One-Carbon Metabolism Associated Vulnerabilities in Glioblastoma: A Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Metabolic Reprogramming in Cancer and Cancer Initiating Cells
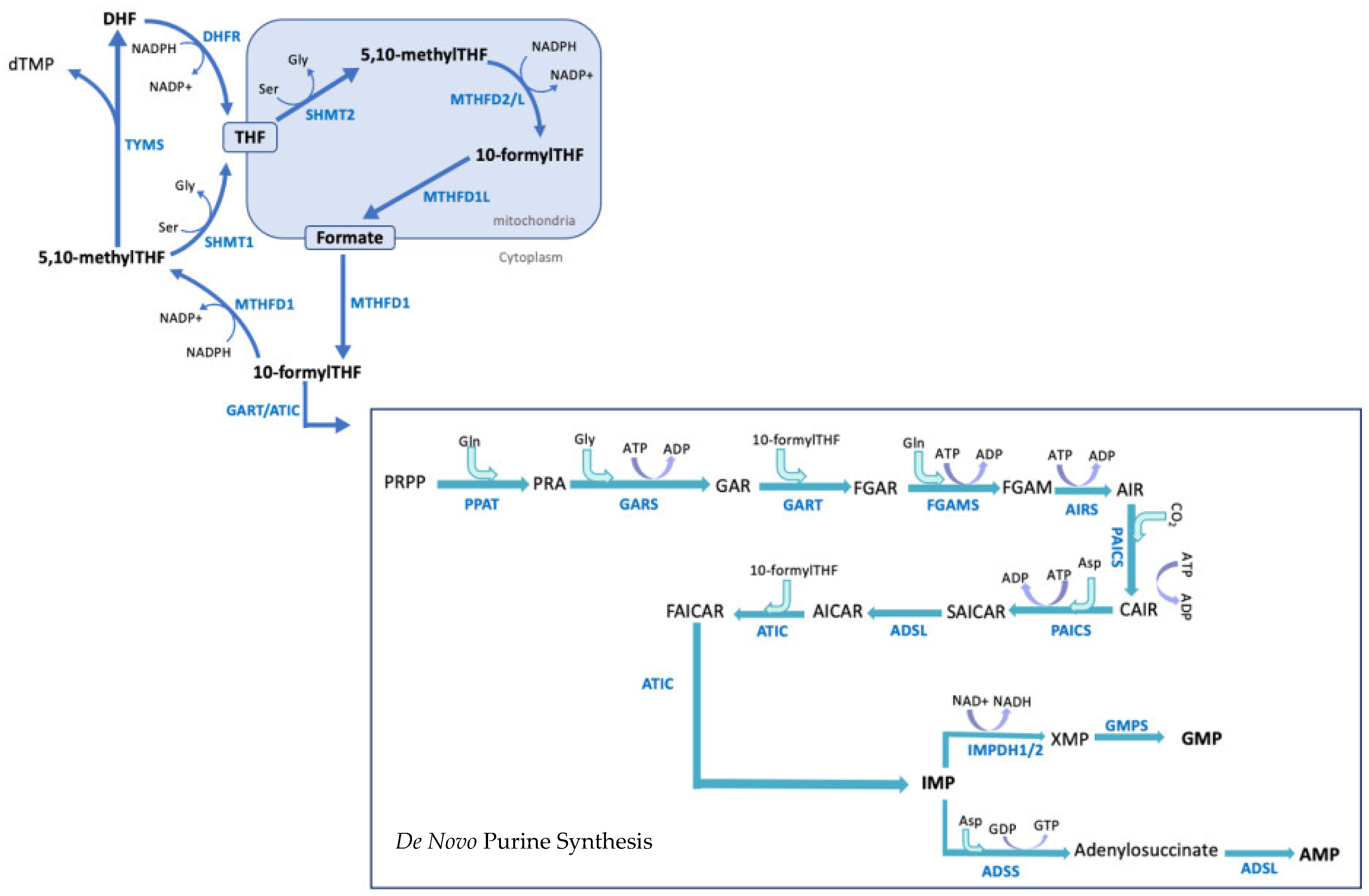
3. 1-C-Mediated de Novo Purine Synthesis: A Brief Overview
4. 1-C-Mediated de Novo Purine Synthesis: Relevance in Cancer and Glioblastoma
5. Signalling Pathways Upstream of Metabolic Reprogramming
6. Treatments Targeting 1-C Metabolism and Purine Synthesis in Cancer
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- NHanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Wirsching, H.-G.; Galanis, E.; Weller, M. Chapter 23—Glioblastoma. In Handbook of Clinical Neurology; Berger, M.S., Weller, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; Volume 134, pp. 381–397. ISBN 9780128029978. [Google Scholar]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Hildebrandt, I.J.; Prins, R.M.; Soto, H.; Mazzotta, M.M.; Dang, J.; Czernin, J.; Shyy, J.Y.J.; Watson, A.D.; Phelps, M.; et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 12932–12937. [Google Scholar] [CrossRef]
- Strickland, M.; Stoll, E.A. Metabolic reprogramming in glioma. Front. Cell Dev. Biol. 2017, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Papa, S.; Choy, P.M.; Bubici, C. The ERK and JNK pathways in the regulation of metabolic reprogramming. Oncogene 2019, 38, 2223–2240. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Poteet, E.; Choudhury, G.R.; Winters, A.; Li, W.; Ryou, M.G.; Liu, R.; Tang, L.; Ghorpade, A.; Wen, Y.; Yuan, F.; et al. Reversing the Warburg effect as a treatment for glioblastoma. J. Biol. Chem. 2013, 288, 9153–9164. [Google Scholar] [CrossRef]
- De Santis, M.C.; Porporato, P.E.; Martini, M.; Morandi, A. Signaling pathways regulating redox balance in cancer metabolism. Front. Oncol. 2018, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, W.; Tao, B.; Wang, X.; Yang, Z.; Zhang, Y.; Wang, C.; Liu, R.; Gao, H.; Liang, J.; et al. Tyrosine phosphorylation activates 6-phosphogluconate dehydrogenase and promotes tumor growth and radiation resistance. Nat. Commun. 2019, 10, 991. [Google Scholar] [CrossRef]
- Kathagen-Buhmann, A.; Schulte, A.; Weller, J.; Holz, M.; Herold-Mende, C.; Glass, R.; Lamszus, K. Glycolysis and the Pentose Phosphate Pathway Are Differentially Associated with the Dichotomous Regulation of Glioblastoma Cell Migration versus Proliferation. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4998991/ (accessed on 23 March 2020).
- Marin-Valencia, I.; Cho, S.K.; Rakheja, D.; Hatanpaa, K.J.; Kapur, P.; Mashimo, T.; Jindal, A.; Vemireddy, V.; Good, L.B.; Raisanen, J.; et al. Glucose metabolism via the pentose phosphate pathway, glycolysis and Krebs cycle in an orthotopic mouse model of human brain tumors. NMR Biomed. 2012, 25, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Zhao, F.; Thompson, C.B. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr. Opin. Genet. Dev. 2009, 19, 32–37. [Google Scholar] [CrossRef]
- Izquierdo-Garcia, J.L.; Cai, L.M.; Chaumeil, M.M.; Eriksson, P.; Robinson, A.E.; Pieper, R.O.; Phillips, J.J.; Ronen, S.M. Glioma cells with the IDH1 mutation modulate metabolic fractional flux through pyruvate carboxylase. PLoS ONE 2014, 9, e108289. [Google Scholar] [CrossRef] [PubMed]
- Goji, T.; Takahara, K.; Negishi, M.; Katoh, H. Cystine uptake through the cystine/glutamate antiporter xCT triggers glioblastoma cell death under glucose deprivation. J. Biol. Chem. 2017, 292, 19721–19732. [Google Scholar] [CrossRef]
- Suh, E.H.; Hackett, E.P.; Wynn, R.M.; Chuang, D.T.; Zhang, B.; Luo, W.; Sherry, A.D.; Park, J.M. In vivo assessment of increased oxidation of branched-chain amino acids in glioblastoma. Sci. Rep. 2019, 9, 340. [Google Scholar] [CrossRef] [PubMed]
- Tönjes, M.; Barbus, S.; Park, Y.J.; Wang, W.; Schlotter, M.; Lindroth, A.M.; Pleier, S.V.; Bai, A.H.C.; Karra, D.; Piro, R.M.; et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat. Med. 2013, 19, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Shim, J.K.; Kang, J.H.; Choi, J.; Chang, J.H.; Kim, S.Y.; Kang, S.G. Regulation of bioenergetics through dual inhibition of aldehyde dehydrogenase and mitochondrial complex i suppresses glioblastoma tumorspheres. Neuro. Oncol. 2018, 20, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Garcia, J.L.; Viswanath, P.; Eriksson, P.; Cai, L.; Radoul, M.; Chaumeil, M.M.; Blough, M.; Luchman, H.A.; Weiss, S.; Cairncross, J.G.; et al. IDH1 mutation induces reprogramming of pyruvate metabolism. Cancer Res. 2015, 75, 2999–3009. [Google Scholar] [CrossRef]
- Miska, J.; Lee-Chang, C.; Rashidi, A.; Muroski, M.E.; Chang, A.L.; Lopez-Rosas, A.; Zhang, P.; Panek, W.K.; Cordero, A.; Han, Y.; et al. HIF-1α Is a Metabolic Switch between Glycolytic-Driven Migration and Oxidative Phosphorylation-Driven Immunosuppression of Tregs in Glioblastoma. Cell Rep. 2019, 27, 226–237.e4. [Google Scholar] [CrossRef]
- Lu, C.-L.; Qin, L.; Liu, H.-C.; Candas, D.; Fan, M.; Li, J.J. Tumor Cells Switch to Mitochondrial Oxidative Phosphorylation under Radiation via mTOR-Mediated Hexokinase II Inhibition - A Warburg-Reversing Effect. PLoS ONE 2015, 10, e0121046. [Google Scholar] [CrossRef] [PubMed]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.A.; Brault, C.; Peck, B.; Bensaad, K.; Griffiths, B.; Mitter, R.; Chakravarty, P.; East, P.; Dankworth, B.; Alibhai, D.; et al. SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene 2015, 34, 5128–5140. [Google Scholar] [CrossRef]
- Geng, F.; Cheng, X.; Wu, X.; Yoo, J.Y.; Cheng, C.; Guo, J.Y.; Mo, X.; Ru, P.; Hurwitz, B.; Kim, S.H.; et al. Inhibition of SOAT1 suppresses glioblastoma growth via blocking SREBP-1-mediated lipogenesis. Clin. Cancer Res. 2016, 22, 5337–5348. [Google Scholar] [CrossRef] [PubMed]
- Pirmoradi, L.; Seyfizadeh, N.; Ghavami, S.; Zeki, A.A.; Shojaei, S. Targeting cholesterol metabolism in glioblastoma: A new therapeutic approach in cancer therapy. J. Investig. Med. 2019, 67, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, Z.; Chai, R.C.; Liu, Y.Q.; Li, G.Z.; Jiang, H.Y.; Jiang, T. Prognostic power of a lipid metabolism gene panel for diffuse gliomas. J. Cell. Mol. Med. 2019, 23, 7741–7748. [Google Scholar] [CrossRef]
- Guo, D. SCAP links glucose to lipid metabolism in cancer cells. Mol. Cell. Oncol. 2016, 3, e1132120. [Google Scholar] [CrossRef] [PubMed]
- Rusu, P.; Shao, C.; Neuerburg, A.; Acikgöz, A.A.; Wu, Y.; Zou, P.; Phapale, P.; Shankar, T.S.; Döring, K.; Dettling, S.; et al. GPD1 Specifically Marks Dormant Glioma Stem Cells with a Distinct Metabolic Profile. Cell Stem Cell 2019, 25, 241–257. [Google Scholar] [CrossRef] [PubMed]
- Zgheib, R.; Battaglia-Hsu, S.F.; Hergalant, S.; Quéré, M.; Alberto, J.M.; Chéry, C.; Rouyer, P.; Gauchotte, G.; Guéant, J.L.; Namour, F. Folate can promote the methionine-dependent reprogramming of glioblastoma cells towards pluripotency. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Wang, X.; Yang, K.; Xie, Q.; Wu, Q.; Mack, S.C.; Shi, Y.; Kim, L.J.Y.Y.; Prager, B.C.; Flavahan, W.A.; Liu, X.; et al. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat. Neurosci. 2017, 20, 661–673. [Google Scholar] [CrossRef]
- Lamb, R.; Harrison, H.; Smith, D.L.; Townsend, P.A.; Jackson, T.; Ozsvari, B.; Martinez-Outschoorn, U.E.; Pestell, R.G.; Howell, A.; Lisanti, M.P.; et al. Targeting tumor-initiating cells: Eliminating anabolic cancer stem cells with inhibitors of protein synthesis or by mimicking caloric restriction. Oncotarget 2015, 6, 4585–4601. [Google Scholar] [CrossRef]
- Zhang, J.; Nuebel, E.; Daley, G.Q.; Koehler, C.M.; Teitell, M.A. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 2012, 11, 589–595. [Google Scholar] [CrossRef]
- Intlekofer, A.M.; Finley, L.W.S. Metabolic signatures of cancer cells and stem cells. Nat. Metab. 2019, 1, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Dzeja, P.P.; Faustino, R.S.; Perez-Terzic, C.; Behfar, A.; Terzic, A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat. Clin. Pract. Cardiovasc. Med. 2007, 4. [Google Scholar] [CrossRef]
- Zhou, W.; Choi, M.; Margineantu, D.; Margaretha, L.; Hesson, J.; Cavanaugh, C.; Blau, C.A.; Horwitz, M.S.; Hockenbery, D.; Ware, C.; et al. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012, 31, 2103–2116. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Kamphorst, J.J.; Mathew, R.; Chung, M.K.; White, E.; Shlomi, T.; Rabinowitz, J.D. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol. Syst. Biol. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Tohyama, S.; Fujita, J.; Hishiki, T.; Matsuura, T.; Hattori, F.; Ohno, R.; Kanazawa, H.; Seki, T.; Nakajima, K.; Kishino, Y.; et al. Glutamine Oxidation Is Indispensable for Survival of Human Pluripotent Stem Cells. Cell Metab. 2016, 23, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; Thompson, C.B. Nutrient acquisition strategies of mammalian cells. Nature 2017, 546, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zheng, Y.; Xia, Y.; Ji, H.; Chen, X.; Guo, F.; Lyssiotis, C.A.; Aldape, K.; Cantley, L.C.; Lu, Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012, 14, 1295–1304. [Google Scholar] [CrossRef]
- Ahmad, F.; Dixit, D.; Sharma, V.; Kumar, A.; Joshi, S.D.; Sarkar, C.; Sen, E. Nrf2-driven TERT regulates pentose phosphate pathway in glioblastoma. Cell Death Dis. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, R.; Wu, M.; Smiljanic, S.; Kaskun, O.; Ghannad-Zadeh, K.; Celebre, A.; Isaev, K.; Morrissy, A.S.; Guan, J.; Tong, J.; et al. ID1 is critical for tumorigenesis and regulates chemoresistance in glioblastoma. Cancer Res. 2019, 79, 4057–4071. [Google Scholar] [CrossRef]
- Sachdeva, R.; Wu, M.; Johnson, K.; Kim, H.; Celebre, A.; Shahzad, U.; Graham, M.S.; Kessler, J.A.; Chuang, J.H.; Karamchandani, J.; et al. BMP signaling mediates glioma stem cell quiescence and confers treatment resistance in glioblastoma. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Vlashi, E.; Lagadec, C.; Vergnes, L.; Matsutani, T.; Masui, K.; Poulou, M.; Popescu, R.; Della Donna, L.; Evers, P.; Dekmezian, C.; et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc. Natl. Acad. Sci. USA 2011, 108, 16062–16067. [Google Scholar] [CrossRef]
- Tibbetts, A.S.; Appling, D.R. Compartmentalization of Mammalian Folate-Mediated One-Carbon Metabolism. Annu. Rev. Nutr. 2010, 30, 57–81. [Google Scholar] [CrossRef]
- Villa, E.; Ali, E.S.; Sahu, U.; Ben-Sahra, I. Cancer cells tune the signaling pathways to empower de novo synthesis of nucleotides. Cancers 2019, 11, 688. [Google Scholar] [CrossRef]
- Yin, J.; Ren, W.; Huang, X.; Deng, J.; Li, T.; Yin, Y. Potential Mechanisms Connecting Purine Metabolism and Cancer Therapy. Front. Immunol. 2018, 9, 1697. [Google Scholar] [CrossRef] [PubMed]
- Di Virgilio, F.; Adinolfi, E. Extracellular purines, purinergic receptors and tumor growth. Oncogene 2017, 36, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.Y.; Zhao, H.; Pugh, R.J.; Pedley, A.M.; French, J.; Jones, S.A.; Zhuang, X.; Jinnah, H.; Huan, T.J.; Benkovic, S.J. Purinosome formation as a function of the cell cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 1368–1373. [Google Scholar] [CrossRef]
- An, S.; Kumar, R.; Sheets, E.D.; Benkovic, S.J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 2008, 320, 103–106. [Google Scholar] [CrossRef]
- Pedley, A.M.; Benkovic, S.J. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Hatefi, Y.; Huennekens, F.M.; Kay, L.D. Manometric assay and cofactor requirements for serine hydroxymethylase. J. Biol. Chem. 1957, 224, 435–444. [Google Scholar] [PubMed]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef]
- Hatefi, Y.; Osborn, M.J.; Kay, L.D.; Huennekens, F.M. Hydroxymethyl tetrahydrofolic dehydrogenase. J. Biol. Chem. 1957, 227, 637–647. [Google Scholar] [CrossRef]
- Nilsson, R.; Jain, M.; Madhusudhan, N.; Sheppard, N.G.; Strittmatter, L.; Kampf, C.; Huang, J.; Asplund, A.; Mootha, V.K. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014, 5, 3128. [Google Scholar] [CrossRef]
- Miyo, M.; Konno, M.; Colvin, H.; Nishida, N.; Koseki, J.; Kawamoto, K.; Tsunekuni, K.; Nishimura, J.; Hata, T.; Takemasa, I.; et al. The importance of mitochondrial folate enzymes in human colorectal cancer. Oncol. Rep. 2017, 37, 417–425. [Google Scholar] [CrossRef]
- Fawal, M.A.; Jungas, T.; Davy, A. Inhibition of DHFR targets the self-renewing potential of brain tumor initiating cells. Cancer Lett. 2021, 503, 129–137. [Google Scholar] [CrossRef]
- Zhang, Z.; Shi, Q.; Sturgis, E.M.; Spitz, M.R.; Wei, Q. Polymorphisms and haplotypes of serine hydroxymethyltransferase and risk of squamous cell carcinoma of the head and neck: A case-control analysis. Pharmacogenet. Genom. 2005, 15, 557–564. [Google Scholar] [CrossRef]
- Noguchi, K.; Konno, M.; Koseki, J.; Nishida, N.; Kawamoto, K.; Yamada, D.; Asaoka, T.; Noda, T.; Wada, H.; Gotoh, K.; et al. The mitochondrial one-carbon metabolic pathway is associated with patient survival in pancreatic cancer. Oncol. Lett. 2018, 16, 1827–1834. [Google Scholar] [CrossRef]
- Park, A.K.; Lee, J.Y.; Cheong, H.; Ramaswamy, V.; Park, S.-H.; Kool, M.; Phi, J.H.; Choi, S.A.; Cavalli, F.; Taylor, M.D.; et al. Subgroup-specific prognostic signaling and metabolic pathways in pediatric medulloblastoma. BMC Cancer 2019, 19, 571. [Google Scholar] [CrossRef]
- Liu, F.; Liu, Y.; He, C.; Tao, L.; He, X.; Song, H.; Zhang, G. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumor Biol. 2014, 35, 8685–8690. [Google Scholar] [CrossRef]
- Koseki, J.; Konno, M.; Asai, A.; Colvin, H.; Kawamoto, K.; Nishida, N.; Sakai, D.; Kudo, T.; Satoh, T.; Doki, Y.; et al. Enzymes of the one-carbon folate metabolism as anticancer targets predicted by survival rate analysis. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Nakata, A.; Chen, X.; Nishi, K.; Meguro-Horike, M.; Sasaki, S.; Kita, K.; Horike, S.-i.; Saitoh, K.; Kato, K.; et al. Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2. Oncogene 2019, 38, 2464–2481. [Google Scholar] [CrossRef]
- Ben-Sahra, I.; Hoxhaj, G.; Ricoult, S.J.H.; Asara, J.M.; Manning, B.D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 2016, 351, 728–733. [Google Scholar] [CrossRef]
- Kong, W.; Wang, J. Hypoxanthine Transport in Human Glioblastoma Cells and Effect on Cell Susceptibility to Methotrexate. Pharm. Res. 2003, 20, 1804–1811. [Google Scholar] [CrossRef]
- Coulthard, S.A.; Redfern, C.P.F.; Vikingsson, S.; Lindqvist-Appell, M.; Skoglund, K.; Jakobsen-Falk, I.; Hall, A.G.; Taylor, G.A.; Hogarth, L.A. Increased sensitivity to thiopurines in methylthioadenosine phosphorylase-deleted cancers. Mol. Cancer Ther. 2011, 10, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Tardito, S.; Oudin, A.; Ahmed, S.U.; Fack, F.; Keunen, O.; Zheng, L.; Miletic, H.; Sakariassen, P.Ø.; Weinstock, A.; Wagner, A.; et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat. Cell Biol. 2015, 17, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Baresova, V.; Skopova, V.; Sikora, J.; Patterson, D.; Sovova, J.; Zikanova, M.; Kmoch, S. Mutations of ATIC and ADSL affect purinosome assembly in cultured skin fibroblasts from patients with AICA-ribosiduria and ADSL deficiency. Hum. Mol. Genet. 2012, 21, 1534–1543. [Google Scholar] [CrossRef]
- Mazzarino, R.C.; Baresova, V.; Zikánová, M.; Duval, N.; Wilkinson, T.G.; Patterson, D.; Vacano, G.N. The CRISPR-Cas9 crADSL HeLa transcriptome: A first step in establishing a model for ADSL deficiency and SAICAR accumulation. Mol. Genet. Metab. Rep. 2019, 21, 100512. [Google Scholar] [CrossRef]
- French, J.B.; Zhao, H.; An, S.; Niessen, S.; Deng, Y.; Cravatt, B.F.; Benkovic, S.J. Hsp70/Hsp90 chaperone machinery is involved in the assembly of the purinosome. Proc. Natl. Acad. Sci. USA 2013, 110, 2528–2533. [Google Scholar] [CrossRef]
- Goswami, M.T.; Chen, G.; Chakravarthi, B.V.S.K.; Pathi, S.S.; Anand, S.K.; Carskadon, S.L.; Giordano, T.J.; Chinnaiyan, A.M.; Thomas, D.G.; Palanisamy, N.; et al. Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer. Oncotarget 2015, 6, 23445–23461. [Google Scholar] [CrossRef]
- Kofuji, S.; Hirayama, A.; Eberhardt, A.O.; Kawaguchi, R.; Sugiura, Y.; Sampetrean, O.; Ikeda, Y.; Warren, M.; Sakamoto, N.; Kitahara, S.; et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat. Cell Biol. 2019, 21, 1003–1014. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.W.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yao, Y.; Scott, A.J.; Wilder-Romans, K.; Dresser, J.J.; Werner, C.K.; Sun, H.; Pratt, D.; Sajjakulnukit, P.; Zhao, S.G.; et al. Purine metabolism regulates DNA repair and therapy resistance in glioblastoma. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Shireman, J.M.; Atashi, F.; Lee, G.; Ali, E.S.; Saathoff, M.R.; Park, C.H.; Savchuk, S.; Baisiwala, S.; Miska, J.; Lesniak, M.S.; et al. De novo purine biosynthesis is a major driver of chemoresistance in glioblastoma. Brain 2021. [Google Scholar] [CrossRef]
- Wink, M.R.; Lenz, G.; Braganhol, E.; Tamajusuku, A.S.K.; Schwartsmann, G.; Sarkis, J.J.F.; Battastini, A.M.O. Altered extracellular ATP, ADP and AMP catabolism in glioma cell lines. Cancer Lett. 2003, 198, 211–218. [Google Scholar] [CrossRef]
- Conti, C.R. Adenosine: Clinical pharmacology and applications. Clin. Cardiol. 1991, 14, 91–93. [Google Scholar]
- Allard, B.; Longhi, M.S.; Robson, S.C.; Stagg, J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 2017, 276, 121–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Fridman, A.; Blackledge, W.; Connely, S.; Wilson, I.A.; Pilz, R.; Boss, G.R. The phosphatidylinositol 3-kinase/Akt cassette regulates purine nucleotide synthesis. J. Biol. Chem. 2009, 284, 3521–3528. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Connelly, S.; Jiang, J.; Zhuang, S.; Amador, D.T.; Phan, T.; Pilz, R.B.; Boss, G.R. Akt phosphorylation and regulation of transketolase is a nodal point for amino acid control of purine synthesis. Mol. Cell 2014, 55, 264–276. [Google Scholar] [CrossRef]
- Jin, N.; Bi, A.; Lan, X.; Xu, J.; Wang, X.; Liu, Y.; Wang, T.; Tang, S.; Zeng, H.; Chen, Z.; et al. Identification of metabolic vulnerabilities of receptor tyrosine kinases-driven cancer. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Lehtinen, L.; Ketola, K.; Mäkelä, R.; Mpindi, J.P.; Viitala, M.; Kallioniemi, O.; Iljin, K. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget 2013, 4, 48–63. [Google Scholar] [CrossRef]
- Tanaka, K.; Sasayama, T.; Nagashima, H.; Irino, Y.; Takahashi, M.; Izumi, Y.; Uno, T.; Satoh, N.; Kitta, A.; Kyotani, K.; et al. Glioma cells require one-carbon metabolism to survive glutamine starvation. Acta Neuropathol. Commun. 2021, 9, 16. [Google Scholar] [CrossRef]
- Kollareddy, M.; Dimitrova, E.; Vallabhaneni, K.C.; Chan, A.; Le, T.; Chauhan, K.M.; Carrero, Z.I.; Ramakrishnan, G.; Watabe, K.; Haupt, Y.; et al. Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Racanelli, A.C.; Rothbart, S.B.; Heyer, C.L.; Moran, R.G. Therapeutics by cytotoxic metabolite accumulation: Pemetrexed causes ZMP accumulation, AMPK activation, and mammalian target of rapamycin inhibition. Cancer Res. 2009, 69, 5467–5474. [Google Scholar] [CrossRef]
- Liu, X.; Chhipa, R.R.; Pooya, S.; Wortman, M.; Yachyshin, S.; Chow, L.M.L.; Kumar, A.; Zhou, X.; Sun, Y.; Quinn, B.; et al. Discrete mechanisms of mTOR and cell cycle regulation by AMPK agonists independent of AMPK. Proc. Natl. Acad. Sci. USA 2014, 111, E435–E444. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, B.; Hirota, Y.; Fujii, Y.; Osaka, N.; Ito, D.; Plas, D.R.; Sasaki, A.T. Targeting Energy Metabolism to Overcome Therapeutic Resistance of Glioblastoma and Tumor-associated Edema. In Gliomas; Exon Publications: Brisbane, Australia, 2021; pp. 121–138. [Google Scholar]
- Chhipa, R.R.; Fan, Q.; Anderson, J.; Muraleedharan, R.; Huang, Y.; Ciraolo, G.; Chen, X.; Waclaw, R.; Chow, L.M.; Khuchua, Z.; et al. AMP kinase promotes glioblastoma bioenergetics and tumour growth. Nat. Cell Biol. 2018, 20, 823–835. [Google Scholar] [CrossRef]
- Laderoute, K.R.; Amin, K.; Calaoagan, J.M.; Knapp, M.; Le, T.; Orduna, J.; Foretz, M.; Viollet, B. 5′-AMP-Activated Protein Kinase (AMPK) Is Induced by Low-Oxygen and Glucose Deprivation Conditions Found in Solid-Tumor Microenvironments. Mol. Cell. Biol. 2006, 26, 5336–5347. [Google Scholar] [CrossRef] [PubMed]
- Adjei, A.A. Pharmacology and mechanism of action of pemetrexed. Clin. Lung Cancer 2004, 5, S51–S55. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Moran, R.G.; Goldman, I.D. Pemetrexed: Biochemical and cellular pharmacology, mechanisms, and clinical applications. Mol. Cancer Ther. 2007, 6, 404–417. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, D.L.; Cheng, Y.; Park, J.; An, S. Sequestration-Mediated Downregulation of de Novo Purine Biosynthesis by AMPK. ACS Chem. Biol. 2016, 11, 1917–1924. [Google Scholar] [CrossRef] [PubMed]
- Asai, A.; Konno, M.; Koseki, J.; Taniguchi, M.; Vecchione, A.; Ishii, H. One-carbon metabolism for cancer diagnostic and therapeutic approaches. Cancer Lett. 2020, 470, 141–148. [Google Scholar] [CrossRef]
- Puig, L. Methotrexate: New Therapeutic Approaches. Actas Dermo-Sifiliográficas English Ed. 2014, 105, 583–589. [Google Scholar] [CrossRef]
- Maksimovic, V.; Pavlovic-Popovic, Z.; Vukmirovic, S.; Cvejic, J.; Mooranian, A.; Al-Salami, H.; Mikov, M.; Golocorbin-Kon, S. Molecular mechanism of action and pharmacokinetic properties of methotrexate. Mol. Biol. Rep. 2020, 47, 4699–4708. [Google Scholar] [CrossRef]
- Walling, J. From methotrexate to pemetrexed and beyond. A review of the pharmacodynamic and clinical properties of antifolates. Invest. New Drugs 2006, 24, 37–77. [Google Scholar] [CrossRef]
- Palanichamy, K.; Thirumoorthy, K.; Kanji, S.; Gordon, N.; Singh, R.; Jacob, J.R.; Sebastian, N.; Litzenberg, K.T.; Patel, D.; Bassett, E.; et al. Methionine and kynurenine activate oncogenic kinases in glioblastoma, and methionine deprivation compromises proliferation. Clin. Cancer Res. 2016, 22, 3513–3523. [Google Scholar] [CrossRef]
- Hori, H.; Tran, P.; Carrera, C.J.; Hori, Y.; Rosenbach, M.D.; Carson, D.A.; Nobori, T. Methylthioadenosine phosphorylase cDNA transfection alters sensitivity to depletion of purine and methionine in A549 lung cancer cells. Cancer Res. 1996, 56, 5653–5658. [Google Scholar]
- de Menezes, W.P.; Silva, V.A.O.; Gomes, I.N.F.; Rosa, M.N.; Spina, M.L.C.; Carloni, A.C.; Alves, A.L.V.; Melendez, M.; Almeida, G.C.; da Silva, L.S.; et al. Loss of 5′-Methylthioadenosine Phosphorylase (MTAP) is Frequent in High-Grade Gliomas; Nevertheless, it is Not Associated with Higher Tumor Aggressiveness. Cells 2020, 9, 492. [Google Scholar] [CrossRef] [PubMed]
- Li, W.W.; Su, D.; Mizobuchi, H.; Martin, D.S.; Gu, B.; Gorlick, R.; Cole, P.; Bertino, J.R. Status of methylthioadenosine phosphorylase and its impact on cellular response to L-alanosine and methylmercaptopurine riboside in human soft tissue sarcoma cells. Oncol. Res. 2004, 14, 373–379. [Google Scholar] [CrossRef]
- Karikari, C.A.; Mullendore, M.; Eshleman, J.R.; Argani, P.; Leoni, L.M.; Chattopadhyay, S.; Hidalgo, M.; Maitra, A. Homozygous deletions of methylthioadenosine phosphorylase in human biliary tract cancers. Mol. Cancer Ther. 2005, 4, 1860–1866. [Google Scholar] [CrossRef]
- Batova, A.; Cottam, H.; Yu, J.; Diccianni, M.B.; Carrera, C.J.; Yu, A.L. EFA(9-β-D-erythrofuranosyladenine) is an effective salvage agent for methylthioadenosine phosphorylase-selective therapy of T-cell acute lymphoblastic leukemia with L-alanosine. Blood 2006, 107, 898–903. [Google Scholar] [CrossRef]
- Hansen, L.J.; Sun, R.; Yang, R.; Singh, S.X.; Chen, L.H.; Pirozzi, C.J.; Moure, C.J.; Hemphill, C.; Carpenter, A.B.; Healy, P.; et al. MTAP loss promotes stemness in glioblastoma and confers unique susceptibility to purine starvation. Cancer Res. 2019, 79, 3383–3394. [Google Scholar] [CrossRef] [PubMed]
- National Library of Medicine (U.S.) Mycophenolate Mofetil Combined With Radiation Therapy in Recurrent Glioblastoma. Available online: https://www.clinicaltrials.gov/ct2/show/NCT04477200 (accessed on 10 May 2021).
- Asai, A.; Koseki, J.; Konno, M.; Nishimura, T.; Gotoh, N.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H. Drug discovery of anticancer drugs targeting methylenetetrahydrofolate dehydrogenase 2. Heliyon 2018, 4, e01021. [Google Scholar] [CrossRef] [PubMed]
- Ducker, G.S.; Ghergurovich, J.M.; Mainolfi, N.; Suri, V.; Jeong, S.K.; Hsin-Jung Li, S.; Friedman, A.; Manfredi, M.G.; Gitai, Z.; Kim, H.; et al. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 2017, 114, 11404–11409. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Tan, B.; Dai, X.; Shao, Y.; He, Q.; Yang, B.; Wang, J.; Weng, Q. DHFR/TYMS are positive regulators of glioma cell growth and modulate chemo-sensitivity to temozolomide. Eur. J. Pharmacol. 2019, 863. [Google Scholar] [CrossRef] [PubMed]
- Kosti, A.; De Araujo, P.R.; Li, W.Q.; Guardia, G.D.A.; Chiou, J.; Yi, C.; Ray, D.; Meliso, F.; Li, Y.M.; Delambre, T.; et al. The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation. Genome Biol. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Niechi, I.; Uribe-Ojeda, A.; Erices, J.I.; Torres, Á.; Uribe, D.; Rocha, J.D.; Silva, P.; Richter, H.G.; San Martín, R.; Quezada, C. Adenosine Depletion as A New Strategy to Decrease Glioblastoma Stem-Like Cells Aggressiveness. Cells 2019, 8, 1353. [Google Scholar] [CrossRef]
| Metabolic Enzyme | Implication | Cancer Type/Cell Type | Reference |
|---|---|---|---|
| MTHFD2 | Cell growth and tumourigenesis; knockdown of MTHFD2 resulted in reduced cell growth and Ki-67 staining | Lung adenocarcinoma | [66] |
| MTHFD2 | Cell migration and invasion; overexpression associated with poor prognosis and increased metastasis | Breast cancer | [85] |
| MTHFD2 | Cell growth and survival; metabolic adaptation to glutamine starvation | Glioblastoma | [86] |
| DHFR, SHMT1, MTHFD1 | Tumour sphere formation, methionine dependency, and stem-like phenotype | Glioblastoma | [31] |
| MTHFD2 | Highly overexpressed; overexpression associated with poor prognosis | Various cancer types | [58] |
| SHMT2 | Polymorphisms associated with increased risk of cancer | Squamous cell carcinoma of the head and neck | [61] |
| SHMT2, MTHFD2, MTHFD1 | Overexpressed and associated with increased proliferation; associated with increased mortality in breast cancer | Various cancer types | [56] |
| MTHFD2, SHMT2, ALDH1L2 1 | Overexpressed; overexpression associated with poor prognosis | Colorectal cancer | [59] |
| MTHFD2 and SHMT2, ALDH1L2 | High expression associated with lower overall survival and shorter progression free survival | Pancreatic cancer | [62] |
| DHFR, TYMS, MTHFD2 | Overexpression associated with poor prognosis | Group 4 Medulloblastoma | [63] |
| PPAT, PAICS | Overexpressed; overexpression associated with aneuploidy and gene amplification in subgroup of patients | Lung adenocarcinoma | [74] |
| DHFR, TYMS, MTHFD2 | Tumourigenesis; overexpression associated with poor prognosis | Brain tumour initiating cells | [32] |
| IMPDH2 | Cell proliferation and tumourigenesis; overexpression associated with poor prognosis | Glioblastoma | [75] |
| IMPDH2 | Chemoresistance | Glioblastoma | [78] |
| IMPDH2 | Resistant to radiotherapy | Glioblastoma | [77] |
| Chemical Compound/Drug | Metabolic Target | Reference |
|---|---|---|
| Mycophenolate Mofetil | IMPDH2 | [107] |
| Mycophenolate Mofetil | IMPDH2; Purine synthesis | [78] |
| Mycophenolate Mofetil | IMPDH2; Purine synthesis | [77] |
| Methotrexate | DHFR; Folate-mediated 1-C metabolism | [60] |
| Pemetrexed | DHFR, TYMS; Folate-mediated 1-C metabolism, nucleotide synthesis | [110] |
| siRNA-mediated knockdown SERBP1 1 | SERBP1 Methionine synthesis and 1-C metabolism | [111] |
| siRNA-mediated knockdown of MTHFD2 | MTHFD2; Purine synthesis | [86] |
| L-Alanosine | ADSS; Purine synthesis | [106] |
| Adenosine Deaminase | Adenosine synthesis | [112] |
| shRNA-mediated knockdown of PRPS1 2, GMPS and ADSL | De novo purine synthesis enzymes | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghannad-Zadeh, K.; Das, S. One-Carbon Metabolism Associated Vulnerabilities in Glioblastoma: A Review. Cancers 2021, 13, 3067. https://doi.org/10.3390/cancers13123067
Ghannad-Zadeh K, Das S. One-Carbon Metabolism Associated Vulnerabilities in Glioblastoma: A Review. Cancers. 2021; 13(12):3067. https://doi.org/10.3390/cancers13123067
Chicago/Turabian StyleGhannad-Zadeh, Kimia, and Sunit Das. 2021. "One-Carbon Metabolism Associated Vulnerabilities in Glioblastoma: A Review" Cancers 13, no. 12: 3067. https://doi.org/10.3390/cancers13123067
APA StyleGhannad-Zadeh, K., & Das, S. (2021). One-Carbon Metabolism Associated Vulnerabilities in Glioblastoma: A Review. Cancers, 13(12), 3067. https://doi.org/10.3390/cancers13123067







