The Role of Extracellular Vesicles in Disease Progression and Detection of Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Nomenclature
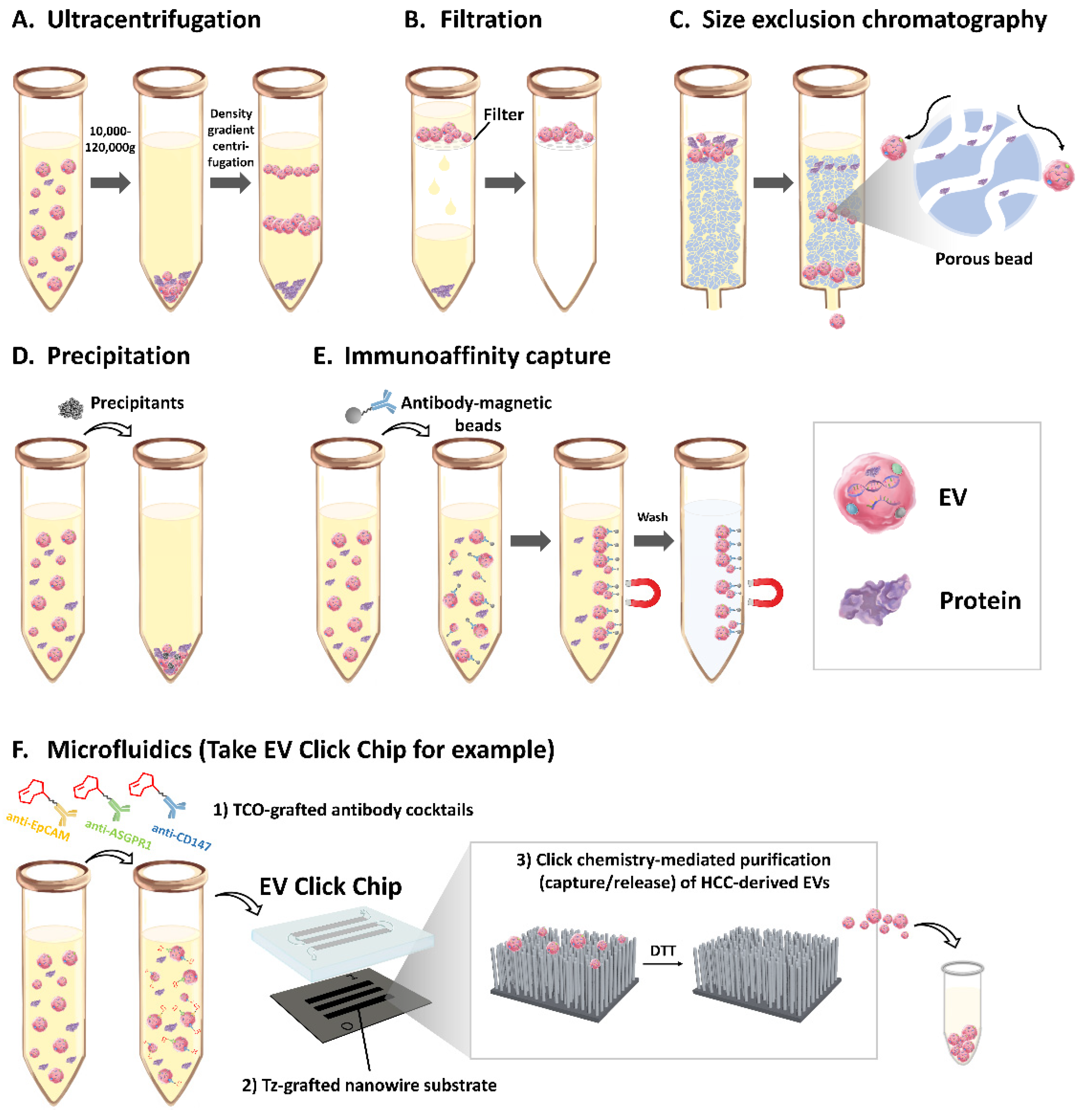
3. EVs isolation method
3.1. Ultracentrifugation
3.2. Filtration
3.3. Size Exclusion Chromatography
3.4. Precipitation
3.5. Immunoaffinity Capture
3.6. Microfluidics
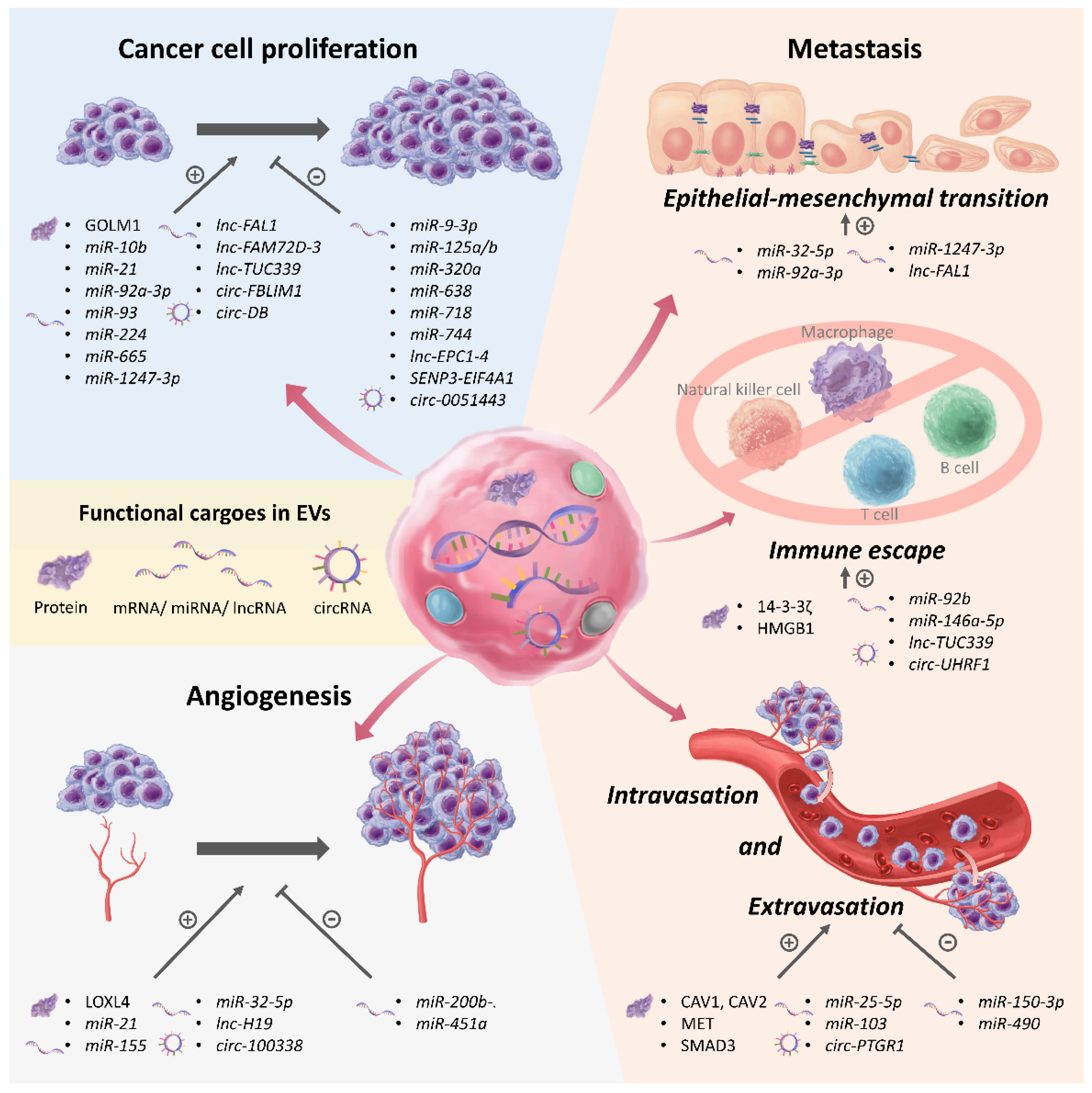
4. EVs as Mediators of Chronic Liver Disease and HCC Progression
4.1. EVs in Promoting Progression of Chronic Liver Disease
4.2. EVs in Regulating Proliferation of HCC
4.3. EVs in Regulating Angiogenesis in HCC
4.4. EVs in Promoting Metastasis, Immune Escape, and Recurrence in HCC
| Name of the Cargo in EVs. | Cargo Type | Level in HCC EVs 1 | EVs isolation Method 2 | Function of the Cargo | Mechanism of the Cargo | Ref |
|---|---|---|---|---|---|---|
| HCC cell proliferation | ||||||
| GOLM1 | Protein | ↑ | Differential ultracentrifugation | Promotes HCC cell proliferation, migration, and invasion in vitro | Activates the GSK-3β/MMP-1 and -9 pathway | [63] |
| miR-9-3p | miRNA | ↓ | Differential ultracentrifugation | Suppresses HCC cell proliferation in vitro | Suppresses the ERK1/2 pathway and HBGF-5 expression | [64] |
| miR-10b, miR-21 (cultured at acidic condition—pH 6.6) | miRNA | ↑ | Differential ultracentrifugation | Promotes HCC cell proliferation, migration, and invasion in vitro; promotes HCC growth and lung metastasis in vivo | – | [65] |
| miR-92a-3p | miRNA | ↑ | Differential ultracentrifugation | Promotes HCC cell proliferation, migration, invasion, and EMT in vitro, promotes EMT and metastasis in vivo | Suppresses PTEN and activates the PI3K/AkT pathway | [66] |
| miR-93 | miRNA | ↑ | Total Exosome Isolation Kit | Promotes HCC cell proliferation and invasion in vitro | Suppresses expression of TP53INP1, TIMP2, and CDKN1A | [67] |
| miR-125a/b (from TAM) | miRNA | - | ExoQuick™ Exosome Precipitation Solution | Suppresses HCC cell proliferation, migration, invasion, and stem cell properties in vitro | Suppresses CD90 expression | [82] |
| miR-224 | miRNA | ↑ | Total Exosome Isolation Kit | Promotes HCC cell proliferation and invasion in vitro | Suppresses GNMT expression | [71] |
| miR-320a (from CAF) | miRNA | - | Total Exosome Isolation Kit | Suppresses HCC cells proliferation, migration and metastasis in vitro and in vivo | Suppresses the PBX3/ERK1/2/CDK2 pathway | [83] |
| miR-638 | miRNA | ↓ | Total Exosome Isolation Kit | Suppresses HCC cell proliferation in vitro | – | [68] |
| miR-665 | miRNA | ↑ | Differential ultracentrifugation | Promotees HCC cell proliferation in vitro, promotees HCC growth in vivo | Activates the MAPK/ERK pathway | [72] |
| miR-718 | miRNA | ↓ | Differential ultracentrifugation | Suppresseses HCC cell proliferation in vitro | Suppresses HOXB8 expression | [69] |
| miR-744 | miRNA | ↓ | Differential ultracentrifugation | Suppresseses HCC cell proliferation and chemoresistance to sorafenib in vitro | Suppresses PAX2 expression | [70] |
| miR-1247-3p | miRNA | ↑ | Differential ultracentrifugation | Promotes proliferation of CAF in vitro, the activated CAF further promotes HCC cell progression, migration, stem cell properties, EMT, and chemoresistance to sorafenib in vitro and in vivo | Suppresses B4GALT3 to activate the NF-κB pathway | [92] |
| lnc-EPC1-4 | lncRNA | ↓ | Differential ultracentrifugation | Suppresses HCC cell proliferation and promotes HCC cell apoptosis | – | [73] |
| lnc-FAL1 | lncRNA | ↑ | ExoQuick-TC™ Exosome Precipitation Solution | Promotees HCC cell proliferation, migration, invasion, and EMT in vitro | Suppresses miR-1236 to activate ZEB1 and AFP expression | [74] |
| lnc-FAM72D-3 | lncRNA | ↑ | Differential ultracentrifugation | Promotes HCC cell proliferation and suppresses HCC cell apoptosis | – | [73] |
| lnc-TUC339 | lncRNA | ↑ | Differential ultracentrifugation | Promotes proliferation and suppresses cell adhesion to extracellular matrix of HCC cell in vitro, suppresses phagocytic activity and promotes M2-polarization of macrophage in vitro | May be involved in several pathways to regulate macrophages | [75] |
| SENP3-EIF4A1 | lncRNA | ↓ | ExoQuick-TC™ Exosome Precipitation Solution | Suppresses HCC cell proliferation and migration in vitro, suppresses HCC growth in vivo | Suppresses miR-9-5p to activates ZFP36 expression | [76] |
| circ-0051443 | circRNA | ↓ | ExoQuick™ Exosome Precipitation Solutio | Suppresses HCC cell proliferation and promotes HCC cell apoptosis in vitro, suppresses HCC growth in vivo | Activates BAK1 expression | [77] |
| circ-FBLIM1 | circRNA | ↑ | Differential ultracentrifugation | Promotes HCC cell proliferation and glycolysis in vitro, promotes HCC growth in vivo | Suppresses miR-338 to activate LRP6 expression | [78] |
| circ-DB (from adipocyte) | circRNA | - | Differential ultracentrifugation | Promotes HCC cell proliferation and reduces DNA damage in vitro, promotes HCC growth in vivo | Suppresses miR-34a and activates expression of USP7 and cyclin A2 | [79] |
| Angiogenesis | ||||||
| LOXL4 | Protein | ↑ | Differential ultracentrifugation | Promotes angiogenesis, HCC cell migration and invasion in vitro, promotes liver and lung metastasis in vivo | Activates the FAK/Src pathway | [84] |
| miR-21 | miRNA | ↑ | Differential ultracentrifugation | Converts hepatic stellate cells into to cancer-associated fibroblasts and promotes angiogenesis in vitro, promotes HCC growth and angiogenesis in vivo | Suppresses PTEN and activates the PI3K/AkT pathway in hepatic stellate cells | [91] |
| miR-32-5p (from multidrug-resistant HCC cell line, Bel/5-FU) | miRNA | ↑ | Differential ultracentrifugation | Promotes angiogenesis, HCC cell migration, invasion, and EMT, causes multidrug resistance in vitro, promotes angiogenesis and EMT, and causes 5-FU resistance in vivo | Suppresses PTEN and activates the PI3K/Akt pathway | [90] |
| miR-155 (cultured at hypoxic condition—1% O2) | miRNA | ↑ | ExoQuick-TC™ Exosome Precipitation Solution | Promotes angiogenesis in vitro | – | [85] |
| miR-200b-3p | miRNA | ↓ | Total Exosome Isolation Kit | Suppresses angiogenesis in vitro | Suppresses ERG expression | [86] |
| miR-451a | miRNA | ↓ | Differential ultracentrifugation | Suppresses cell proliferation and migration, promotes apoptosis of HCC cell and HUVEC in vitro, and suppresses angiogenesis in vitro and in vivo | Suppresses LPIN1 expression | [89] |
| lnc-H19 (from CD90+ HCC cell) | lncRNA | ↑ | Differential ultracentrifugation | Promotes cell–cell adhesion of HCC cells and promotes angiogenesis in vitro | Activates VEGF expression | [87] |
| circ-100338 | circRNA | ↑ | Differential ultracentrifugation | Promotes HCC cell invasion and angiogenesis in vitro, promotes HCC growth, angiogenesis, and lung metastasis in vivo | – | [88] |
| Metastasis | ||||||
| 14-3-3ζ | Protein | ↑ | Differential ultracentrifugation | Suppresses anti-tumor activity of tumor-infiltrating T lymphocytes | – | [93] |
| CAV1, CAV2, MET | Protein | ↑ | Differential ultracentrifugation | Promotes migration and invasion of non-motile immortalized hepatocyte cells in vitro | Activates the PI3K/AkT and MAPK/ERK pathways | [98] |
| SMAD3 | Protein | ↑ | ExoQuick™ Exosome Precipitation Solution | Promotes HCC cells adhesion in vitro | Activates ROS expression | [99] |
| HMGB1 | Protein | ↑ | Differential ultracentrifugation | Promotes TIM-1+ B cell expansion and suppresses CD8+ T cells activity in vitro | Activates the TLR2/4-MAPK pathway | [94] |
| miR-25-5p | miRNA | ↑ | Differential ultracentrifugation | Promotes transendothelial migration of HCC cell in vitro, promotes HCC tumor self-seeding in vivo | Suppresses LRRC7 expression | [100] |
| miR-92b | miRNA | ↑ | ExoQuick™ Exosome Precipitation Solution | Promotes HCC cell migration and suppresses NK cells cytotoxicity in vitro | Mechanism regarding HCC migration is not mentioned Suppresses CD69 on NK cells | [95] |
| miR-103 | miRNA | ↑ | Differential ultracentrifugation | Increases vascular permeability in vitro and in vivo, promotes liver and lung metastasis in vivo | Suppresses expression of VE-cadherin, p120-catenin, and ZO-1 | [101] |
| miR-146a-5p | miRNA | ↑ | Differential ultracentrifugation | Promotes M2-polarization of tumor-associated macrophages and suppresses T cells activity in vitro and in vivo | – | [96] |
| miR-150-3p (from CAF) | miRNA | – | Total Exosome Isolation Reagent | Suppresses HCC cell migration and invasion in vitro | – | [102] |
| miR-490 (from mast cells) | miRNA | – | Total Exosome Isolation Reagent | Suppresses HCC cell migration and invasion in vitro | Suppresses the EGFR/AkT/ERK1/2 pathway | [103] |
| circ-PTGR1 | circRNA | ↑ | ExoQuick-TC™ Exosome Precipitation Solution | Promotees HCC cell migration and invasion in vitro, promotes mesenteric lymph node metastasis in vivo | Competes with MET and suppresses miR449a expression | [104] |
| circ-UHRF1 | circRNA | ↑ | ExoQuick™ Exosome Precipitation Solution | Suppresses NK cell secretion of IFN-γ and TNF-α in vitro and in vivo, promotes metastasis in vivo | Suppresses miR-449c-5p to upregulate TIM-3 | [97] |
5. EVs as Biomarkers for Detection of HCC
5.1. EV Protein for Detection of HCC
5.2. EV miRNA for Detection of HCC
| Biomarkers/ Diagnostic Model | Biomarker Type | Expression Level in HCC | EV isolation Method 1 | Number of Patients | Sen/Spe (%) | AUROC | Study Type | Restricts HCC to Early-Stage? | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Amount of total EVs | – | ↑ | Ultracentrifugation | 28 TNM stage I HCC vs. 40 cirrhosis | 63/89 | 0.83 | Case-control | Yes | [12] |
| Amount of AnnexinV+ EpCAM+ ASGPR1+ EV | – | ↑ | Ultracentrifugation | 86 HCC vs. 49 cirrhosis | 81/47 | 0.73 | Case-control | No | [13] |
| FIBG | Protein | – (↑in iCCA) | Filtration and Ultracentrifugation | 29 HCC vs. 12 iCCA | 83/90 | 0.89 | Case-control | No | [113] |
| SMAD3 | Protein | ↑ | ExoQuick™ Exosome Precipitation Solution | 29 HCC vs. 37 HD + benign hepatoma | –/– | 0.70 | Case-control | No | [99] |
| A panel combining miR-122, miR-148a, and AFP | miRNA + AFP | ↑ | Ultracentrifugation, filtration, and precipitation | 50 HCC vs. 40 cirrhosis | 86/88 | 0.93 | Case-control | No | [114] |
| A panel combining miR-10b-5p, miR-221-3p, miR-223-3p, and miR-21-5p | miRNA | ↑ | ExoEnrich™ instant exosome isolation kit and immunoaffinity capture (anti-ASGR2) | 38 HCC vs. 35 CH + 25 cirrhosis | 59/95 | 0.80 | Case-control | No | [115] |
| miR-18a, miR-101, miR-106b, miR-122, miR-195, miR-221, miR-222, miR-224 | miRNA | ↑(18a, 221, 222, 224) ↓(101, 106b, 122, 195) | Ultracentrifugation | 20 HCC vs. 20 cirrhosis vs. 20 CH B | –/– | – | Case-control | No | [116] |
| LINC00853 | lncRNA | ↑ | ExoQuick™ Exosome Precipitation Solution | 32 early-stage HCC (single, <2 cm) vs. 28 CH + 35 cirrhosis | 94/85 | 0.96 | Case-control | Yes | [117] |
| Lnc85 | lncRNA | ↑ | Ribo™ Exosome Isolation Reagent | 122 HCC vs. 43 cirrhosis | 80/74 | 0.89 | Case-control | No | [118] |
| RN7SL1 S fragment | lncRNA | ↑ | Ultracentrifugation and Filtration | 25 HCC vs. 25 healthy donors | –/– | 0.75 | Case-control | No | [119] |
| A risk score panel combining AFP and ENSG00000248932.1, ENST00000440688.1, ENST00000457302.2 | lncRNA + AFP | ↑ | ExoQuick™ Exosome Precipitation Solution | Training set: 20 HCC vs. 20 CH Validation set: 180 HCC vs. 180 CH | –/– | 0.97 0.87 | Case-control | No | [120] |
| A panel combining circ_0004001, circ_0004123, and circ_0075792 | circRNA | ↑ | Ultracentrifugation | 71 HCC vs. 40 HD | 91/78 | 0.89 | Case-control | No | [121] |
| A panel combining 8 long RNAs | – | ↑ | exoRNeasy Maxi Kit | Training set: 44 HCC vs. 78 HD 1st Validation set: 27 HCC vs. 53 HD 2nd Validation set: 33 HCC vs. 33 HD + 6 hepatic benign disorders | 84/94 89/91 –/– | 0.95 0.96 0.96 | Case-control | No | [122] |
| LDHC | mRNA | ↑ | exoRNeasy Midi Kit | 50 TNM stage I/II HCC vs. 100 HD | 88/93 | 0.95 | Case-control | Yes | [123] |
| A panel combining AFP, GPC3, ALB, APOH, FABP1, FGB, FGG, AHSG, RBP4,TF | mRNA | ↑ | EV Click Chip (immunoaffinity + microfluidic device) | 36 BCLC stage 0-A HCC vs. 26 cirrhosis | 84/88 | 0.93 | Case-control | Yes | [38] |
5.3. EV lncRNA and EV circRNA for Detection of HCC
5.4. EV mRNA for Detection of HCC
6. Conclusion and Future Direction
Author Contributions
Funding
Conflicts of Interest
References
- Global Burden of Disease Cancer Collaboration; Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017, 3, 524–548. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Wang, J.J.; Luu, M.; Noureddin, M.; Kosari, K.; Agopian, V.G.; Rich, N.E.; Lu, S.C.; Tseng, H.R.; Nissen, N.N.; et al. The Mortality and Overall Survival Trends of Primary Liver Cancer in the United States. J. Natl. Cancer Inst. 2021. [Google Scholar] [CrossRef]
- Parikh, N.D.; Mehta, A.S.; Singal, A.G.; Block, T.; Marrero, J.A.; Lok, A.S. Biomarkers for the Early Detection of Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2495–2503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhanasekaran, R.; Bandoh, S.; Roberts, L.R. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, G.; Ignatiadis, M. Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine. Cancer Res. 2019, 79, 2798–2804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Chen, J.F.; Smalley, M.; Zhao, M.; Ke, Z.; Zhu, Y.; Tseng, H.R. Nanostructured Substrates for Detection and Characterization of Circulating Rare Cells: From Materials Research to Clinical Applications. Adv. Mater. 2020, 32, e1903663. [Google Scholar] [CrossRef]
- Krebs, M.G.; Metcalf, R.L.; Carter, L.; Brady, G.; Blackhall, F.H.; Dive, C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat. Rev. Clin. Oncol. 2014, 11, 129–144. [Google Scholar] [CrossRef]
- Kosaka, N.; Kogure, A.; Yamamoto, T.; Urabe, F.; Usuba, W.; Prieto-Vila, M.; Ochiya, T. Exploiting the message from cancer: The diagnostic value of extracellular vesicles for clinical applications. Exp. Mol. Med. 2019, 51, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Wang, W.; Li, H.; Zhou, Y.; Jie, S. Peripheral blood microvesicles are potential biomarkers for hepatocellular carcinoma. Cancer Biomark. 2013, 13, 351–357. [Google Scholar] [CrossRef]
- Julich-Haertel, H.; Urban, S.K.; Krawczyk, M.; Willms, A.; Jankowski, K.; Patkowski, W.; Kruk, B.; Krasnodebski, M.; Ligocka, J.; Schwab, R.; et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol. 2017, 67, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Zijlstra, A.; Di Vizio, D. Size matters in nanoscale communication. Nat. Cell Biol. 2018, 20, 228–230. [Google Scholar] [CrossRef]
- Gardiner, C.; Di Vizio, D.; Sahoo, S.; Thery, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. J. Extracell. Vesicles 2016, 5, 32945. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006. [Google Scholar] [CrossRef]
- Witwer, K.W.; Buzas, E.I.; Bemis, L.T.; Bora, A.; Lasser, C.; Lotvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Poliakov, A.; Spilman, M.; Dokland, T.; Amling, C.L.; Mobley, J.A. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate 2009, 69, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Ridinger, J.; Rupp, A.K.; Janssen, J.W.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef] [Green Version]
- Cantin, R.; Diou, J.; Belanger, D.; Tremblay, A.M.; Gilbert, C. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J. Immunol. Methods 2008, 338, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, M.L.; Ilmer, M.; Silva, L.P.; Hawke, D.H.; Recio, A.; Vorontsova, M.A.; Alt, E.; Vykoukal, J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A 2014, 1371, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [Google Scholar] [CrossRef]
- Zeringer, E.; Barta, T.; Li, M.; Vlassov, A.V. Strategies for isolation of exosomes. Cold Spring Harb. Protoc. 2015, 2015, 319–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamez-Valero, A.; Monguio-Tortajada, M.; Carreras-Planella, L.; Franquesa, M.; Beyer, K.; Borras, F.E. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef] [Green Version]
- Mol, E.A.; Goumans, M.J.; Doevendans, P.A.; Sluijter, J.P.G.; Vader, P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine 2017, 13, 2061–2065. [Google Scholar] [CrossRef] [PubMed]
- Takov, K.; Yellon, D.M.; Davidson, S.M. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: Yield, purity and functional potential. J. Extracell. Vesicles 2019, 8, 1560809. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.S.; Funk, S.; Muller, L.; Boyiadzis, M.; Whiteside, T.L. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J. Extracell. Vesicles 2016, 5, 29289. [Google Scholar] [CrossRef]
- Welton, J.L.; Webber, J.P.; Botos, L.A.; Jones, M.; Clayton, A. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J. Extracell. Vesicles 2015, 4, 27269. [Google Scholar] [CrossRef]
- Vogel, R.; Coumans, F.A.; Maltesen, R.G.; Boing, A.N.; Bonnington, K.E.; Broekman, M.L.; Broom, M.F.; Buzas, E.I.; Christiansen, G.; Hajji, N.; et al. A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing. J. Extracell. Vesicles 2016, 5, 31242. [Google Scholar] [CrossRef] [PubMed]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef]
- Rider, M.A.; Hurwitz, S.N.; Meckes, D.G., Jr. ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles. Sci. Rep. 2016, 6, 23978. [Google Scholar] [CrossRef] [PubMed]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [Green Version]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Moller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Yoo, C.E.; Kim, M.; Kang, H.J.; Park, D.; Lee, M.; Huh, N. Noble polymeric surface conjugated with zwitterionic moieties and antibodies for the isolation of exosomes from human serum. Bioconjug. Chem. 2012, 23, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.E.; Kim, G.; Kim, M.; Park, D.; Kang, H.J.; Lee, M.; Huh, N. A direct extraction method for microRNAs from exosomes captured by immunoaffinity beads. Anal. Biochem. 2012, 431, 96–98. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Lim, J.W.; Tauro, B.J.; Ji, H.; Moritz, R.L.; Simpson, R.J. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell Proteom. 2010, 9, 197–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, N.; Lee, Y.T.; Zhang, R.Y.; Kao, R.; Teng, P.C.; Yang, Y.; Yang, P.; Wang, J.J.; Smalley, M.; Chen, P.J.; et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat. Commun. 2020, 11, 4489. [Google Scholar] [CrossRef]
- Chen, C.; Skog, J.; Hsu, C.H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 2010, 10, 505–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Yue, S.; Stadel, D.; Zoller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef]
- He, M.; Zeng, Y. Microfluidic Exosome Analysis toward Liquid Biopsy for Cancer. J. Lab. Autom. 2016, 21, 599–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, W.; Jeong, D.; Kim, J.; Cho, S.; Jang, S.C.; Han, C.; Kang, J.Y.; Gho, Y.S.; Park, J. Microfluidic fabrication of cell-derived nanovesicles as endogenous RNA carriers. Lab Chip 2014, 14, 1261–1269. [Google Scholar] [CrossRef]
- Davies, R.T.; Kim, J.; Jang, S.C.; Choi, E.J.; Gho, Y.S.; Park, J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip 2012, 12, 5202–5210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, A.; Sandhu, S.; Lai, J.P.; Sandhu, D.S. Hepatocellular carcinoma in non-cirrhotic liver: A comprehensive review. World J. Hepatol. 2019, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Rios-Colon, L.; Arthur, E.; Niture, S.; Qi, Q.; Moore, J.T.; Kumar, D. The Role of Exosomes in the Crosstalk between Adipocytes and Liver Cancer Cells. Cells 2020, 9, 1988. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, Z.; Zhao, X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J. Hematol. Oncol. 2019, 12, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, R.; Wang, Z.; Shi, W.; Yu, L.; Xia, H.; Huang, Z.; Liu, S.; Zhao, X.; Xu, Y.; Yam, J.W.P.; et al. Exosomes in hepatocellular carcinoma microenvironment and their potential clinical application value. Biomed. Pharmacother. 2021, 138. [Google Scholar] [CrossRef]
- Ramakrishnaiah, V.; Thumann, C.; Fofana, I.; Habersetzer, F.; Pan, Q.; de Ruiter, P.E.; Willemsen, R.; Demmers, J.A.; Stalin Raj, V.; Jenster, G.; et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc. Natl. Acad. Sci. USA 2013, 110, 13109–13113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Han, Q.; Hou, Z.; Zhang, C.; Tian, Z.; Zhang, J. Exosomes mediate hepatitis B virus (HBV) transmission and NK-cell dysfunction. Cell Mol. Immunol. 2017, 14, 465–475. [Google Scholar] [CrossRef]
- Devhare, P.B.; Sasaki, R.; Shrivastava, S.; Di Bisceglie, A.M.; Ray, R.; Ray, R.B. Exosome-Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, N.R.; Chadha, R.; Kumar, S.; Choedon, T.; Reddy, V.S.; Kumar, V. The HBx gene of hepatitis B virus can influence hepatic microenvironment via exosomes by transferring its mRNA and protein. Virus Res. 2017, 240, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, Y.; Takagi, R.; Naito, Y.; Kiniwa, T.; Tanaka, Y.; Hamada-Tsutsumi, S.; Kawano, M.; Matsushita, S.; Ochiya, T.; Miyajima, A. Identification of the novel 3’ UTR sequences of human IL-21 mRNA as potential targets of miRNAs. Sci. Rep. 2017, 7, 7780. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, A.; Arab, J.P.; Reyes, D.; Lapitz, A.; Moshage, H.; Banales, J.M.; Arrese, M. Extracellular Vesicles in NAFLD/ALD: From Pathobiology to Therapy. Cells 2020, 9, 817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsova, P.; Ibrahim, S.H.; Krishnan, A.; Verma, V.K.; Bronk, S.F.; Werneburg, N.W.; Charlton, M.R.; Shah, V.H.; Malhi, H.; Gores, G.J. Lipid-Induced Signaling Causes Release of Inflammatory Extracellular Vesicles From Hepatocytes. Gastroenterology 2016, 150, 956–967. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.Y.; Song, M.J.; Gao, Y.; Mauer, A.S.; Revzin, A.; Malhi, H. Hepatocyte-Derived Lipotoxic Extracellular Vesicle Sphingosine 1-Phosphate Induces Macrophage Chemotaxis. Front. Immunol. 2018, 9, 2980. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.L.; Pan, Q.; Cao, H.X.; Xin, F.Z.; Zhao, Z.H.; Yang, R.X.; Zeng, J.; Zhou, H.; Fan, J.G. Lipotoxic Hepatocyte-Derived Exosomal MicroRNA 192-5p Activates Macrophages Through Rictor/Akt/Forkhead Box Transcription Factor O1 Signaling in Nonalcoholic Fatty Liver Disease. Hepatology 2020, 72, 454–469. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhong, L.; Li, P.; He, K.; Qiu, C.; Zhao, L.; Gong, J. Cholesterol impairs hepatocyte lysosomal function causing M1 polarization of macrophages via exosomal miR-122-5p. Exp. Cell Res. 2020, 387, 111738. [Google Scholar] [CrossRef]
- Ibrahim, S.H.; Hirsova, P.; Tomita, K.; Bronk, S.F.; Werneburg, N.W.; Harrison, S.A.; Goodfellow, V.S.; Malhi, H.; Gores, G.J. Mixed lineage kinase 3 mediates release of C-X-C motif ligand 10-bearing chemotactic extracellular vesicles from lipotoxic hepatocytes. Hepatology 2016, 63, 731–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, D.; Nakao, Y.; Mauer, A.S.; Thompson, J.M.; Sehrawat, T.S.; Liao, C.Y.; Krishnan, A.; Lucien, F.; Guo, Q.; Liu, M.; et al. IRE1A Stimulates Hepatocyte-Derived Extracellular Vesicles That Promote Inflammation in Mice With Steatohepatitis. Gastroenterology 2020, 159, 1487–1503.e17. [Google Scholar] [CrossRef]
- Kakazu, E.; Mauer, A.S.; Yin, M.; Malhi, H. Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1alpha-dependent manner. J. Lipid Res. 2016, 57, 233–245. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Furuta, K.; Lucien, F.; Gutierrez Sanchez, L.H.; Hirsova, P.; Krishnan, A.; Kabashima, A.; Pavelko, K.D.; Madden, B.; Alhuwaish, H.; et al. Integrin beta1-enriched extracellular vesicles mediate monocyte adhesion and promote liver inflammation in murine NASH. J. Hepatol. 2019, 71, 1193–1205. [Google Scholar] [CrossRef]
- Gai, X.; Tang, B.; Liu, F.; Wu, Y.; Wang, F.; Jing, Y.; Huang, F.; Jin, D.; Wang, L.; Zhang, H. mTOR/miR-145-regulated exosomal GOLM1 promotes hepatocellular carcinoma through augmented GSK-3beta/MMPs. J. Genet. Genom. 2019, 46, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Li, Y.; Liu, K.; Zhu, Q.; Yang, W.H.; Xiong, L.K.; Guo, D.L. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Med. 2018, 109, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.P.; Wang, C.Y.; Jin, X.H.; Li, M.; Wang, F.W.; Huang, W.J.; Yun, J.P.; Xu, R.H.; Cai, Q.Q.; Xie, D. Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis. Theranostics 2019, 9, 1965–1979. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Feng, X.; Liu, H.; Tong, R.; Wu, J.; Li, C.; Yu, H.; Chen, Y.; Cheng, Q.; Chen, J.; et al. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene 2020, 39, 6529–6543. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Wang, X.; Zhao, Y.; Hu, R.; Qin, L. Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem. Biophys. Res. Commun. 2018, 502, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Jiang, Y.; Yang, L.; Yan, S.; Wang, Y.G.; Lu, X.J. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell Biochem. 2018, 119, 4711–4716. [Google Scholar] [CrossRef] [PubMed]
- Sugimachi, K.; Matsumura, T.; Hirata, H.; Uchi, R.; Ueda, M.; Ueo, H.; Shinden, Y.; Iguchi, T.; Eguchi, H.; Shirabe, K.; et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer 2015, 112, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, W.; Wang, H.; Qiu, G.; Jiang, Z.; Wei, G.; Li, X. Exosomal MiR-744 Inhibits Proliferation and Sorafenib Chemoresistance in Hepatocellular Carcinoma by Targeting PAX2. Med. Sci. Monit. 2019, 25, 7209–7217. [Google Scholar] [CrossRef]
- Cui, Y.; Xu, H.F.; Liu, M.Y.; Xu, Y.J.; He, J.C.; Zhou, Y.; Cang, S.D. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J. Gastroenterol. 2019, 25, 1890–1898. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Wu, J.; Wu, J.; Ji, A.; Qiang, G.; Jiang, Y.; Jiang, C.; Ding, Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 2017, 8, 80666–80678. [Google Scholar] [CrossRef] [Green Version]
- Yao, Z.; Jia, C.; Tai, Y.; Liang, H.; Zhong, Z.; Xiong, Z.; Deng, M.; Zhang, Q. Serum exosomal long noncoding RNAs lnc-FAM72D-3 and lnc-EPC1-4 as diagnostic biomarkers for hepatocellular carcinoma. Aging (Albany NY) 2020, 12, 11843–11863. [Google Scholar] [CrossRef]
- Li, B.; Mao, R.; Liu, C.; Zhang, W.; Tang, Y.; Guo, Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018, 197, 122–129. [Google Scholar] [CrossRef]
- Li, X.; Lei, Y.; Wu, M.; Li, N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018, 19, 2958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Pu, J.; Zhang, Y.; Yao, T.; Luo, Z.; Li, W.; Xu, G.; Liu, J.; Wei, W.; Deng, Y. Exosome-transmitted long non-coding RNA SENP3-EIF4A1 suppresses the progression of hepatocellular carcinoma. Aging (Albany NY) 2020, 12, 11550–11567. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Quan, Y.; Fan, S.; Wang, H.; Liang, J.; Huang, L.; Chen, L.; Liu, Q.; He, P.; Ye, Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020, 475, 119–128. [Google Scholar] [CrossRef]
- Lai, Z.; Wei, T.; Li, Q.; Wang, X.; Zhang, Y.; Zhang, S. Exosomal circFBLIM1 Promotes Hepatocellular Carcinoma Progression and Glycolysis by Regulating the miR-338/LRP6 Axis. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Deng, T.; Ge, S.; Liu, Y.; Bai, M.; Zhu, K.; Fan, Q.; Li, J.; Ning, T.; Tian, F.; et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 2019, 38, 2844–2859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Yu, J.; Xiong, L.; Liu, Y.; Fan, L.; Li, Y.; Chen, B.; Chen, J.; Xu, X. Exosomes derived from liver cancer cells reprogram biological behaviors of LO2 cells by transferring Linc-ROR. Gene 2019, 719, 144044. [Google Scholar] [CrossRef]
- Amann, T.; Hellerbrand, C. GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin. Ther. Targets 2009, 13, 1411–1427. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, B.; Xiao, S.; Li, Y.; Chen, Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J. Cell Biochem. 2019, 120, 3046–3055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.; Sun, W.; Yue, S.; Yang, J.; Li, J.; Ma, B.; Wang, J.; Yang, X.; Pu, M.; et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017, 397, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, Y.; Zhang, X.; Feng, M.; Ma, J.; Li, J.; Yang, X.; Fang, F.; Xia, Q.; Zhang, Z.; et al. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol. Cancer 2019, 18, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuura, Y.; Wada, H.; Eguchi, H.; Gotoh, K.; Kobayashi, S.; Kinoshita, M.; Kubo, M.; Hayashi, K.; Iwagami, Y.; Yamada, D.; et al. Exosomal miR-155 Derived from Hepatocellular Carcinoma Cells Under Hypoxia Promotes Angiogenesis in Endothelial Cells. Dig. Dis. Sci. 2019, 64, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Moh-Moh-Aung, A.; Fujisawa, M.; Ito, S.; Katayama, H.; Ohara, T.; Ota, Y.; Yoshimura, T.; Matsukawa, A. Decreased miR-200b-3p in cancer cells leads to angiogenesis in HCC by enhancing endothelial ERG expression. Sci Rep. 2020, 10, 10418. [Google Scholar] [CrossRef]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Huang, Z.L.; Huang, J.; Xu, B.; Huang, X.Y.; Xu, Y.H.; Zhou, J.; Tang, Z.Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Li, J.; Zhang, G.; Wang, Q.; Wu, C.; Zhang, Q.; Wang, H.; Sun, P.; Xiang, R.; Yang, S. Exosomal miR-451a Functions as a Tumor Suppressor in Hepatocellular Carcinoma by Targeting LPIN1. Cell Physiol. Biochem. 2019, 53, 19–35. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Liu, M.; Qu, S.; Ma, J.; Zhang, Y.; Shi, T.; Wen, H.; Yang, Y.; Wang, S.; Wang, J.; et al. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 2018, 37, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Ren, H.; Dai, B.; Li, J.; Shang, L.; Huang, J.; Shi, X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J. Exp. Clin. Cancer Res. 2018, 37, 324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, T.; Lv, H.; Lv, G.; Li, T.; Wang, C.; Han, Q.; Yu, L.; Su, B.; Guo, L.; Huang, S.; et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018, 9, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Shen, H.; Zhangyuan, G.; Huang, R.; Zhang, W.; He, Q.; Jin, K.; Zhuo, H.; Zhang, Z.; Wang, J.; et al. 14-3-3zeta delivered by hepatocellular carcinoma-derived exosomes impaired anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death Dis. 2018, 9, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, L.; Zhang, Q.; Cheng, Y.; Chen, X.; Wang, G.; Shi, M.; Zhang, T.; Cao, Y.; Pan, H.; Zhang, L.; et al. Tumor-derived exosomal HMGB1 fosters hepatocellular carcinoma immune evasion by promoting TIM-1(+) regulatory B cell expansion. J. Immunother. Cancer 2018, 6, 145. [Google Scholar] [CrossRef] [Green Version]
- Nakano, T.; Chen, I.H.; Wang, C.C.; Chen, P.J.; Tseng, H.P.; Huang, K.T.; Hu, T.H.; Li, L.C.; Goto, S.; Cheng, Y.F.; et al. Circulating exosomal miR-92b: Its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am. J. Transplant. 2019, 19, 3250–3262. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Han, Q.; Xu, D.; Zheng, B.; Zhao, X.; Zhang, J. SALL4-mediated upregulation of exosomal miR-146a-5p drives T-cell exhaustion by M2 tumor-associated macrophages in HCC. Oncoimmunology 2019, 8, 1601479. [Google Scholar] [CrossRef]
- Zhang, P.F.; Gao, C.; Huang, X.Y.; Lu, J.C.; Guo, X.J.; Shi, G.M.; Cai, J.B.; Ke, A.W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Qin, H.; Poon, T.C.; Sze, S.C.; Ding, X.; Co, N.N.; Ngai, S.M.; Chan, T.F.; Wong, N. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis 2015, 36, 1008–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Q.; Zhang, Q.; Lou, Y.; Yang, J.; Nie, G.; Chen, Q.; Chen, Y.; Zhang, J.; Wang, J.; Wei, T.; et al. Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene 2018, 37, 6105–6118. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Chen, W.; Zhi, X.; Chen, E.J.; Wei, T.; Zhang, J.; Shen, J.; Hu, L.Q.; Zhao, B.; Feng, X.H.; et al. Tumor-derived exosomes promote tumor self-seeding in hepatocellular carcinoma by transferring miRNA-25-5p to enhance cell motility. Oncogene 2018, 37, 4964–4978. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.H.; Zhang, Z.J.; Shang, L.R.; Luo, Y.W.; Lin, Y.F.; Yuan, Y.; Zhuang, S.M. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology 2018, 68, 1459–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yugawa, K.; Yoshizumi, T.; Mano, Y.; Itoh, S.; Harada, N.; Ikegami, T.; Kohashi, K.; Oda, Y.; Mori, M. Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur. J. Surg. Oncol. 2021, 47, 384–393. [Google Scholar] [CrossRef]
- Xiong, L.; Zhen, S.; Yu, Q.; Gong, Z. HCV-E2 inhibits hepatocellular carcinoma metastasis by stimulating mast cells to secrete exosomal shuttle microRNAs. Oncol. Lett. 2017, 14, 2141–2146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Liu, W.; Zou, Y.; Wang, G.; Deng, Y.; Luo, J.; Zhang, Y.; Li, H.; Zhang, Q.; Yang, Y.; et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 2019, 40, 432–445. [Google Scholar] [CrossRef] [Green Version]
- Giannelli, G.; Koudelkova, P.; Dituri, F.; Mikulits, W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatol. 2016, 65, 798–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capece, D.; Fischietti, M.; Verzella, D.; Gaggiano, A.; Cicciarelli, G.; Tessitore, A.; Zazzeroni, F.; Alesse, E. The inflammatory microenvironment in hepatocellular carcinoma: A pivotal role for tumor-associated macrophages. Biomed. Res. Int. 2013, 2013, 187204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Zijl, F.; Krupitza, G.; Mikulits, W. Initial steps of metastasis: Cell invasion and endothelial transmigration. Mutat. Res. 2011, 728, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Guo, P.; He, Y.; Chen, Z.; Chen, L.; Luo, Y.; Qi, L.; Liu, Y.; Wu, Q.; Cui, Y.; et al. HCC-derived exosomes elicit HCC progression and recurrence by epithelial-mesenchymal transition through MAPK/ERK signalling pathway. Cell Death Dis. 2018, 9, 513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef] [Green Version]
- Roberts, L.R.; Sirlin, C.B.; Zaiem, F.; Almasri, J.; Prokop, L.J.; Heimbach, J.K.; Murad, M.H.; Mohammed, K. Imaging for the diagnosis of hepatocellular carcinoma: A systematic review and meta-analysis. Hepatology 2018, 67, 401–421. [Google Scholar] [CrossRef] [Green Version]
- Taleb, R.S.Z.; Moez, P.; Younan, D.; Eisenacher, M.; Tenbusch, M.; Sitek, B.; Bracht, T. Quantitative proteome analysis of plasma microparticles for the characterization of HCV-induced hepatic cirrhosis and hepatocellular carcinoma. Proteom. Clin. Appl. 2017, 11, 1700014. [Google Scholar] [CrossRef] [PubMed]
- Arbelaiz, A.; Azkargorta, M.; Krawczyk, M.; Santos-Laso, A.; Lapitz, A.; Perugorria, M.J.; Erice, O.; Gonzalez, E.; Jimenez-Aguero, R.; Lacasta, A.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017, 66, 1125–1143. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Zhang, P.; Guo, G.; Jiang, T.; Zhao, X.; Jiang, J.; Huang, X.; Tong, H.; Tian, Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018, 7, 1670–1679. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Bhowmik, S.; Majumdar, S.; Goswami, A.; Chakraborty, J.; Gupta, S.; Aggarwal, S.; Ray, S.; Chatterjee, R.; Bhattacharyya, S.; et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int. J. Cancer 2020, 147, 2934–2947. [Google Scholar] [CrossRef] [PubMed]
- Sohn, W.; Kim, J.; Kang, S.H.; Yang, S.R.; Cho, J.Y.; Cho, H.C.; Shim, S.G.; Paik, Y.H. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 2015, 47, e184. [Google Scholar] [CrossRef]
- Kim, S.S.; Baek, G.O.; Ahn, H.R.; Sung, S.; Seo, C.W.; Cho, H.J.; Nam, S.W.; Cheong, J.Y.; Eun, J.W. Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol. Oncol. 2020, 14, 2646–2659. [Google Scholar] [CrossRef]
- Huang, X.; Sun, L.; Wen, S.; Deng, D.; Wan, F.; He, X.; Tian, L.; Liang, L.; Wei, C.; Gao, K.; et al. RNA sequencing of plasma exosomes revealed novel functional long noncoding RNAs in hepatocellular carcinoma. Cancer Sci. 2020, 111, 3338–3349. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Cao, J.; Chen, L.; Xi, X.; Wang, S.; Zhu, Y.; Yang, L.; Ma, L.; Wang, D.; Yin, J.; et al. Noncoding RNAs Serve as Diagnosis and Prognosis Biomarkers for Hepatocellular Carcinoma. Clin. Chem. 2019, 65, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Duan, Y.; Xu, Q.; Zhang, L.; Chen, W.; Qu, Z.; Wu, B.; Liu, W.; Shi, L.; Wu, D.; et al. Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J. Cell Mol. Med. 2020, 24, 1311–1318. [Google Scholar] [CrossRef]
- Sun, X.H.; Wang, Y.T.; Li, G.F.; Zhang, N.; Fan, L. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020, 20, 226. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, J.; Yu, S.; Wang, Z.; He, X.; Su, Y.; Guo, T.; Sheng, H.; Chen, J.; Zheng, Q.; et al. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin. Chem. 2019, 65, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Li, Y.; Gao, Y.; Kong, L.; Lin, Y.; Chen, Y. Cancer-testis antigen lactate dehydrogenase C4 in hepatocellular carcinoma: A promising biomarker for early diagnosis, efficacy evaluation and prognosis prediction. Aging (Albany NY) 2020, 12, 19455–19467. [Google Scholar] [CrossRef] [PubMed]
- Kalinich, M.; Bhan, I.; Kwan, T.T.; Miyamoto, D.T.; Javaid, S.; LiCausi, J.A.; Milner, J.D.; Hong, X.; Goyal, L.; Sil, S.; et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, 1123–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhang, H.; Gu, J.; Zhang, J.; Shi, H.; Qian, H.; Wang, D.; Xu, W.; Pan, J.; Santos, H.A. Engineered Extracellular Vesicles for Cancer Therapy. Adv. Mater. 2021, 33, e2005709. [Google Scholar] [CrossRef] [PubMed]
- Thietart, S.; Rautou, P.E. Extracellular vesicles as biomarkers in liver diseases: A clinician’s point of view. J. Hepatol. 2020, 73, 1507–1525. [Google Scholar] [CrossRef]
- Liangsupree, T.; Multia, E.; Riekkola, M.L. Modern isolation and separation techniques for extracellular vesicles. J. Chromatogr. A 2021, 1636, 461773. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Hoshida, Y.; Pinato, D.J.; Marrero, J.; Nault, J.C.; Paradis, V.; Tayob, N.; Sherman, M.; Lim, Y.S.; Feng, Z.; et al. International Liver Cancer Association (ILCA) White Paper on Biomarker Development for Hepatocellular Carcinoma. Gastroenterology 2021. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.-T.; Tran, B.V.; Wang, J.J.; Liang, I.Y.; You, S.; Zhu, Y.; Agopian, V.G.; Tseng, H.-R.; Yang, J.D. The Role of Extracellular Vesicles in Disease Progression and Detection of Hepatocellular Carcinoma. Cancers 2021, 13, 3076. https://doi.org/10.3390/cancers13123076
Lee Y-T, Tran BV, Wang JJ, Liang IY, You S, Zhu Y, Agopian VG, Tseng H-R, Yang JD. The Role of Extracellular Vesicles in Disease Progression and Detection of Hepatocellular Carcinoma. Cancers. 2021; 13(12):3076. https://doi.org/10.3390/cancers13123076
Chicago/Turabian StyleLee, Yi-Te, Benjamin V. Tran, Jasmine J. Wang, Icy Y. Liang, Sungyong You, Yazhen Zhu, Vatche G. Agopian, Hsian-Rong Tseng, and Ju Dong Yang. 2021. "The Role of Extracellular Vesicles in Disease Progression and Detection of Hepatocellular Carcinoma" Cancers 13, no. 12: 3076. https://doi.org/10.3390/cancers13123076
APA StyleLee, Y.-T., Tran, B. V., Wang, J. J., Liang, I. Y., You, S., Zhu, Y., Agopian, V. G., Tseng, H.-R., & Yang, J. D. (2021). The Role of Extracellular Vesicles in Disease Progression and Detection of Hepatocellular Carcinoma. Cancers, 13(12), 3076. https://doi.org/10.3390/cancers13123076






