, M.F. A survey on the explainability of supervised machine learning. J. Artif. Intell. Res. 2021, 70, 245–317. [Google Scholar] [CrossRef]
- Montavon, G.; Binder, A.; Lapuschkin, S.; Samek, W.; Müller, K.R. Layer-wise relevance propagation: An overview. In Explainable AI: Interpreting, Explaining and Visualizing Deep Learning; Springer International Publishing: Berlin, Germany, 2019; pp. 193–209. [Google Scholar]
- Lei, T.; Barzilay, R.; Jaakkola, T. Rationalizing neural predictions. arXiv 2016, arXiv:1606.04155. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Goncalves, D.M.; Henriques, R.; Santos, L.; Costa, R.S. On the predictability of postoperative complications for cancer patients: A Portuguese cohort study. BMC Med Inform. Decis. Mak. 2021. [Google Scholar] [CrossRef]

| Study | Surgical Cohort | Model | Data Type | Data Size | Validation | Outcome |
|---|---|---|---|---|---|---|
| Saklad [10] | General | N/A | N/A | N/A | N/A | Morbidity, Mortality |
| Knaus et al. [30] | General | LR | Clinical | 5815 | Yes | In-Hospital Death |
| Charlson et al. [31] | General | WI | Clinical | 559 | Yes | 1-Year Mortality |
| Copeland et al. [13] | General | LR | Clinical | 1372 | N/A | Morbidity, Mortality |
| Marcantonio et al. [32] | Noncardiac | LR | Clinical | 876 | Yes | Postoperative Delirium |
| Whiteley et al. [33] | General | LR | Clinical | 10,000 | Yes | Morbidity, Mortality |
| Roques et al. [34] | Cardiac | LR | Clinical | 19,030 | N/A | Mortality |
| Dupuis et al. [35] | Cardiac | LR | Clinical | 3548 | N/A | Morbidity, Mortality |
| Arozullah et al. [36] | Noncardiac | LR | Clinical | 160,805 | Yes | Postoperative Pneumonia |
| Sutton et al. [37] | General | LR | Clinical | 3144 | Yes | Morbidity |
| Donati et al. [38] | Cardiac | LR | Clinical | 1936 | Yes | Mortality |
| Gawande et al. [39] | General | PS | Clinical | 303 | Yes | Morbidity, Mortality |
| Canet et al. [40] | General | LR | Clinical | 2464 | Yes | Pulmonary Complications |
| Gupta et al. [41] | General | LR | Clinical, demographic | 211,410 | Yes | Cardiac Complications |
| Vaid et al. [42] | General | LR | Clinical, demographic | 202,741 | Yes | Mortality |
| Bilimoria et al. [14] | General | LR | Clinical, demographic | 1,414,006 | Yes | Morbidity, Mortality |
| Study | Surgical Cohort | Model | Data Type | Data Size | Validation | Outcome |
|---|---|---|---|---|---|---|
| Khan et al. [45] | Breast | Fuzzy DT | Clinical, Biological | 162,500 | Yes | 5-year mortality |
| Chang et al. [46] | Oral | NN, Fuzzy NN, SVM, LR | Clinical, histopathological, genetic | 31 | Yes | 3-year mortality |
| Zięba et al. [47] | Lung | Boosted SVM | Clinical, histopathological | 1200 | N/A | 1-year survival |
| Danjuma [48] | Lung | MLP, DT, NB | Clinical | 470 | Yes | 1-year mortality |
| Parmar et al. [49] | Head and neck | NB, RF, NN | Radiomics | 101 | Yes | 3-year mortality |
| Wang et al. [50] | Bladder | NB, SVM, kNN, NN | Clinical, histopathological | 117 | Yes | 5-year mortality |
| Thottakkara et al. [51] | Major surgery | LR, GAM, SVM, NB | Demographic, socioeconomic, clinical, laboratory | 50,318 | Yes | Postoperative sepsis and kidney injury |
| Soguero-Ruiz et al. [52] | Colorectal | SVM | Physiological, clinical | 402 | Yes | Anastomosis leakage |
| Kim et al. [53] | Oral | NN | Clinical, histopathological | 255 | Yes | 5-year mortality |
| Parikh et al. [54] | General oncology | LR, GB, RF | Demographic, laboratory, comorbidities | 26,525 | Yes | 180-day and 500-day mortality |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, D.M.; Henriques, R.; Costa, R.S. Predicting Postoperative Complications in Cancer Patients: A Survey Bridging Classical and Machine Learning Contributions to Postsurgical Risk Analysis. Cancers 2021, 13, 3217. https://doi.org/10.3390/cancers13133217
Gonçalves DM, Henriques R, Costa RS. Predicting Postoperative Complications in Cancer Patients: A Survey Bridging Classical and Machine Learning Contributions to Postsurgical Risk Analysis. Cancers. 2021; 13(13):3217. https://doi.org/10.3390/cancers13133217
Chicago/Turabian StyleGonçalves, Daniel M., Rui Henriques, and Rafael S. Costa. 2021. "Predicting Postoperative Complications in Cancer Patients: A Survey Bridging Classical and Machine Learning Contributions to Postsurgical Risk Analysis" Cancers 13, no. 13: 3217. https://doi.org/10.3390/cancers13133217
APA StyleGonçalves, D. M., Henriques, R., & Costa, R. S. (2021). Predicting Postoperative Complications in Cancer Patients: A Survey Bridging Classical and Machine Learning Contributions to Postsurgical Risk Analysis. Cancers, 13(13), 3217. https://doi.org/10.3390/cancers13133217







