The Prevalence of TET2 Gene Mutations in Patients with BCR-ABL-Negative Myeloproliferative Neoplasms (MPN): A Systematic Review and Meta-Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources and Searches
2.2. Study Selection
2.3. Extraction of Data
2.4. Quality Assessment
2.5. Publication Bias
2.6. Data Synthesis and Sensitivity Analysis
3. Results
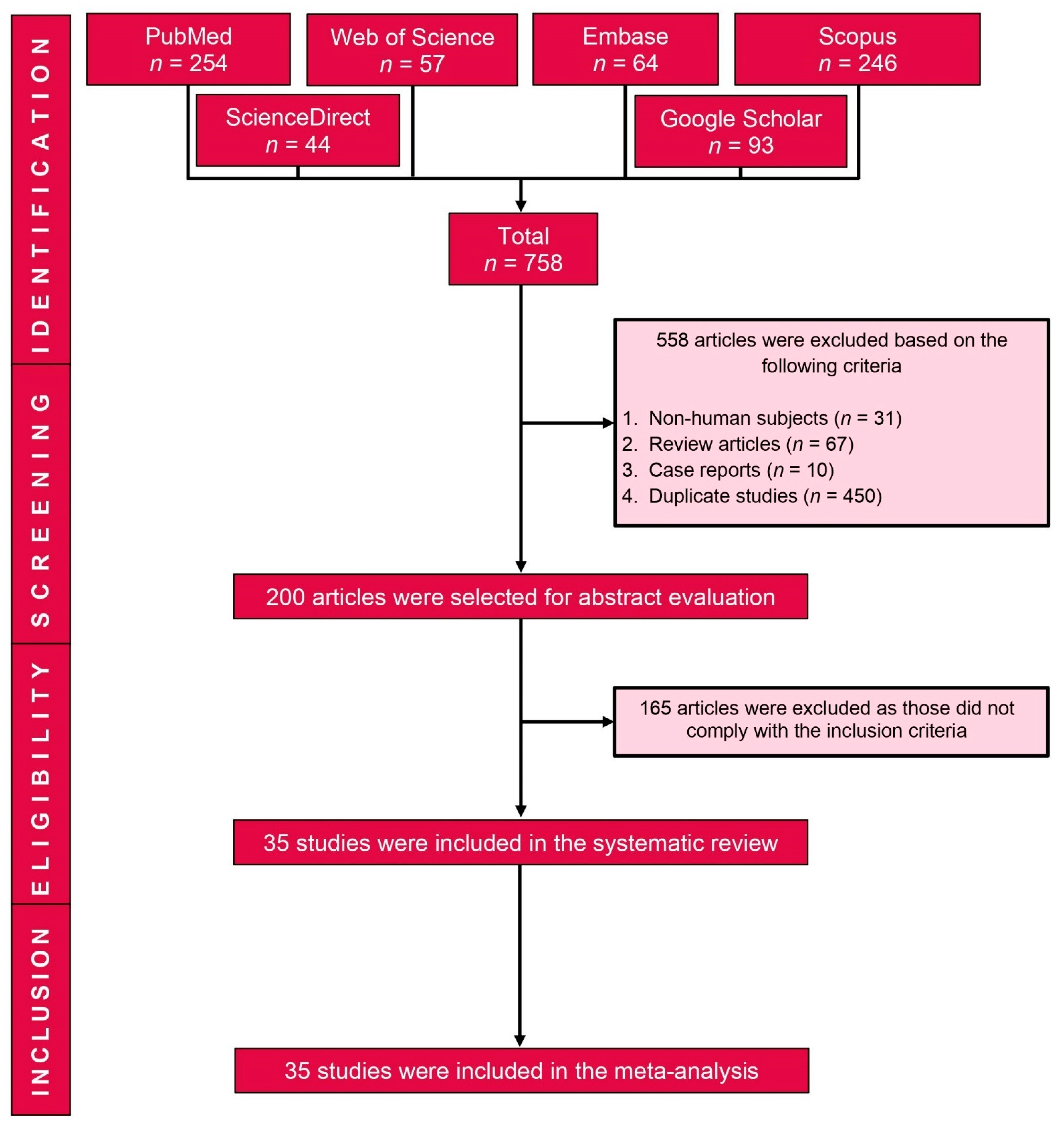
3.1. Study Selection
3.2. Characteristics of Included Studies
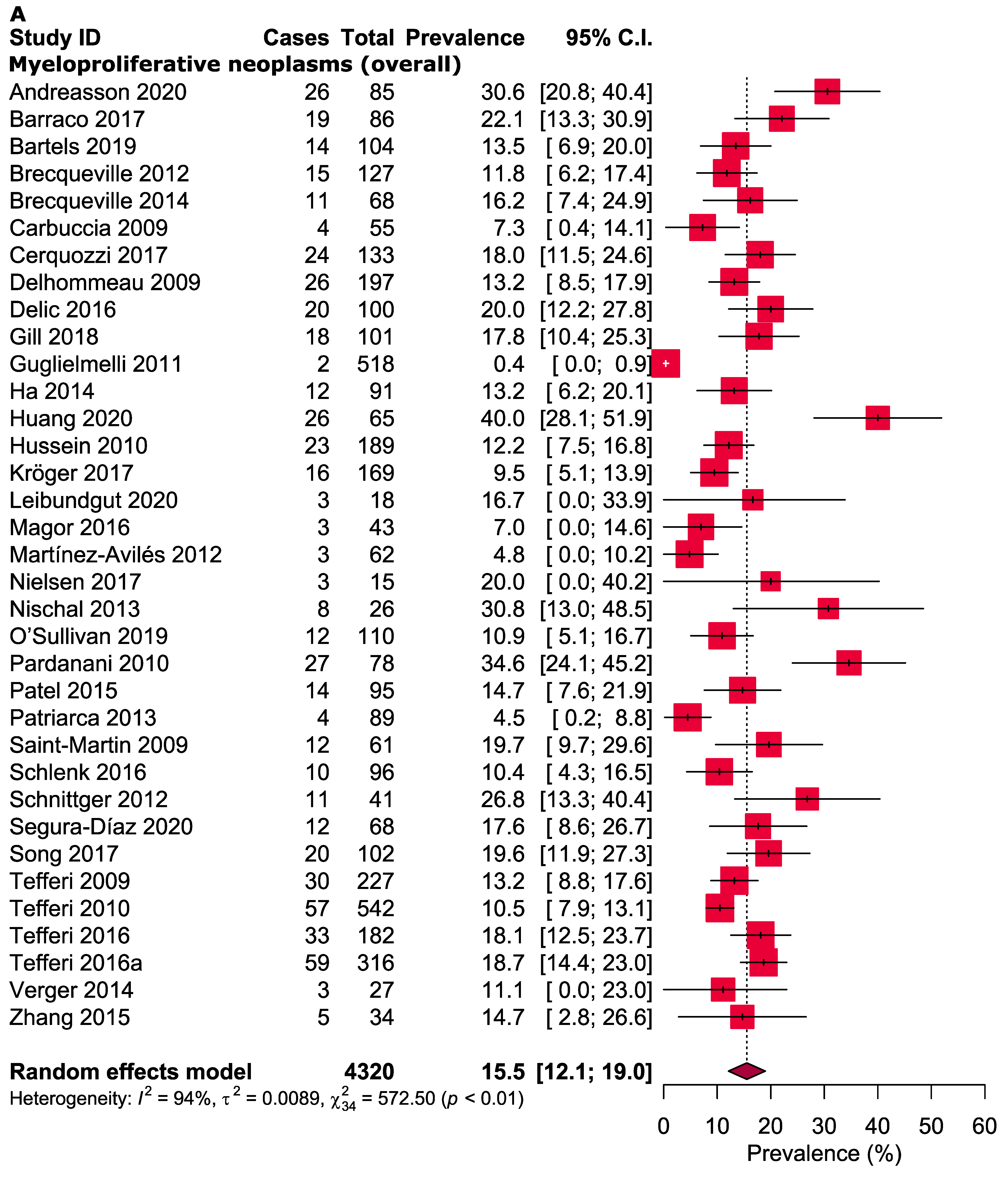
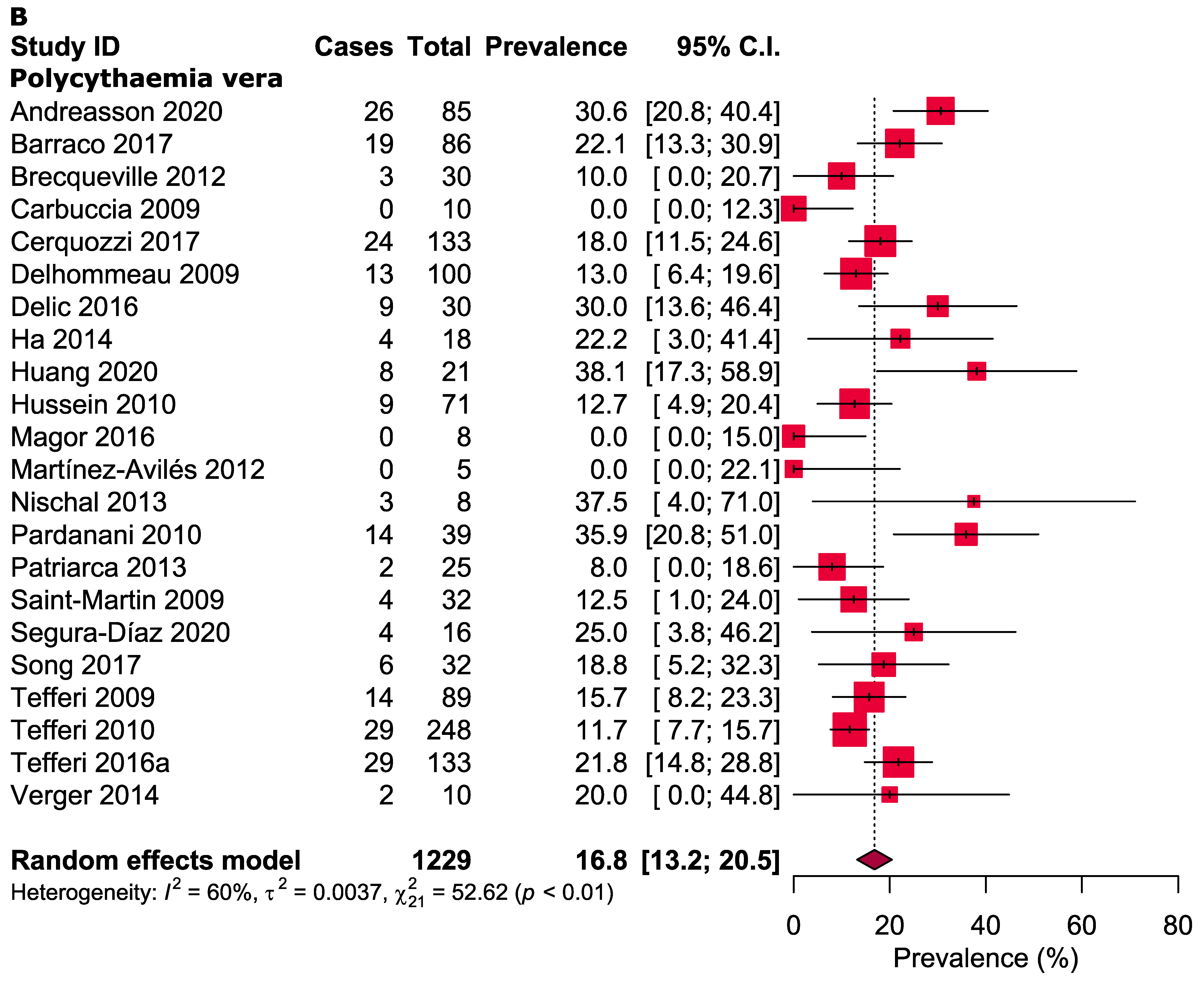
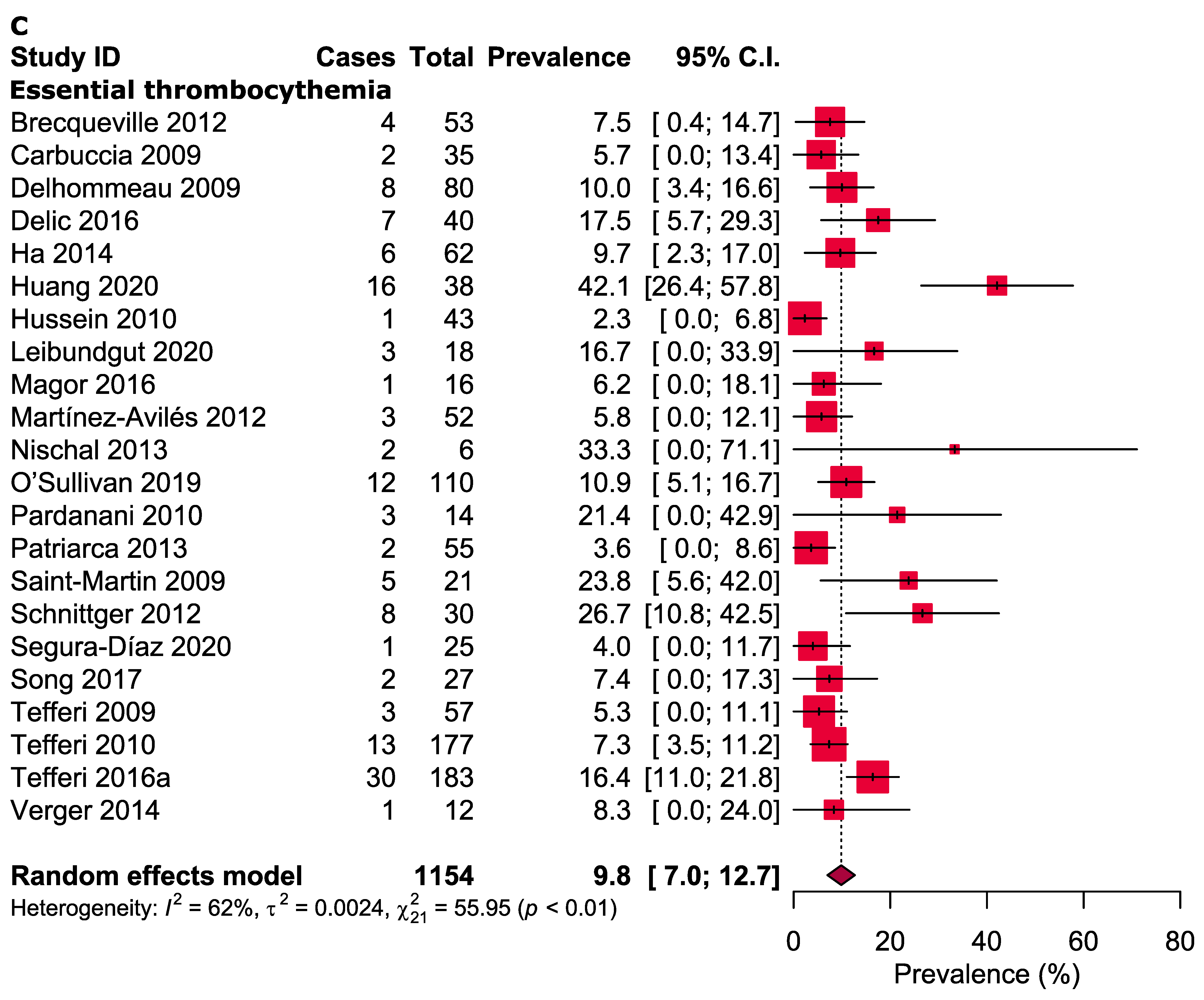
3.3. Meta-Analysis
3.4. Quality Assessment
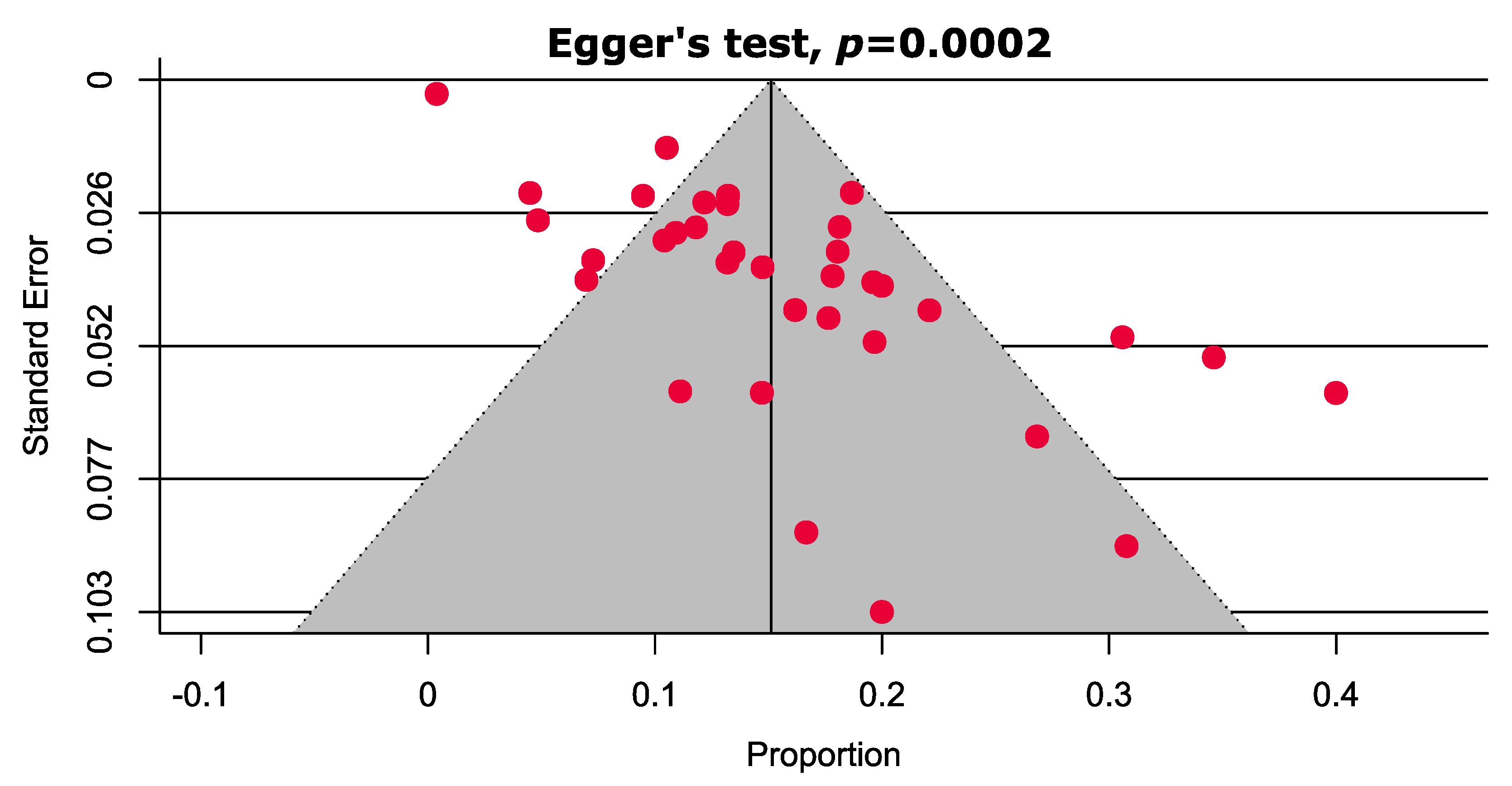
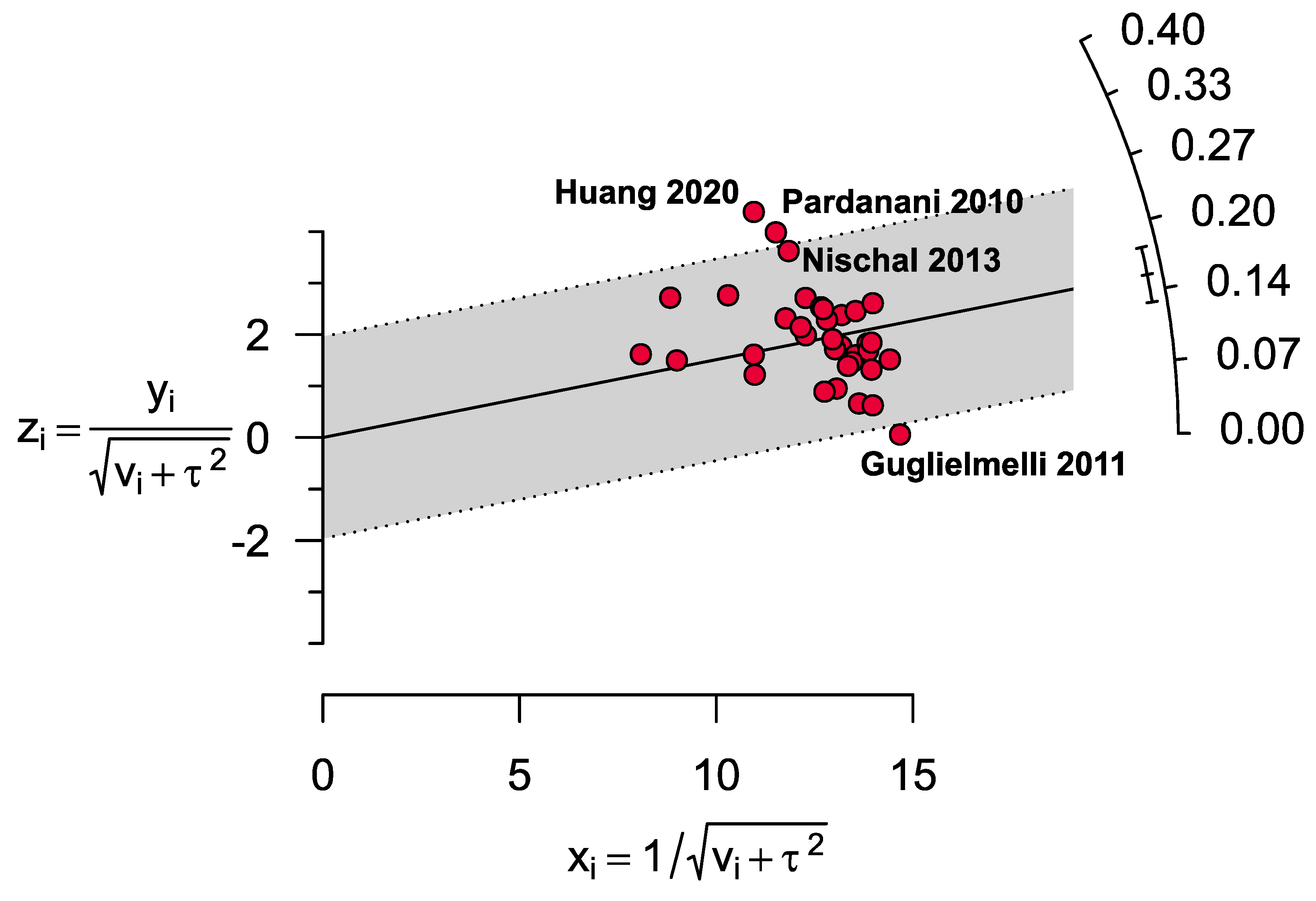
3.5. Publication Bias
3.6. Sensitivity Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Campregher, P.V.; de Souza Santos, F.P.; Perini, G.F.; Hamerschlak, N. Molecular biology of Philadelphia-negative myeloproliferative neoplasms. Rev. Bras. Hematol. Hemoter. 2012, 34, 150–155. [Google Scholar] [CrossRef][Green Version]
- Tefferi, A.; Thiele, J.; Vardiman, J.W. The 2008 World Health Organization classification system for myeloproliferative neoplasms: Order out of chaos. Cancer 2009, 115, 3842–3847. [Google Scholar] [CrossRef]
- Tefferi, A.; Elliott, M. Thrombosis in myeloproliferative disorders: Prevalence, prognostic factors, and the role of leukocytes and JAK2V617F. In Seminars in Thrombosis and Hemostasis; Thieme Medical Publishers, Inc.: New York, NY, USA, 2007; pp. 313–320. [Google Scholar]
- Nangalia, J.; Massie, C.E.; Baxter, E.J.; Nice, F.L.; Gundem, G.; Wedge, D.C.; Avezov, E.; Li, J.; Kollmann, K.; Kent, D.G. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 2013, 369, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Tenedini, E.; Bernardis, I.; Artusi, V.; Artuso, L.; Roncaglia, E.; Guglielmelli, P.; Pieri, L.; Bogani, C.; Biamonte, F.; Rotunno, G. Targeted cancer exome sequencing reveals recurrent mutations in myeloproliferative neoplasms. Leukemia 2014, 28, 1052–1059. [Google Scholar] [CrossRef]
- Reinig, E.; Yang, F.; Traer, E.; Arora, R.; Brown, S.; Rattray, R.; Braziel, R.; Fan, G.; Press, R.; Dunlap, J. Targeted next-generation sequencing in myelodysplastic syndrome and chronic myelomonocytic leukemia aids diagnosis in challenging cases and identifies frequent spliceosome mutations in transformed acute myeloid leukemia. Am. J. Clin. Pathol. 2016, 145, 497–506. [Google Scholar] [CrossRef]
- Vainchenker, W.; Kralovics, R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood 2017, 129, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Chia, Y.C.; Islam, M.A.; Woon, P.Y.; Johan, M.F.; Hassan, R.; Ramli, M. Molecular genetics of thrombotic myeloproliferative neoplasms: Implications in precision oncology. Genes Dis. 2021. [Google Scholar] [CrossRef]
- Scott, L.M.; Rebel, V.I. JAK2 and genomic instability in the myeloproliferative neoplasms: A case of the chicken or the egg? Am. J. Hematol. 2012, 87, 1028–1036. [Google Scholar] [CrossRef]
- Kameda, T.; Shide, K.; Yamaji, T.; Kamiunten, A.; Sekine, M.; Taniguchi, Y.; Hidaka, T.; Kubuki, Y.; Shimoda, H.; Marutsuka, K.; et al. Loss of TET2 has dual roles in murine myeloproliferative neoplasms: Disease sustainer and disease accelerator. Blood 2015, 125, 304–315. [Google Scholar] [CrossRef]
- Delhommeau, F.; Dupont, S.; Della Valle, V.; James, C.; Trannoy, S.; Massé, A.; Kosmider, O.; Le Couedic, J.P.; Robert, F.; Alberdi, A.; et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009, 360, 2289–2301. [Google Scholar] [CrossRef]
- Swierczek, S.I.; Yoon, D.; Bellanné-Chantelot, C.; Kim, S.J.; Saint-Martin, C.; Delhommeau, F.; Najman, A.; Prchal, J.T. Extent of hematopoietic involvement by TET2 mutations in JAK2V617F polycythemia vera. Haematologica 2011, 96, 775–778. [Google Scholar] [CrossRef]
- Moola, S.; Munn, Z.; Tufanaru, C.; Aromataris, E.; Sears, K.; Sfetcu, R.; Currie, M.; Lisy, K.; Qureshi, R.; Mattis, P.; et al. Chapter 7: Systematic reviews of etiology and risk. In JBI Manual for Evidence Synthesis; Aromataris, E., Munn, Z., Eds.; JBI: Adelaide, Australia, 2020; Available online: https://synthesismanual.jbi.global (accessed on 12 December 2020).
- Ha, J.S.; Jeon, D.S.; Kim, J.R.; Ryoo, N.H.; Suh, J.S. Analysis of the ten-eleven translocation 2 (TET2) gene mutation in myeloproliferative neoplasms. Ann. Clin. Lab. Sci. 2014, 44, 173–179. [Google Scholar] [PubMed]
- Abdel-Wahab, O.; Levine, R.L. EZH2 mutations: Mutating the epigenetic machinery in myeloid malignancies. Cancer Cell 2010, 18, 105–107. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Islam, M.A.; Alam, S.S.; Kundu, S.; Hossan, T.; Kamal, M.A.; Cavestro, C. Prevalence of Headache in Patients with Coronavirus Disease 2019 (COVID-19): A Systematic Review and Meta-Analysis of 14,275 Patients. Front. Neurol. 2020, 11, 562634. [Google Scholar] [CrossRef]
- Viechtbauer, W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef]
- Ihle, M.A.; Fassunke, J.; König, K.; Grünewald, I.; Schlaak, M.; Kreuzberg, N.; Tietze, L.; Schildhaus, H.-U.; Büttner, R.; Merkelbach-Bruse, S. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p. V600E and non-p. V600E BRAF mutations. BMC Cancer 2014, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Andréasson, B.; Pettersson, H.; Wasslavik, C.; Johansson, P.; Palmqvist, L.; Asp, J. ASXL1 mutations, Pevious vascular complications and age at diagnosis predict survival in 85 WHO-defined polycythaemia vera patients. Br. J. Haematol. 2020, 189, 913–919. [Google Scholar] [CrossRef]
- Barraco, D.; Cerquozzi, S.; Gangat, N.; Patnaik, M.M.; Lasho, T.; Finke, C.; Hanson, C.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. Monocytosis in polycythemia vera: Clinical and molecular correlates. Am. J. Hematol. 2017, 92, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Bartels, S.; Faisal, M.; Büsche, G.; Schlue, J.; Hasemeier, B.; Schipper, E.; Vogtmann, J.; Westphal, L.; Lehmann, U.; Kreipe, H. Mutations associated with age-related clonal hematopoiesis in PMF patients with rapid progression to myelofibrosis. Leukemia 2019, 34, 1364–1372. [Google Scholar] [CrossRef]
- Brecqueville, M.; Rey, J.; Bertucci, F.; Coppin, E.; Finetti, P.; Carbuccia, N.; Cervera, N.; Gelsi-Boyer, V.; Arnoulet, C.; Gisserot, O.; et al. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer 2012, 51, 743–755. [Google Scholar] [CrossRef]
- Brecqueville, M.; Rey, J.; Devillier, R.; Guille, A.; Gillet, R.; Adélaide, J.; Gelsi-Boyer, V.; Arnoulet, C.; Chaffanet, M.; Mozziconacci, M.J.; et al. Array comparative genomic hybridization and sequencing of 23 genes in 80 patients with myelofibrosis at chronic or acute phase. Haematologica 2014, 99, 37–45. [Google Scholar] [CrossRef]
- Carbuccia, N.; Murati, A.; Trouplin, V.; Brecqueville, M.; Adelaide, J.; Rey, J.; Vainchenker, W.; Bernard, O.; Chaffanet, M.; Vey, N. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia 2009, 23, 2183–2186. [Google Scholar] [CrossRef]
- Cerquozzi, S.; Barraco, D.; Lasho, T.; Finke, C.; Hanson, C.A.; Ketterling, R.P.; Pardanani, A.; Gangat, N.; Tefferi, A. Risk factors for arterial versus venous thrombosis in polycythemia vera: A single center experience in 587 patients. Blood Cancer J. 2017, 7, 662. [Google Scholar] [CrossRef]
- Delic, S.; Rose, D.; Kern, W.; Nadarajah, N.; Haferlach, C.; Haferlach, T.; Meggendorfer, M. Application of an NGS-based 28-gene panel in myeloproliferative neoplasms reveals distinct mutation patterns in essential thrombocythaemia, primary myelofibrosis and polycythaemia vera. Br. J. Haematol. 2016, 175, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Gill, H.; Ip, H.W.; Yim, R.; Tang, W.F.; Pang, H.H.; Lee, P.; Leung, G.M.K.; Li, J.; Tang, K.; So, J.C.C.; et al. Next-generation sequencing with a 54-gene panel identified unique mutational profile and prognostic markers in Chinese patients with myelofibrosis. Ann. Hematol. 2019, 98, 869–879. [Google Scholar] [CrossRef]
- Guglielmelli, P.; Biamonte, F.; Score, J.; Hidalgo-Curtis, C.; Cervantes, F.; Maffioli, M.; Fanelli, T.; Ernst, T.; Winkelman, N.; Jones, A.V.; et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood 2011, 118, 5227–5234. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wu, J.; Deng, X.; Xu, X.; Zhang, X.; Ma, W.; Hu, T.; Yang, J.; Guan, M.; Tang, G. Mutation profiles of classic myeloproliferative neoplasms detected by a customized next-generation sequencing-based 50-gene panel. J. Bio-X Res. 2020, 3, 13–20. [Google Scholar] [CrossRef]
- Hussein, K.; Abdel-Wahab, O.; Lasho, T.L.; Van Dyke, D.L.; Levine, R.L.; Hanson, C.A.; Pardanani, A.; Tefferi, A. Cytogenetic correlates of TET2 mutations in 199 patients with myeloproliferative neoplasms. Am. J. Hematol. 2010, 85, 81. [Google Scholar] [CrossRef]
- Kröger, N.; Panagiota, V.; Badbaran, A.; Zabelina, T.; Triviai, I.; Araujo Cruz, M.M.; Shahswar, R.; Ayuk, F.; Gehlhaar, M.; Wolschke, C.; et al. Impact of Molecular Genetics on Outcome in Myelofibrosis Patients after Allogeneic Stem Cell Transplantation. Biol. Blood Marrow Transplant. 2017, 23, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Oppliger Leibundgut, E.; Haubitz, M.; Burington, B.; Ottmann, O.G.; Spitzer, G.; Odenike, O.; McDevitt, M.A.; Röth, A.; Snyder, D.S.; Baerlocher, G.M. Dynamics of mutations in patients with essential thrombocythemia treated with imetelstat. Haematologica 2020. [Google Scholar] [CrossRef] [PubMed]
- Magor, G.W.; Tallack, M.R.; Klose, N.M.; Taylor, D.; Korbie, D.; Mollee, P.; Trau, M.; Perkins, A.C. Rapid Molecular Profiling of Myeloproliferative Neoplasms Using Targeted Exon Resequencing of 86 Genes Involved in JAK-STAT Signaling and Epigenetic Regulation. J. Mol. Diagn. 2016, 18, 707–718. [Google Scholar] [CrossRef]
- Martinez-Aviles, L.; Besses, C.; Alvarez-Larran, A.; Torres, E.; Serrano, S.; Bellosillo, B. TET2, ASXL1, IDH1, IDH2, And c-CBL genes in JAK2-and MPL-negative myeloproliferative neoplasms. Ann. Hematol. 2012, 91, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Myrtue Nielsen, H.; Lykkegaard Andersen, C.; Westman, M.; Sommer Kristensen, L.; Asmar, F.; Arvid Kruse, T.; Thomassen, M.; Stauffer Larsen, T.; Skov, V.; Lotte Hansen, L.; et al. Epigenetic changes in myelofibrosis: Distinct methylation changes in the myeloid compartments and in cases with ASXL1 mutations. Sci. Rep. 2017, 7, 6774. [Google Scholar] [CrossRef]
- Nischal, S.; Bhattacharyya, S.; Christopeit, M.; Yu, Y.; Zhou, L.; Bhagat, T.D.; Sohal, D.; Will, B.; Mo, Y.; Suzuki, M.; et al. Methylome profiling reveals distinct alterations in phenotypic and mutational subgroups of myeloproliferative neoplasms. Cancer Res. 2013, 73, 1076–1085. [Google Scholar] [CrossRef]
- O’Sullivan, J.M.; Hamblin, A.; Yap, C.; Fox, S.; Boucher, R.; Panchal, A.; Alimam, S.; Dreau, H.; Howard, K.; Ware, P.; et al. The poor outcome in high molecular risk, hydroxycarbamide-resistant/intolerant ET is not ameliorated by ruxolitinib. Blood 2019, 134, 2107–2111. [Google Scholar] [CrossRef] [PubMed]
- Pardanani, A.; Lasho, T.; Finke, C.; Oh, S.T.; Gotlib, J.; Tefferi, A. LNK mutation studies in blast-phase myeloproliferative neoplasms, and in chronic-phase disease with TET2, IDH, JAK2 or MPL mutations. Leukemia 2010, 24, 1713–1718. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.P.; Newberry, K.J.; Luthra, R.; Jabbour, E.; Pierce, S.; Cortes, J.; Singh, R.; Mehrotra, M.; Routbort, M.J.; Luthra, M.; et al. Correlation of mutation profile and response in patients with myelofibrosis treated with ruxolitinib. Blood 2015, 126, 790–797. [Google Scholar] [CrossRef]
- Patriarca, A.; Colaizzo, D.; Tiscia, G.; Spadano, R.; Di Zacomo, S.; Spadano, A.; Villanova, I.; Margaglione, M.; Grandone, E.; Dragani, A. TET2 mutations in Ph-negative myeloproliferative neoplasms: Identification of three novel mutations and relationship with clinical and laboratory findings. BioMed Res. Int. 2013, 2013, 929840. [Google Scholar] [CrossRef]
- Saint-Martin, C.; Leroy, G.; Delhommeau, F.; Panelatti, G.; Dupont, S.; James, C.; Plo, I.; Bordessoule, D.; Chomienne, C.; Delannoy, A.; et al. Analysis of the Ten-Eleven Translocation 2 (TET2) gene in familial myeloproliferative neoplasms. Blood 2009, 114, 1628–1632. [Google Scholar] [CrossRef]
- Schlenk, R.F.; Stegelmann, F.; Reiter, A.; Jost, E.; Gattermann, N.; Hebart, H.; Waller, C.; Hochhaus, A.; Platzbecker, U.; Schafhausen, P.; et al. Pomalidomide in myeloproliferative neoplasm-associated myelofibrosis. Leukemia 2017, 31, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, S.; Bacher, U.; Eder, C.; Dicker, F.; Alpermann, T.; Grossmann, V.; Kohlmann, A.; Kern, W.; Haferlach, C.; Haferlach, T. Molecular analyses of 15,542 patients with suspected BCR-ABL1-negative myeloproliferative disorders allow to develop a stepwise diagnostic workflow. Haematologica 2012, 97, 1582–1585. [Google Scholar] [CrossRef] [PubMed]
- Segura-Díaz, A.; Stuckey, R.; Florido, Y.; González-Martín, J.M.; López-Rodríguez, J.F.; Sánchez-Sosa, S.; González-Pérez, E.; Perdomo, M.N.S.; del Mar Perera, M.; de la Iglesia, S.; et al. Thrombotic risk detection in patients with polycythemia vera: The predictive role of DNMT3A/TET2/ASXL1 mutations. Cancers 2020, 12, 934. [Google Scholar] [CrossRef]
- Song, J.; Hussaini, M.; Zhang, H.; Shao, H.; Qin, D.; Zhang, X.; Ma, Z.; Hussnain Naqvi, S.M.; Zhang, L.; Moscinski, L.C. Comparison of the Mutational Profiles of Primary Myelofibrosis, Polycythemia Vera, and Essential Thrombocytosis. Am. J. Clin. Pathol. 2017, 147, 444–452. [Google Scholar] [CrossRef]
- Tefferi, A.; Pardanani, A.; Lim, K.H.; Abdel-Wahab, O.; Lasho, T.L.; Patel, J.; Gangat, N.; Finke, C.M.; Schwager, S.; Mullally, A.; et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia 2009, 23, 905–911. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Abdel-Wahab, O.; Guglielmelli, P.; Patel, J.; Caramazza, D.; Pieri, L.; Finke, C.M.; Kilpivaara, O.; Wadleigh, M.; et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia 2010, 24, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Lasho, T.L.; Finke, C.M.; Elala, Y.; Hanson, C.A.; Ketterling, R.P.; Gangat, N.; Pardanani, A. Targeted deep sequencing in primary myelofibrosis. Blood Adv. 2016, 1, 105–111. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Guglielmelli, P.; Finke, C.M.; Rotunno, G.; Elala, Y.; Pacilli, A.; Hanson, C.A.; Pancrazzi, A.; Ketterling, R.P.; et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood Adv. 2016, 1, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Verger, E.; Andreoli, A.; Chomienne, C.; Kiladjian, J.J.; Cassinat, B. TET2 gene sequencing may be helpful for myeloproliferative neoplasm diagnosis. Br. J. Haematol. 2014, 165, 416–419. [Google Scholar] [CrossRef]
- Chunxia, Z.; Li, G.; Qianqian, J.; Zhenling, L.; Fanzhou, H.; Yayue, G.; Ming, G.; Shaohua, X.; Yin, T.; Yanrong, C.; et al. Symptom burden and its relationships with risk assessment and gene mutations in patients with primary myelofibrosis. J. Leuk. Lymphoma 2015, 24, 453–456. [Google Scholar]
- Tefferi, A.; Lim, K.H.; Abdel-Wahab, O.; Lasho, T.L.; Patel, J.; Patnaik, M.M.; Hanson, C.A.; Pardanani, A.; Gilliland, D.G.; Levine, R.L. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia 2009, 23, 1343–1345. [Google Scholar] [CrossRef] [PubMed]
- Shih, A.H.; Abdel-Wahab, O.; Patel, J.P.; Levine, R.L. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer 2012, 12, 599–612. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, S.-K.; Zou, Z.; Fan, R.-H.; Lyu, X.-D. Prognostic significance of TET2 mutations in myelodysplastic syndromes: A meta-analysis. Leukemia Res. 2017, 58, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011, 364, 2496–2506. [Google Scholar] [CrossRef] [PubMed]
- Kosmider, O.; Gelsi-Boyer, V.; Cheok, M.; Grabar, S.; Della-Valle, V.; Picard, F.; Viguié, F.; Quesnel, B.; Beyne-Rauzy, O.; Solary, E. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs). Blood 2009, 114, 3285–3291. [Google Scholar] [CrossRef]
- Itzykson, R.; Kosmider, O.; Cluzeau, T.; Mansat-De Mas, V.; Dreyfus, F.; Beyne-Rauzy, O.; Quesnel, B.; Vey, N.; Gelsi-Boyer, V.; Raynaud, S. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia 2011, 25, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Kohlmann, A.; Grossmann, V.; Klein, H.U.; Schindela, S.; Weiss, T.; Kazak, B.; Dicker, F.; Schnittger, S.; Dugas, M.; Kern, W.; et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J. Clin. Oncol. 2010, 28, 3858–3865. [Google Scholar] [CrossRef]
- Itzykson, R.; Kosmider, O.; Renneville, A.; Gelsi-Boyer, V.; Meggendorfer, M.; Morabito, M.; Berthon, C.; Adès, L.; Fenaux, P.; Beyne-Rauzy, O. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J. Clin. Oncol. 2013, 31, 2428–2436. [Google Scholar] [CrossRef]
- Grossmann, V.; Kohlmann, A.; Eder, C.; Haferlach, C.; Kern, W.; Cross, N.; Haferlach, T.; Schnittger, S. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia 2011, 25, 877–879. [Google Scholar] [CrossRef]
- Tefferi, A.; Levine, R.L.; Lim, K.H.; Abdel-Wahab, O.; Lasho, T.L.; Patel, J.; Finke, C.M.; Mullally, A.; Li, C.Y.; Pardanani, A.; et al. Frequent TET2 mutations in systemic mastocytosis: Clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia 2009, 23, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Soucie, E.; Hanssens, K.; Mercher, T.; Georgin-Lavialle, S.; Damaj, G.; Livideanu, C.; Chandesris, M.O.; Acin, Y.; Létard, S.; de Sepulveda, P. In aggressive forms of mastocytosis, TET2 loss cooperates with c-KITD816V to transform mast cells. Blood 2012, 120, 4846–4849. [Google Scholar] [CrossRef] [PubMed]
- Jardin, F.; Ruminy, P.; Parmentier, F.; Troussard, X.; Vaida, I.; Stamatoullas, A.; Leprêtre, S.; Penther, D.; Duval, A.B.; Picquenot, J.M. TET2 and TP53 mutations are frequently observed in blastic plasmacytoid dendritic cell neoplasm. Br. J. Haematol. 2011, 153, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Menezes, J.; Acquadro, F.; Wiseman, M.; Gomez-Lopez, G.; Salgado, R.; Talavera-Casanas, J.; Buno, I.; Cervera, J.; Montes-Moreno, S.; Hernandez-Rivas, J. Exome sequencing reveals novel and recurrent mutations with clinical impact in blastic plasmacytoid dendritic cell neoplasm. Leukemia 2014, 28, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Mejía-Ochoa, M.; Toro, P.A.A.; Cardona-Arias, J.A. Systematization of analytical studies of polycythemia vera, essential thrombocythemia and primary myelofibrosis, and a meta-analysis of the frequency of JAK2, CALR and MPL mutations: 2000–2018. BMC Cancer 2019, 19, 590. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.A.; Duncombe, A.S.; Hughes, M.; Mills, M.E.; Wilson, J.C.; McMullin, M.F. Environmental, Lifestyle, And familial/ethnic factors associated with myeloproliferative neoplasms. Am. J. Hematol. 2012, 87, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Wadleigh, M.; Cools, J.; Ebert, B.L.; Wernig, G.; Huntly, B.J.; Boggon, T.J.; Wlodarska, I.; Clark, J.J.; Moore, S. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005, 7, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Passamonti, F.; Maffioli, M. Update from the latest WHO classification of MPNs: A user’s manual. Hematology 2016, 2016, 534–542. [Google Scholar] [CrossRef]
- Tefferi, A.; Vardiman, J.W. Classification and diagnosis of myeloproliferative neoplasms: The 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia 2008, 22, 14–22. [Google Scholar] [CrossRef]
| No | Study ID [References] | Study Design | Country | Type of MPN | Total Number of MPN Patients (Female) | Age (Years) [Mean ± SD/Median (IQR)/Range] | Haemoglobin (g/dL) [Mean ± SD/Median (IQR)/Range] | Leucocyte Count (109/L) [Mean ± SD/Range/Median (IQR)] | Platelet Count (109/L) [Mean ± SD/Range/Median (IQR)] | Total Number of Mutated ASXL1 (%) | Screening Method for TET2 Gene Mutations | Diagnostic Criteria |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Andreasson 2020 [20] | Cross-sectional | Sweden | PV | 85 (41) | 71.0 (37.0–94.0) | NR | NR | NR | 8.2 | NGS | 2008 WHO |
| 2 | Barraco 2017 [21] | Cross-sectional | USA | PV | 267 (125) | 64.0 (17.0–94.0) | 18.0 (14.8–24.3) | 11.5 (4.3–59.3) | 439.0 (37.0–2747.0) | 8.1 | NR | 2016 WHO |
| 3 | Bartels 2019 [22] | Case–control | Germany | MF | 104 (53) | NR | NR | NR | NR | 9.6 | NGS | 2016 WHO |
| 4 | Brecqueville 2012 [23] | Cross-sectional | France | PV, ET & MF | 127 (57) | NR (29.0–97.0) | NR | NR | NR | 11.0 | SS | 2008 WHO |
| 5 | Brecqueville 2014 [24] | Cross-sectional | France | MF | 68 (NR) | 69.0 (30.0–86.0) | 11.4 (5.8–17.8) | 8.9 (1.3–120.0) | 256.0 (5.0–1188.0) | 26.5 | SS | 2008 WHO |
| 6 | Carbuccia 2009 [25] | Cross-sectional | France | PV, ET & MF | NR | NR | NR | NR | NR | 7.3 | SS | NR |
| 7 | Cerquozzi 2017 [26] | Cross-sectional | USA | PV | 587 (302) | 60.0 (17.0–94.0) | NR | NR | 476.0 (41.0–2747.0) | 10.5 | NGS | 2016 WHO, ELN |
| 8 | Delhommeau 2009 [11] | Cross-sectional | France | PV, ET & MF | 203 (41) | NR | NR | NR | NR | NR | SS, SNP array, CGH | 2001 WHO |
| 9 | Delic 2016 [27] | Cross-sectional | Germany | PV, ET & MF | 100 (NR) | 69.0 (28.0–87.0) | NR | NR | NR | 21.0 | NGS | 2008 WHO |
| 10 | Gill 2018 [28] | Cross-sectional | China | MF | 101 (39) | 60.0 (26.0–89.0) | 10.3 (3.0–18.5) | 12.1 (1.5–177.4) | 344.0 (19.0–1720.0) | 30.7 | NGS | 2016 WHO, IWG-MRT |
| 11 | Guglielmelli 2011 [29] | Cross-sectional | Italy | MF | 518 (303) | NR | NR | NR | NR | 22.2 | HRM | 2008 WHO, IWG-MRT |
| 12 | Ha 2014 [14] | Cross-sectional | Korea | PV, ET & MF | 99 (50) | 63.7 ± 13.0 | 13.7 ± 3.8 | 16.5 ± 15.4 | 825.4 ± 490.0 | NR | SS, SNP array, CGH | 2008 WHO |
| 13 | Huang 2020 [30] | Cross-sectional | China | PV, ET & MF | 65 (32) | 62.0 (NR) | NR | NR | NR | 10.8 | NGS | 2016 WHO |
| 14 | Hussein 2010 [31] | Cross-sectional | USA | PV, ET & MF | 199 (96) | 58.0 (19.0–93.0) | NR | NR | NR | NR | NGS | 2001 WHO |
| 15 | Kröger 2017 [32] | Cross-sectional | Germany | MF | 169 (73) | 58.0 (18.0–75.0) | NR | NR | NR | 29.0 | SS | NR |
| 16 | Leibundgut 2020 [33] | Cross-sectional | Switzerland | ET | 18 (10) | 59.5 (21.0–83.0) | NR | 7.8 (3.0–14.6) | 788.0 (521.0–1359.0) | 11.1 | NGS | 2016 WHO |
| 17 | Magor 2016 [34] | Cross-sectional | Australia | PV, ET & MF | 43 (16) | 61.0 (24.0–91.0) | NR | NR | NR | 9.3 | Targeted exon resequencing | 2008 WHO |
| 18 | Martínez-Avilés 2012 [35] | Cross-sectional | Spain | PV, ET & MF | 62 (43) | NR | NR | NR | NR | 4.8 | HRM, SS | 2008 WHO |
| 19 | Nielsen 2017 [36] | Case–control | Denmark | MF | 16 (3) | 66.0 (52.0–80.0) | 10.3 (7.9–13.4) | 5.9 (2.3–64.4) | 155.5 (56.0–357.0) | 50.0 | PCR-DGGE | NR |
| 20 | Nischal 2013 [37] | Cross-sectional | USA | PV, ET & MF | 25 (14) | 68.0 (54.0–72.0) | NR | NR | NR | 24.0 | SS | NR |
| 21 | O’Sullivan 2019 [38] | Cross-sectional | UK | ET | NR | NR | NR | NR | NR | NR | NGS | NR |
| 22 | Pardanani 2010 [39] | Cross-sectional | USA | PV, ET & MF | 78 (34) | 64.0 (22.0–95.0) | NR | NR | NR | NR | NGS | 2008 WHO |
| 23 | Patel 2015 [40] | Cross-sectional | USA | MF | 95 (44) | 66.0 (40.0–84.0) | 10.7 (7.2–16.9) | 25.0 (2.5–159.0) | 339.0 (13.0–969.0) | 21.1 | NGS | IWG-MRT |
| 24 | Patriarca 2013 [41] | Cross-sectional | Italy | PV, ET & MF | 97 (44) | NR | NR | NR | NR | NR | NGS | 2008 WHO |
| 25 | Saint-Martin 2009 [42] | Cross-sectional | France | PV, ET & MF | NR | NR | NR | NR | NR | NR | SS | 2008 WHO |
| 26 | Schlenk 2016 [43] | Cross-sectional | Germany | MF | 96 (33) | NR | NR | NR | NR | 30.2 | SS | 2008 WHO, IWG-MRT |
| 27 | Schnittger 2012 [44] | Cross-sectional | Germany | ET & MF | NR | NR | NR | NR | NR | NR | SS, HRM | NR |
| 28 | Segura-Díaz 2020 [45] | Cross-sectional | Spain | PV, ET & MF | 68 (40) | 68.0 (43.0–90.0) | NR | NR | NR | 8.8 | NGS | 2016 WHO |
| 29 | Song 2017 [46] | Cross-sectional | USA | PV, ET & MF | 135 (64) | NR | NR | NR | NR | 21.2 | NGS | 2008 WHO |
| 30 | Tefferi 2009 [47] | Cross-sectional | USA | PV, ET & MF | 227 (111) | NR | NR | NR | NR | NR | NGS | 2001 WHO |
| 31 | Tefferi 2010 [48] | Cross-sectional | USA | PV, ET & MF | 908 (487) | NR | NR | NR | NR | NR | NGS | 2008 WHO, IWG-MRT |
| 32 | Tefferi 2016 [49] | Cross-sectional | USA | MF | 182 (64) | 63.0 (22.0–87.0) | 10.1 (5.8–16.0) | 10.5 (1.9–219.0) | 224.0 (11.0–1493.0) | 35.7 | NGS | 2008 WHO |
| 33 | Tefferi 2016a [50] | Cross-sectional | USA | PV & ET | 316 (177) | NR | NR | NR | NR | 11.4 | NGS | 2008 WHO |
| 34 | Verger 2014 [51] | Cross-sectional | France | PV, ET & MF | 27 (NR) | NR | NR | NR | NR | NR | SS | NR |
| 35 | Zhang 2015 [52] | Cross-sectional | China | MF | 36 (15) | 65.0 (46.0–93.0) | 10.9 (3.0–16.0) | 22.3 (1.4–54.5) | 215.0 (3.0–1157.0) | 11.1 | WGS | 2008 WHO |
| Subgroups | Prevalence [95% CIs] (%) | Number of Studies Analysed | Total Number of Patients | Heterogeneity | Publication Bias, Egger’s Test (p-Value) | |
|---|---|---|---|---|---|---|
| I2 | p-Value | |||||
| Overall myeloproliferative neoplasms | ||||||
| Europe | 13.0 [8.8–17.2] | 19 | 2010 | 92% | <0.0001 | 0.004 |
| North America | 17.4 [14.0–20.9] | 11 | 1976 | 74% | <0.0001 | 0.0005 |
| Asia | 20.8 [10.5–31.1] | 4 | 291 | 80% | 0.001 | NA |
| Australia | 7.0 [0.0–14.6] | 1 | 43 | NA | NA | NA |
| China | 23.9 [9.6–38.1] | 3 | 200 | 82% | 0.003 | NA |
| France | 13.6 [10.6–16.7] | 5 | 480 | 0% | 0.67 | NA |
| Germany | 14.2 [9.2–19.1] | 5 | 510 | 61% | 0.03 | NA |
| Italy | 1.9 [0.0–5.7] | 2 | 607 | 71% | 0.06 | NA |
| Spain | 10.7 [0.0–23.2] | 2 | 130 | 82% | 0.01 | NA |
| USA | 17.4 [14.0–20.9] | 11 | 1976 | 74% | <0.0001 | 0.0005 |
| WHO criteria reported | 15.7 [11.8–19.7] | 27 | 3782 | 95% | <0.0001 | 0.0002 |
| WHO criteria not reported | 13.1 [8.9–17.3] | 8 | 538 | 47% | 0.06 | 0.005 |
| WHO 2001 criteria | 12.9 [10.2–15.5] | 3 | 613 | 0% | 0.93 | NA |
| WHO 2008 criteria | 14.5 [9.7–19.3] | 17 | 2594 | 95% | <0.0001 | 0.0004 |
| WHO 2016 criteria | 20.1 [14.7–25.4] | 7 | 575 | 61% | 0.01 | 0.40 |
| NGS method | 17.2 [14.0–20.4] | 18 | 2604 | 80% | <0.0001 | 0.0007 |
| SS method | 12.7 [9.6–15.9] | 11 | 965 | 52% | 0.02 | 0.001 |
| HRM method | 7.7 [0.0–16.6] | 3 | 621 | 88% | 0.0002 | NA |
| Polycythaemia vera | ||||||
| Europe | 14.6 [8.0–21.1] | 10 | 343 | 63% | 0.01 | 0.58 |
| North America | 18.2 [14.2–22.5] | 9 | 839 | 57% | 0.01 | NA |
| Asia | 29.6 [14.1–45.2] | 2 | 39 | 17% | 0.27 | NA |
| Australia | 0.0 [0.0–15.0] | 1 | 8 | NA | NA | NA |
| France | 12.5 [7.6–17.5] | 4 | 172 | 0% | 0.90 | NA |
| Spain | 12.7 [0.0–37.2] | 2 | 21 | 61% | 0.28 | NA |
| USA | 18.2 [14.0–22.5] | 9 | 839 | 57% | 0.01 | NA |
| WHO 2001 criteria | 13.7 [9.6–17.9] | 3 | 260 | 0% | 0.40 | NA |
| WHO 2008 criteria | 16.9 [11.3–22.6] | 12 | 685 | 69% | 0.0009 | 0.77 |
| WHO 2016 criteria | 21.4 [15.6–27.3] | 4 | 256 | 16% | 0.31 | NA |
| NGS method | 19.8 [15.1–24.6] | 12 | 922 | 67% | 0.0005 | 0.009 |
| SS method | 13.0 [8.4–17.7] | 7 | 203 | 0% | 0.71 | NA |
| HRM method | 0.0 [0.0–22.1] | 1 | 5 | NA | NA | NA |
| Essential thrombocythaemia | ||||||
| Europe | 8.8 [5.7–12.0] | 12 | 531 | 39% | 0.08 | 0.002 |
| North America | 8.7 [3.8–13.6] | 7 | 507 | 69% | 0.003 | NA |
| Asia | 25.1 [0.0–56.9] | 2 | 100 | 93% | 0.0002 | NA |
| Australia | 6.2 [0.0–18.1] | 1 | 16 | NA | NA | NA |
| France | 9.7 [5.3–14.2] | 4 | 166 | 0% | 0.44 | NA |
| Spain | 12.1 [0.0–21.2] | 2 | 46 | 74% | 0.04 | NA |
| USA | 8.7 [3.8–13.6] | 7 | 507 | 69% | 0.003 | NA |
| WHO 2001 criteria | 5.3 [1.1–9.6] | 3 | 180 | 44% | 0.16 | NA |
| WHO 2008 criteria | 9.4 [6.1–12.6] | 11 | 700 | 49% | 0.03 | 0.06 |
| WHO 2016 criteria | 20.3 [0.0–43.7] | 3 | 81 | 89% | <0.0001 | 0.41 |
| NGS method | 10.2 [6.1–14.4] | 12 | 787 | 75% | <0.0001 | 0.003 |
| SS method | 10.4 [6.2–14.6] | 8 | 316 | 31% | 0.18 | NA |
| HRM method | 14.9 [0.0–35.2] | 2 | 82 | 83% | 0.01 | NA |
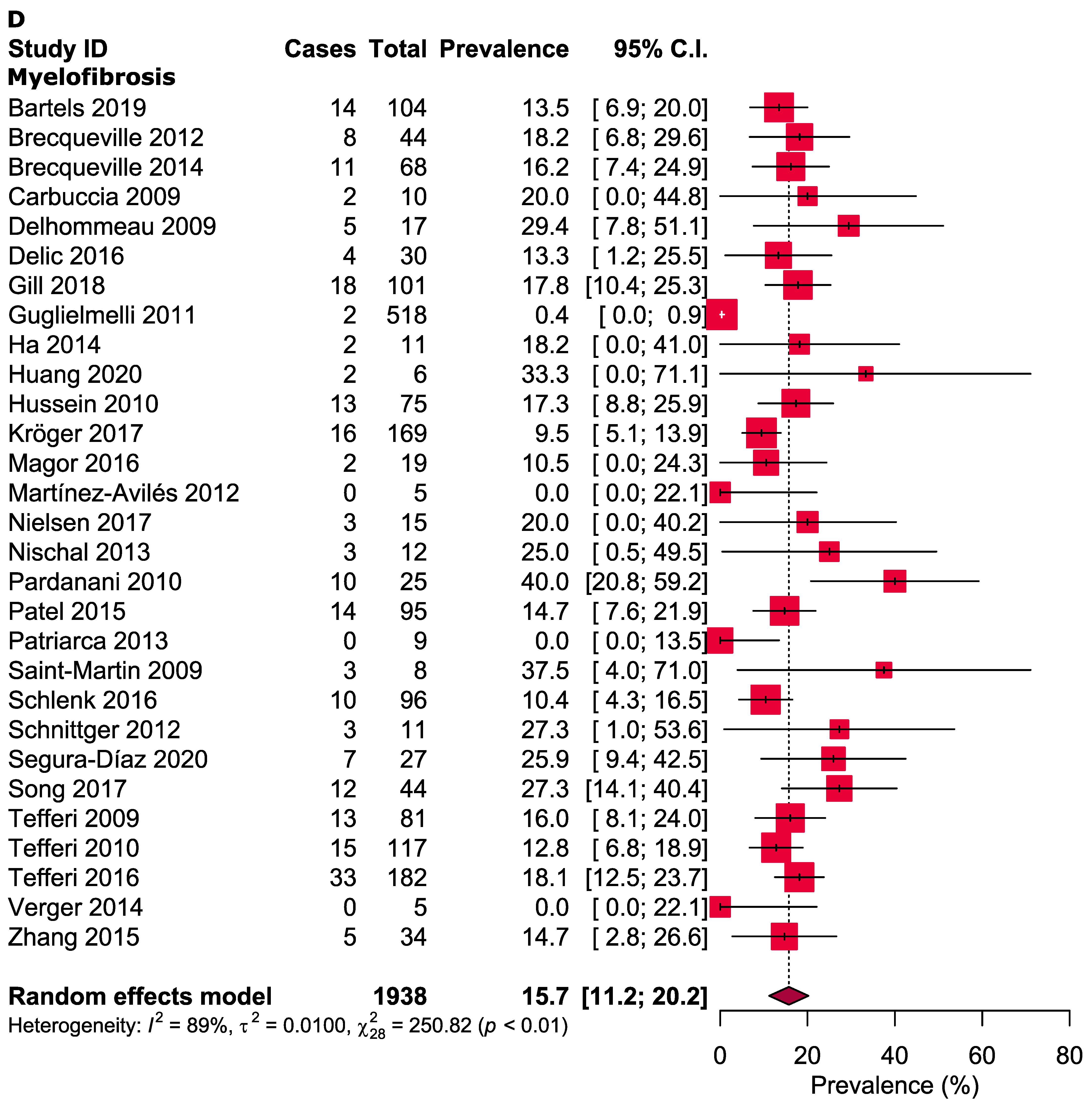
| Myelofibrosis | ||||||
| Europe | 13.7 [7.9–19.5] | 15 | 1127 | 85% | <0.0001 | 0.008 |
| North America | 16.8 [12.3–23.7] | 9 | 640 | 52% | 0.09 | NA |
| Asia | 17.4 [11.4–23.5] | 4 | 152 | 0% | 0.82 | NA |
| Australia | 10.5 [0.0–24.3] | 1 | 19 | NA | NA | NA |
| China | 17.4 [11.2–23.6] | 3 | 141 | 0% | 0.63 | NA |
| France | 17.6 [9.9–25.3] | 5 | 142 | 20% | 0.51 | NA |
| Germany | 11.0 [8.0–14.0] | 5 | 410 | 0% | 0.61 | NA |
| Italy | 0.4 [0.0–0.9] | 2 | 527 | 0% | 0.50 | NA |
| Spain | 14.0 [0.0–39.4] | 2 | 32 | 70% | 0.21 | NA |
| USA | 17.7 [13.8–21.6] | 8 | 631 | 35% | 0.15 | NA |
| WHO 2001 criteria | 17.5 [11.9–23.3] | 3 | 173 | 0% | 0.52 | NA |
| WHO 2008 criteria | 14.4 [8.1–20.7] | 15 | 1210 | 90% | <0.0001 | 0.04 |
| WHO 2016 criteria | 16.5 [11.8–21.2] | 4 | 238 | 0% | 0.39 | 0.20 |
| NGS method | 16.5 [13.2–19.8] | 13 | 896 | 38% | 0.17 | 0.053 |
| SS method | 13.3 [9.1–17.5] | 11 | 446 | 24% | 0.35 | 0.01 |
| HRM method | 5.0 [0.0–18.2] | 3 | 534 | 50% | 0.10 | NA |
| Different types of myelofibrosis | ||||||
| PMF | 16.7 [13.6–19.8] | 20 | 853 | 24% | 0.41 | 0.06 |
| SMF | 14.8 [9.3–20.2] | 9 | 158 | 0% | 0.95 | NA |
| Strategies of Sensitivity Analyses | Prevalence [95% CIs] (%) | Difference of Pooled Prevalence Compared to the Main Result | Number of Studies Analysed | Total Number of Subjects | Heterogeneity | |
|---|---|---|---|---|---|---|
| I2 | p-Value | |||||
| Myeloproliferative neoplasms (overall) | ||||||
| Excluding small studies | 13.6 [8.8–18.4] | 1.9% lower | 15 | 3117 | 96% | <0.0001 |
| Excluding low- and moderate-quality studies | 15.4 [11.3–19.6] | 0.1% lower | 24 | 3485 | 95% | <0.0001 |
| Excluding outlier studies | 13.9 [12.0–15.9] | 1.6% lower | 31 | 3633 | 63% | <0.0001 |
| Polycythaemia vera | ||||||
| Excluding small studies | 15.6 [11.0–20.3] | 1.2% lower | 4 | 614 | 59% | 0.06 |
| Excluding low- and moderate-quality studies | 18.6 [14.4–22.7] | 1.8% higher | 13 | 946 | 59% | 0.003 |
| Excluding outlier studies | 15.4 [12.0–18.7] | 1.4% lower | 19 | 1161 | 54% | 0.01 |
| Essential thrombocythaemia | ||||||
| Excluding small studies | 11.3 [5.9–16.8] | 1.5% higher | 3 | 470 | 72% | 0.02 |
| Excluding low- and moderate-quality studies | 11.1 [7.1–15.0] | 1.3% higher | 12 | 839 | 72% | <0.0001 |
| Excluding outlier studies | 8.4 [6.0–10.8] | 4.4% lower | 19 | 1096 | 48% | 0.01 |
| Myelofibrosis | ||||||
| Excluding small studies | 11.7 [4.0–19.5] | 4.0% lower | 6 | 1191 | 95% | <0.0001 |
| Excluding low- and moderate-quality studies | 14.0 [8.9–19.1] | 1.7% lower | 18 | 1700 | 91% | <0.0001 |
| Excluding outlier studies | 14.5 [12.4–16.7] | 1.2% lower | 25 | 1377 | 18% | 0.46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chia, Y.C.; Islam, M.A.; Hider, P.; Woon, P.Y.; Johan, M.F.; Hassan, R.; Ramli, M. The Prevalence of TET2 Gene Mutations in Patients with BCR-ABL-Negative Myeloproliferative Neoplasms (MPN): A Systematic Review and Meta-Analysis. Cancers 2021, 13, 3078. https://doi.org/10.3390/cancers13123078
Chia YC, Islam MA, Hider P, Woon PY, Johan MF, Hassan R, Ramli M. The Prevalence of TET2 Gene Mutations in Patients with BCR-ABL-Negative Myeloproliferative Neoplasms (MPN): A Systematic Review and Meta-Analysis. Cancers. 2021; 13(12):3078. https://doi.org/10.3390/cancers13123078
Chicago/Turabian StyleChia, Yuh Cai, Md Asiful Islam, Phil Hider, Peng Yeong Woon, Muhammad Farid Johan, Rosline Hassan, and Marini Ramli. 2021. "The Prevalence of TET2 Gene Mutations in Patients with BCR-ABL-Negative Myeloproliferative Neoplasms (MPN): A Systematic Review and Meta-Analysis" Cancers 13, no. 12: 3078. https://doi.org/10.3390/cancers13123078
APA StyleChia, Y. C., Islam, M. A., Hider, P., Woon, P. Y., Johan, M. F., Hassan, R., & Ramli, M. (2021). The Prevalence of TET2 Gene Mutations in Patients with BCR-ABL-Negative Myeloproliferative Neoplasms (MPN): A Systematic Review and Meta-Analysis. Cancers, 13(12), 3078. https://doi.org/10.3390/cancers13123078







