Nectin-1 Expression Correlates with the Susceptibility of Malignant Melanoma to Oncolytic Herpes Simplex Virus In Vitro and In Vivo
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Generation of Viral Stocks
2.3. Infection, Cell Viability, and Toxicity Experiments
2.4. FACS Analysis
2.5. Western Blot
2.6. Biopsies
2.7. Immunohistochemistry (IHC)
2.8. CRISPR/Cas9 Knockout
2.9. HSV-1 DNA qPCR
2.10. Statistics
3. Results
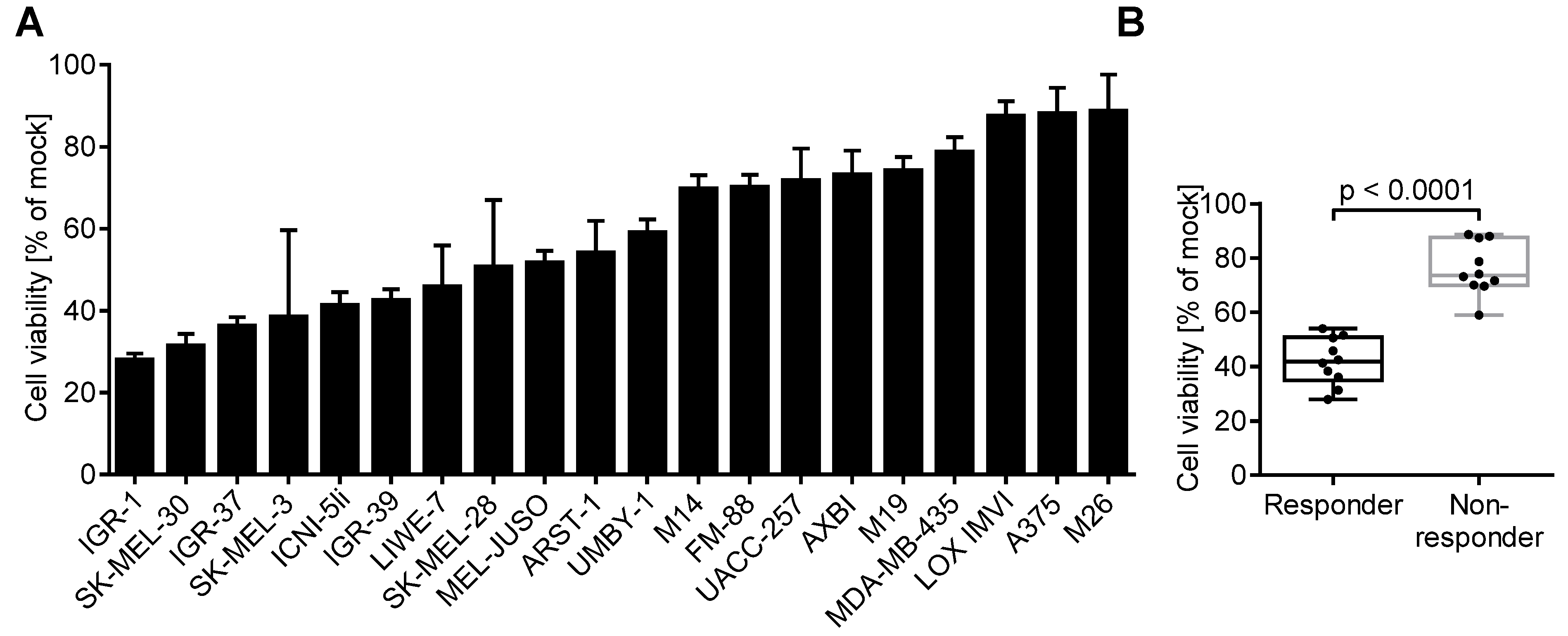
3.1. Melanoma Cell Lines Differ in Their Susceptibility to Herpes Virus-Induced Oncolysis
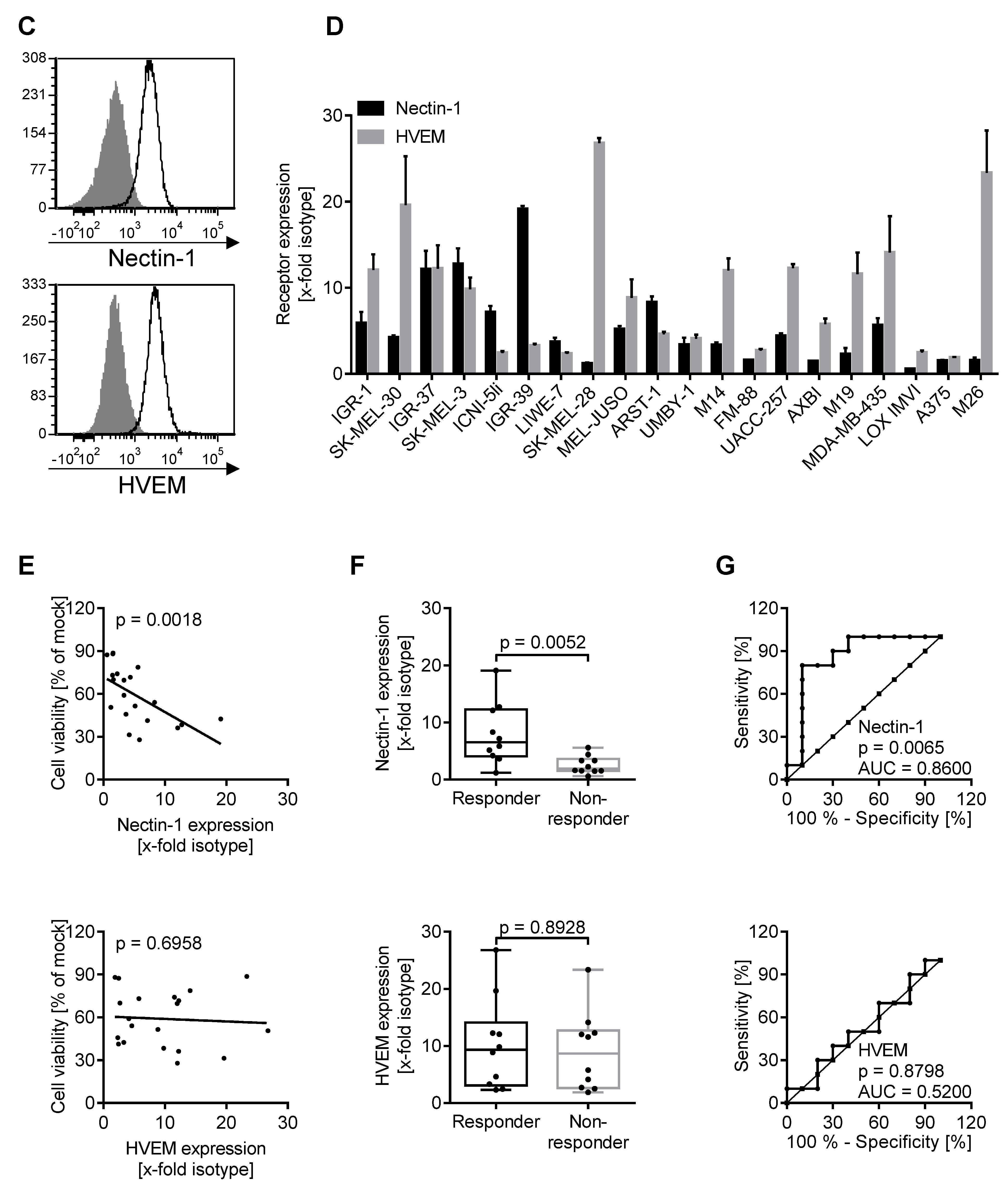
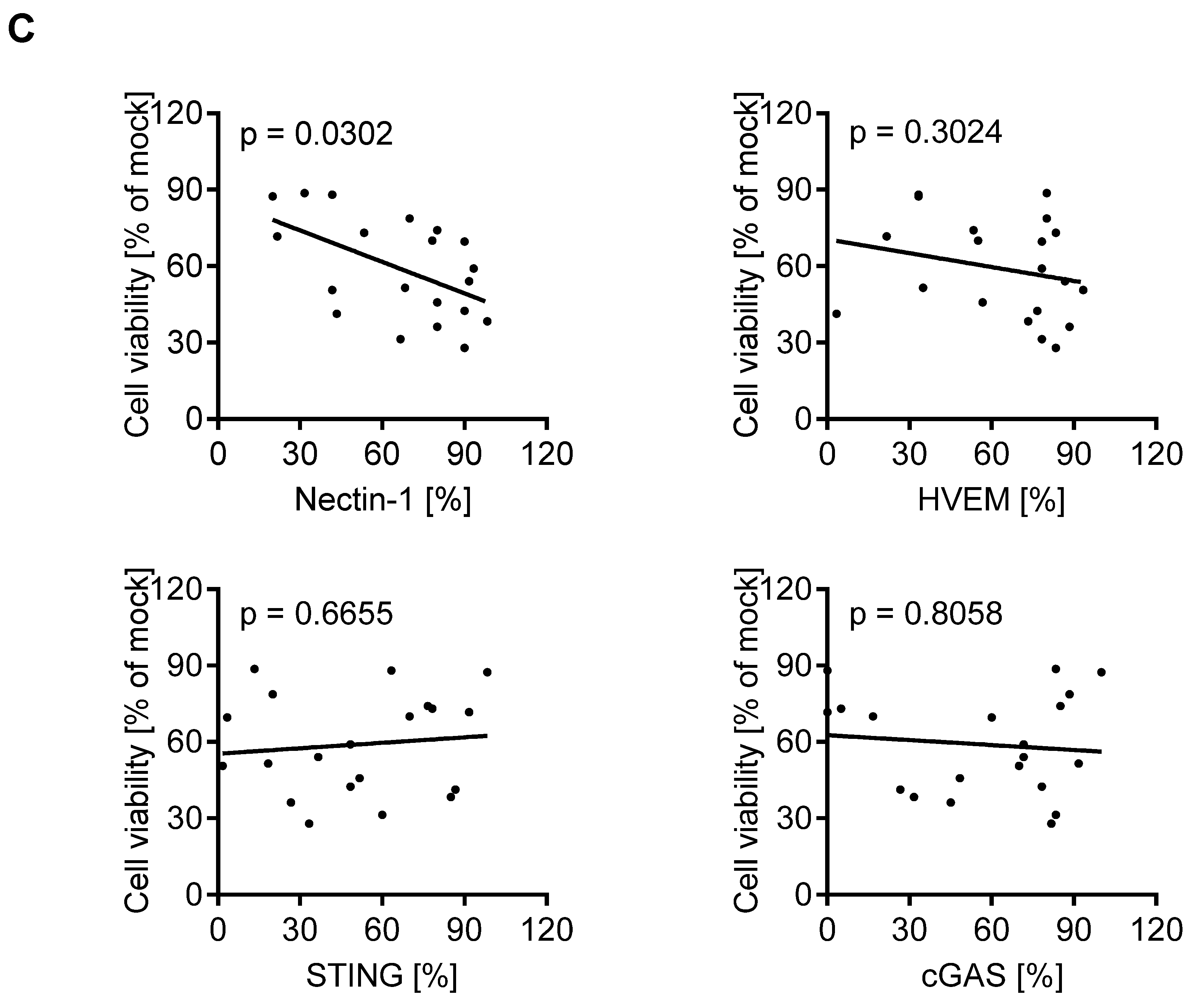
3.2. Oncolytic Activity of T-VEC Correlates with Nectin-1 Expression In Vitro
3.3. Nectin-1 Knockout Mediates Resistance to Viral Oncolysis
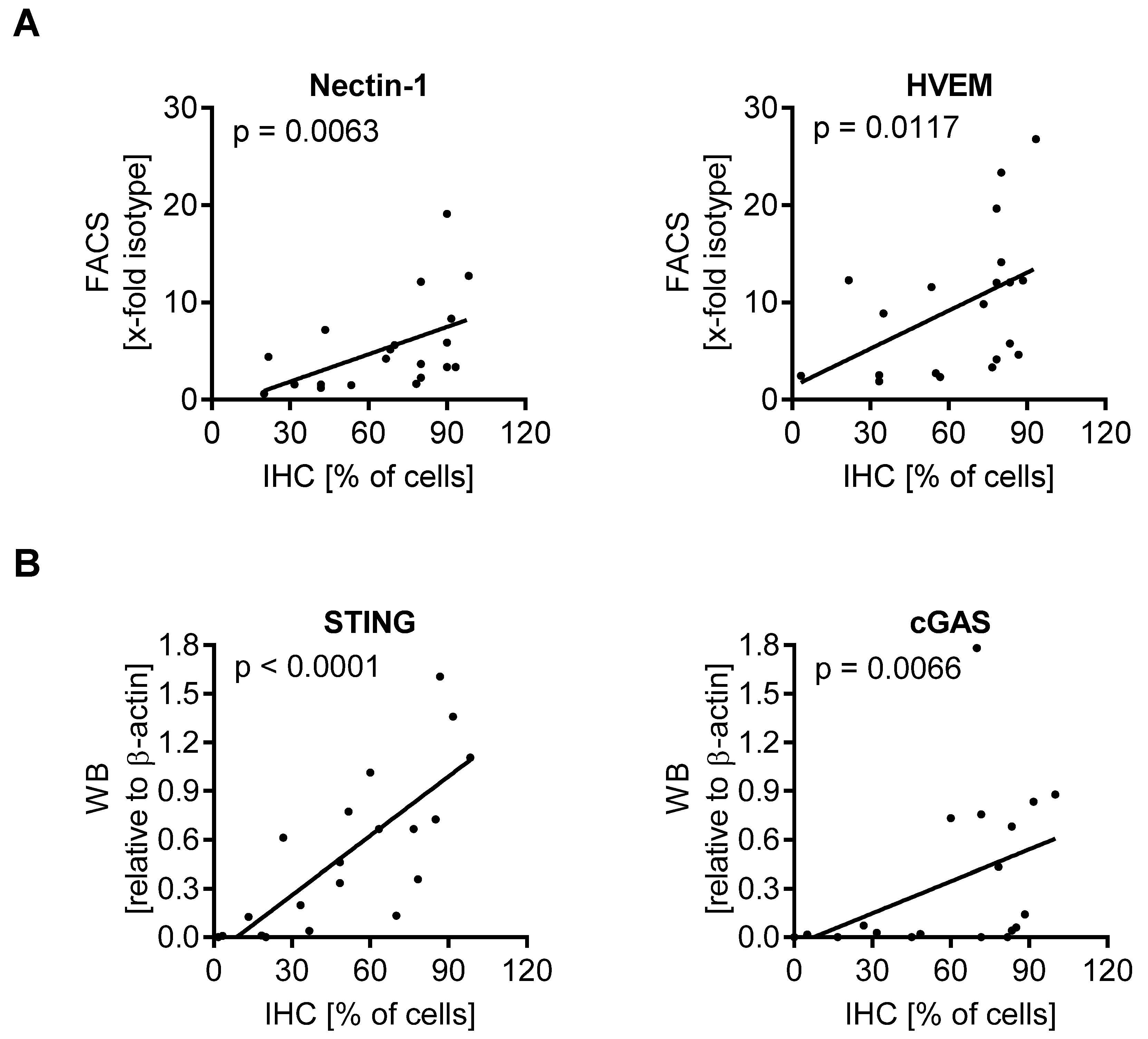
3.4. cGAS/STING Play a Subordinate Role for the Oncolytic Activity of T-VEC
3.5. Expression of Biomarkers Is Consistent between Different Methodological Approaches
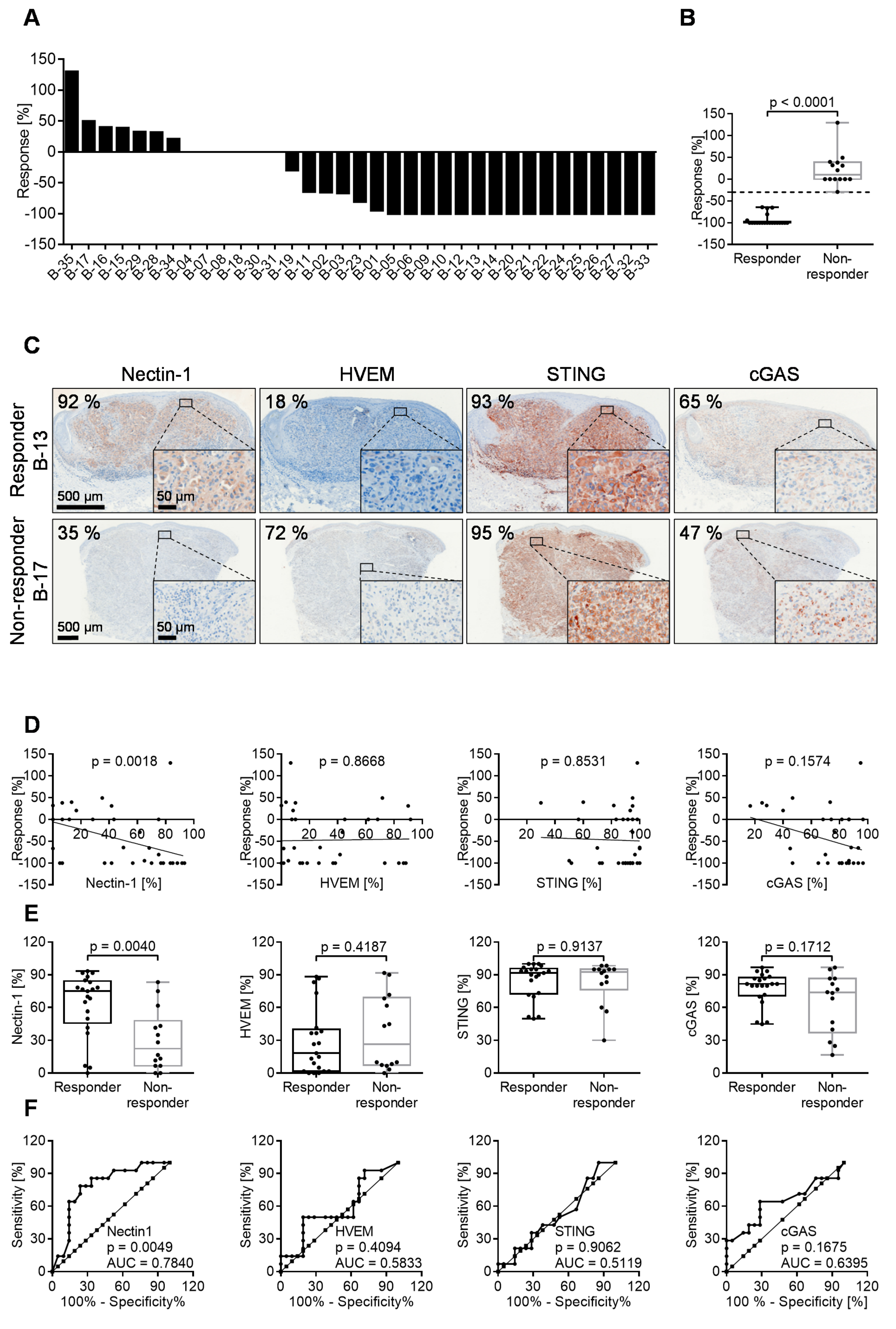
3.6. 60% of Melanoma Lesions in Patients Respond to Intratumoral T-VEC Injection
3.7. Oncolytic Activity of T-VEC Correlates with Nectin-1 Expression In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, B.L.; Robinson, M.; Han, Z.Q.; Branston, R.H.; English, C.; Reay, P.; McGrath, Y.; Thomas, S.K.; Thornton, M.; Bullock, P.; et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003, 10, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Cassady, K.A.; Gross, M.; Roizman, B. The second-site mutation in the herpes simplex virus recombinants lacking the gamma134.5 genes precludes shutoff of protein synthesis by blocking the phosphorylation of eIF-2alpha. J. Virol. 1998, 72, 7005–7011. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, K.; Chen, W.; Johnson, D.C.; Hendricks, R.L. Infected cell protein (ICP)47 enhances herpes simplex virus neurovirulence by blocking the CD8+ T cell response. J. Exp. Med. 1998, 187, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Fruh, K.; Ahn, K.; Djaballah, H.; Sempe, P.; van Endert, P.M.; Tampe, R.; Peterson, P.A.; Yang, Y. A viral inhibitor of peptide transporters for antigen presentation. Nature 1995, 375, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, H.L.; Kim, D.W.; DeRaffele, G.; Mitcham, J.; Coffin, R.S.; Kim-Schulze, S. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann. Surg. Oncol. 2010, 17, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Andtbacka, R.H.I.; Collichio, F.; Harrington, K.J.; Middleton, M.R.; Downey, G.; Öhrling, K.; Kaufman, H.L. Final analyses of OPTiM: A randomized phase III trial of talimogene laherparepvec versus granulocyte-macrophage colony-stimulating factor in unresectable stage III-IV melanoma. J. Immunother. Cancer 2019, 7, 145. [Google Scholar] [CrossRef]
- Ribas, A.; Dummer, R.; Puzanov, I.; VanderWalde, A.; Andtbacka, R.H.I.; Michielin, O.; Olszanski, A.J.; Malvehy, J.; Cebon, J.; Fernandez, E.; et al. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy. Cell 2017, 170, 1109–1119 e10. [Google Scholar] [CrossRef]
- Bommareddy, P.K.; Shettigar, M.; Kaufman, H.L. Integrating oncolytic viruses in combination cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 498–513. [Google Scholar] [CrossRef]
- Xia, T.; Konno, H.; Barber, G.N. Recurrent Loss of STING Signaling in Melanoma Correlates with Susceptibility to Viral Oncolysis. Cancer Res. 2016, 76, 6747–6759. [Google Scholar] [CrossRef]
- Friedman, G.K.; Bernstock, J.D.; Chen, D.; Nan, L.; Moore, B.P.; Kelly, V.M.; Youngblood, S.L.; Langford, C.P.; Han, X.; Ring, E.K.; et al. Enhanced Sensitivity of Patient-Derived Pediatric High-Grade Brain Tumor Xenografts to Oncolytic HSV-1 Virotherapy Correlates with Nectin-1 Expression. Sci. Rep. 2018, 8, 13930. [Google Scholar] [CrossRef]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Houben, R.; Hesbacher, S.; Schmid, C.P.; Kauczok, C.S.; Flohr, U.; Haferkamp, S.; Muller, C.S.; Schrama, D.; Wischhusen, J.; Becker, J.C. High-level expression of wild-type p53 in melanoma cells is frequently associated with inactivity in p53 reporter gene assays. PLoS ONE 2011, 6, e22096. [Google Scholar] [CrossRef]
- Thomann, S.; Boscheinen, J.B.; Vogel, K.; Knipe, D.M.; DeLuca, N.; Gross, S.; Schuler-Thurner, B.; Schuster, P.; Schmidt, B. Combined cytotoxic activity of an infectious, but non-replicative herpes simplex virus type 1 and plasmacytoid dendritic cells against tumour cells. Immunology 2015, 146, 327–338. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Koh, C.M. Chapter Eighteen—Preparation of Cells for Microscopy using ‘Cell Blocks’. In Methods in Enzymology; Lorsch, J., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 533, pp. 249–255. [Google Scholar]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Boscheinen, J.B.; Thomann, S.; Knipe, D.M.; DeLuca, N.; Schuler-Thurner, B.; Gross, S.; Dorrie, J.; Schaft, N.; Bach, C.; Rohrhofer, A.; et al. Generation of an Oncolytic Herpes Simplex Virus 1 Expressing Human MelanA. Front. Immunol. 2019, 10, 2. [Google Scholar] [CrossRef]
- Mashal, R.D.; Koontz, J.; Sklar, J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat. Genet. 1995, 9, 177–183. [Google Scholar] [CrossRef]
- Vouillot, L.; Thélie, A.; Pollet, N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3: Genes Genomes Genet. 2015, 5, 407–415. [Google Scholar] [CrossRef]
- Ablasser, A.; Goldeck, M.; Cavlar, T.; Deimling, T.; Witte, G.; Rohl, I.; Hopfner, K.P.; Ludwig, J.; Hornung, V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013, 498, 380–384. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Thomas, S.; Kuncheria, L.; Roulstone, V.; Kyula, J.N.; Mansfield, D.; Bommareddy, P.K.; Smith, H.; Kaufman, H.L.; Harrington, K.J.; Coffin, R.S. Development of a new fusion-enhanced oncolytic immunotherapy platform based on herpes simplex virus type 1. J. Immunother. Cancer 2019, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Geraghty, R.J.; Krummenacher, C.; Cohen, G.H.; Eisenberg, R.J.; Spear, P.G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 1998, 280, 1618–1620. [Google Scholar] [CrossRef] [PubMed]
- Kopp, S.J.; Banisadr, G.; Glajch, K.; Maurer, U.E.; Grünewald, K.; Miller, R.J.; Osten, P.; Spear, P.G. Infection of neurons and encephalitis after intracranial inoculation of herpes simplex virus requires the entry receptor nectin-1. Proc. Natl. Acad. Sci. USA 2009, 106, 17916–17920. [Google Scholar] [CrossRef] [PubMed]
- Karaba, A.H.; Kopp, S.J.; Longnecker, R. Herpesvirus entry mediator and nectin-1 mediate herpes simplex virus 1 infection of the murine cornea. J. Virol. 2011, 85, 10041–10047. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Y.; Yu, Z.; Lin, S.F.; Li, S.; Fong, Y.; Wong, R.J. Nectin-1 is a marker of thyroid cancer sensitivity to herpes oncolytic therapy. J. Clin. Endocrinol. Metab. 2007, 92, 1965–1970. [Google Scholar] [CrossRef]
- Yu, Z.; Adusumilli, P.S.; Eisenberg, D.P.; Darr, E.; Ghossein, R.A.; Li, S.; Liu, S.; Singh, B.; Shah, J.P.; Fong, Y.; et al. Nectin-1 expression by squamous cell carcinoma is a predictor of herpes oncolytic sensitivity. Mol. Ther. 2007, 15, 103–113. [Google Scholar] [CrossRef]
- Schuster, P.; Lindner, G.; Thomann, S.; Haferkamp, S.; Schmidt, B. Prospect of Plasmacytoid Dendritic Cells in Enhancing Anti-Tumor Immunity of Oncolytic Herpes Viruses. Cancers 2019, 11, 651. [Google Scholar] [CrossRef]
- Montgomery, R.I.; Warner, M.S.; Lum, B.J.; Spear, P.G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 1996, 87, 427–436. [Google Scholar] [CrossRef]
- Krummenacher, C.; Nicola, A.V.; Whitbeck, J.C.; Lou, H.; Hou, W.; Lambris, J.D.; Geraghty, R.J.; Spear, P.G.; Cohen, G.H.; Eisenberg, R.J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J. Virol. 1998, 72, 7064–7074. [Google Scholar] [CrossRef]
- Krummenacher, C.; Baribaud, F.; Ponce de Leon, M.; Baribaud, I.; Whitbeck, J.C.; Xu, R.; Cohen, G.H.; Eisenberg, R.J. Comparative usage of herpesvirus entry mediator A and nectin-1 by laboratory strains and clinical isolates of herpes simplex virus. Virology 2004, 322, 286–299. [Google Scholar] [CrossRef]
- Petermann, P.; Rahn, E.; Thier, K.; Hsu, M.J.; Rixon, F.J.; Kopp, S.J.; Knebel-Morsdorf, D. Role of Nectin-1 and Herpesvirus Entry Mediator as Cellular Receptors for Herpes Simplex Virus 1 on Primary Murine Dermal Fibroblasts. J. Virol. 2015, 89, 9407–9416. [Google Scholar] [CrossRef][Green Version]
- Sciortino, M.T.; Medici, M.A.; Marino-Merlo, F.; Zaccaria, D.; Giuffre-Cuculletto, M.; Venuti, A.; Grelli, S.; Bramanti, P.; Mastino, A. Involvement of gD/HVEM interaction in NF-kB-dependent inhibition of apoptosis by HSV-1 gD. Biochem. Pharmacol. 2008, 76, 1522–1532. [Google Scholar] [CrossRef]
- Cheung, T.C.; Steinberg, M.W.; Oborne, L.M.; Macauley, M.G.; Fukuyama, S.; Sanjo, H.; D’Souza, C.; Norris, P.S.; Pfeffer, K.; Murphy, K.M.; et al. Unconventional ligand activation of herpesvirus entry mediator signals cell survival. Proc. Natl. Acad. Sci. USA 2009, 106, 6244–6249. [Google Scholar] [CrossRef]
- Yu, X.; He, S. The interplay between human herpes simplex virus infection and the apoptosis and necroptosis cell death pathways. Virol. J. 2016, 13, 77. [Google Scholar] [CrossRef]
- Bommareddy, P.K.; Zloza, A.; Rabkin, S.D.; Kaufman, H.L. Oncolytic virus immunotherapy induces immunogenic cell death and overcomes STING deficiency in melanoma. Oncoimmunology 2019, 8, 1591875. [Google Scholar] [CrossRef]
- De Queiroz, N.; Xia, T.; Konno, H.; Barber, G.N. Ovarian Cancer Cells Commonly Exhibit Defective STING Signaling Which Affects Sensitivity to Viral Oncolysis. Mol. Cancer Res. 2019, 17, 974–986. [Google Scholar] [CrossRef]
- Ma, Z.; Damania, B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016, 19, 150–158. [Google Scholar] [CrossRef]
- Gehrke, N.; Mertens, C.; Zillinger, T.; Wenzel, J.; Bald, T.; Zahn, S.; Tuting, T.; Hartmann, G.; Barchet, W. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 2013, 39, 482–495. [Google Scholar] [CrossRef]
- Brunnstrom, H.; Johansson, A.; Westbom-Fremer, S.; Backman, M.; Djureinovic, D.; Patthey, A.; Isaksson-Mettavainio, M.; Gulyas, M.; Micke, P. PD-L1 immunohistochemistry in clinical diagnostics of lung cancer: Inter-pathologist variability is higher than assay variability. Mod. Pathol. 2017, 30, 1411–1421. [Google Scholar] [CrossRef]


| Characteristics | Patients (n = 21) |
|---|---|
| Sex, n (%) | |
| Female | 11 (53) |
| Male | 10 (48) |
| Median age, years (range) | 72 (56–91) |
| ECOG performance status, n (%) | |
| 0 | 19 (86) |
| 1 | 3 (14) |
| Disease substage, n (%) | |
| IIIB | 2 (10) |
| IIIC | 19 (90) |
| LDH, n (%) | |
| ≤ULN | 21 (100) |
| >ULN | 0 |
| Prior anticancer therapy, n (%) | |
| Interferon | 4 (19) |
| No prior therapy | 17 (81) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwertner, B.; Lindner, G.; Toledo Stauner, C.; Klapproth, E.; Magnus, C.; Rohrhofer, A.; Gross, S.; Schuler-Thurner, B.; Öttl, V.; Feichtgruber, N.; et al. Nectin-1 Expression Correlates with the Susceptibility of Malignant Melanoma to Oncolytic Herpes Simplex Virus In Vitro and In Vivo. Cancers 2021, 13, 3058. https://doi.org/10.3390/cancers13123058
Schwertner B, Lindner G, Toledo Stauner C, Klapproth E, Magnus C, Rohrhofer A, Gross S, Schuler-Thurner B, Öttl V, Feichtgruber N, et al. Nectin-1 Expression Correlates with the Susceptibility of Malignant Melanoma to Oncolytic Herpes Simplex Virus In Vitro and In Vivo. Cancers. 2021; 13(12):3058. https://doi.org/10.3390/cancers13123058
Chicago/Turabian StyleSchwertner, Barbara, Georg Lindner, Camila Toledo Stauner, Elisa Klapproth, Clara Magnus, Anette Rohrhofer, Stefanie Gross, Beatrice Schuler-Thurner, Veronika Öttl, Nicole Feichtgruber, and et al. 2021. "Nectin-1 Expression Correlates with the Susceptibility of Malignant Melanoma to Oncolytic Herpes Simplex Virus In Vitro and In Vivo" Cancers 13, no. 12: 3058. https://doi.org/10.3390/cancers13123058
APA StyleSchwertner, B., Lindner, G., Toledo Stauner, C., Klapproth, E., Magnus, C., Rohrhofer, A., Gross, S., Schuler-Thurner, B., Öttl, V., Feichtgruber, N., Drexler, K., Evert, K., Krahn, M. P., Berneburg, M., Schmidt, B., Schuster, P., & Haferkamp, S. (2021). Nectin-1 Expression Correlates with the Susceptibility of Malignant Melanoma to Oncolytic Herpes Simplex Virus In Vitro and In Vivo. Cancers, 13(12), 3058. https://doi.org/10.3390/cancers13123058






