Albumin-Based Nanoparticles for the Delivery of Doxorubicin in Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
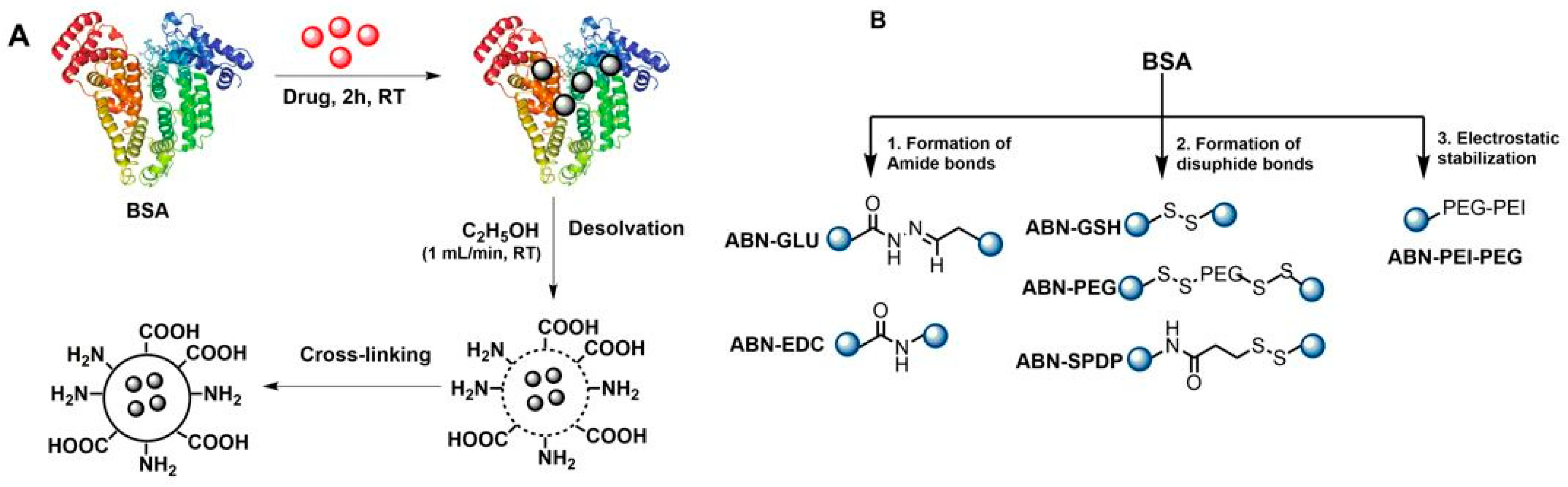
2.2. Preparation of BSA Nanoparticles (ABNs)
2.2.1. Formation of Amide Bonds
- (a)
- Use of glutaraldehyde: For the preparation of nanoparticles using GLU (ABN-GLU), 20 mg/mL of BSA in an aqueous solution was incubated with 0.5 mg/mL doxorubicin Hcl for 2 h at room temperature. To this solution, 2.7 mL of anhydrous ethanol was added dropwise with the syringe pump at the constant flow rate of 1 mL/min. After the solution became turbid, 7 µL of 8% glutaraldehyde was added for cross-linking. The solution was stirred at 550 rpm for 18 h. Then, the free albumin, unbound Dox, ethanol, and excess glutaraldehyde, were removed by 3 cycles of centrifugations at 13,200 rpm for 15 min. After each centrifugation cycle, the pellets were redispersed in 1 mL of water.
- (b)
- Use of EDC: For the preparation of ABNs using EDC (ABN-EDC), a freshly prepared aqueous solution of EDC (2 mg/mL) was added to the turbid solution of BSA after the desolvation process. The mixture was left rotating at 550 rpm for 3 h and purified by three cycles of centrifugation to remove the unreacted EDC and ethanol.
2.2.2. Formation of Disulfide Bonds
- (a)
- Use of glutathione: Firstly, the intramolecular disulfide bonds in albumin were cleaved by using glutathione (GSH), which is one of the major endogenous antioxidants in vivo. After the pre-treatment, it was purified using NAP-10 column and incubated with 0.5 mg/mL doxorubicin Hcl for 2 h, followed by desolvation with ethanol to precipitate albumin into Dox-loaded ABNs (ABN-GSH). Then, ethanol and unbound Dox were removed through centrifugation at 13,200 rpm for 15 min.
- (b)
- Use of N-succinimidyl 3-(2-pyridyldithio) propionate (SPDP): The schematic representation for the preparation of ABNs by using SPDP (ABN-SPDP) is depicted in Figure S1. Firstly, thiol groups were introduced in BSA with 2-iminothiolane, commonly known as Traut’s reagent. In parallel, the same amount of BSA was modified with SPDP. The modified BSAs were purified with NAP-10 column and the resulting solutions combined. The mixture was incubated with DOX for 2 h, followed by preparation of NPs by desolvation method as described earlier in Section 2.2.1.
- (c)
- Use of modified polyethylene glycol (PEG): The schematic representation of the synthesis of this derivative is provided in SI 2. The preparation of this modified PEG is as follows: to a solution of PEG(NH2)2 (3000 M.W.) (1) (100 mg, 33 µmol) in THF (6 mL) at 0 °C, a solution of SPDP (41.5 mg, 0.13 mmol) in THF (3 mL) was added. The reaction was allowed to warm up to room temperature under vigorous stirring for 16 h. Then, the solvent was removed under vacuum and re-dissolved in methanol (3 mL). The product was purified by dialysis using a 3.5 KDa. dialysis membrane for 16 h at 4 °C against distilled water. After this time, the solution turned cloudy, and the solvent was removed under vacuum. The desired product (2) was isolated as a greyish oil (44% of yield, 48.3 mg) (linker/polymer ratio 2:1). The product was characterized by NMR and MS (Figure S3). 1H NMR (D2O, 400 MHz): δ 8.26 (d, 2H), 7.71 (m, 4H), 7.17 (td, 2H), 3.56 (m, 264H), 3.21 (t, 4H), 2.93 (t, 4H). 13C NMR (D2O, 101 MHz): δ 173.74, 158.67, 149.03, 138.61, 121.69, 120.08, 38.83, 34.37, 33.56. MS (MALDI): theoretical mass: 3296.97, calculated mass: 3295.8.
2.2.3. Electrostatic Stabilization
2.3. Surface Charge and Size Characterization of NPs
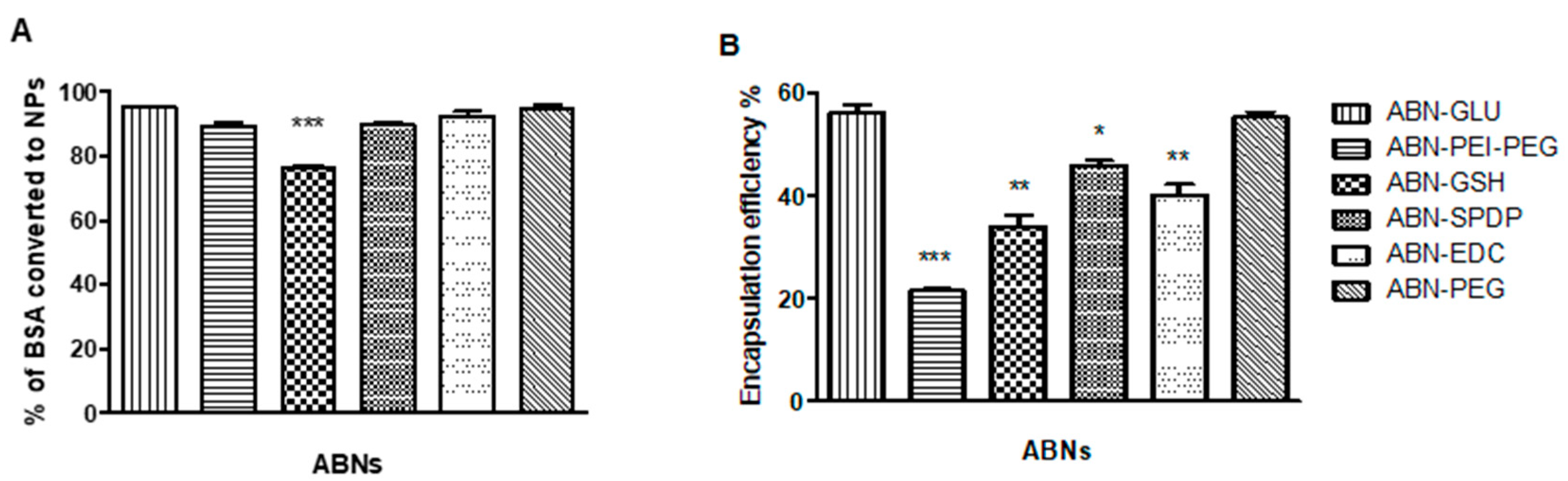
2.4. Quantification of Nanoparticle Formation
2.5. Drug Loading of NPs
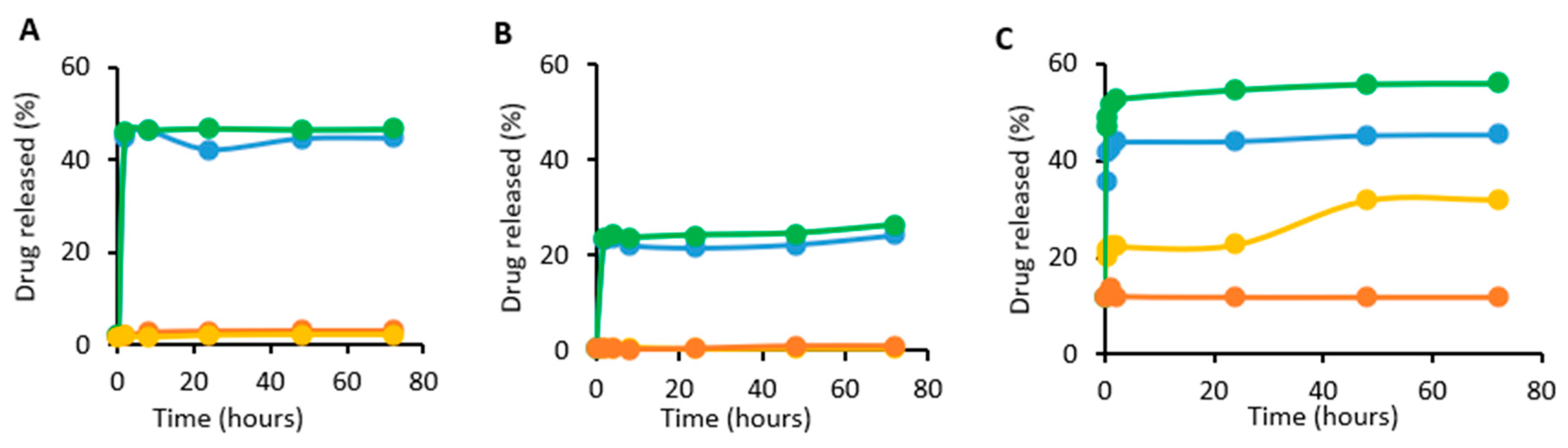
2.6. In Vitro Release Studies
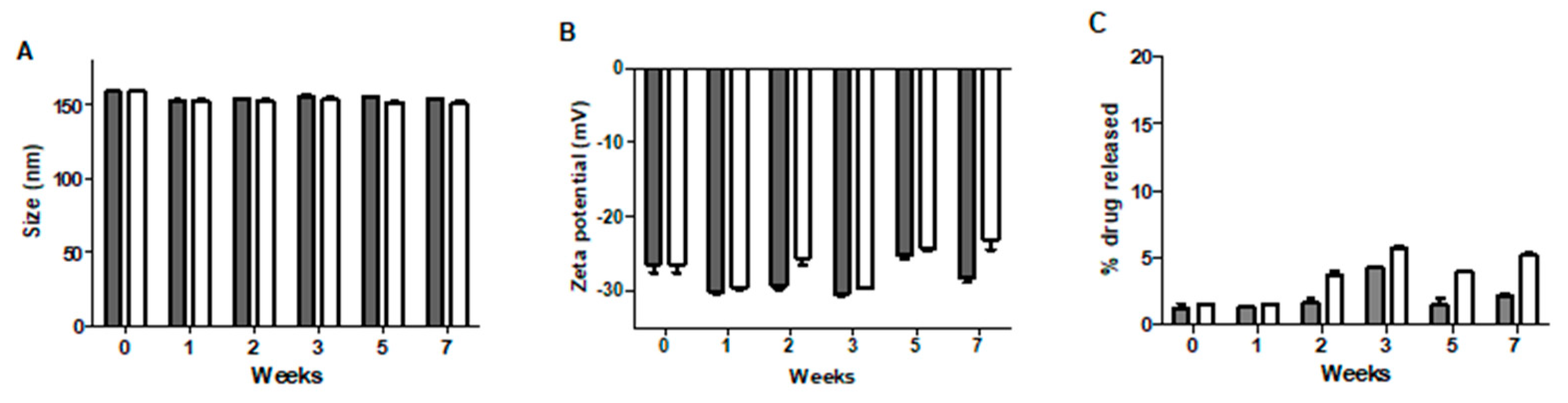
2.7. Stability Studies
2.8. Cell Culture
2.8.1. Cytotoxicity Assay
2.8.2. In Vitro Transfection
2.8.3. Determination of Cell Cycle Phase
2.8.4. Determination of Induction of Apoptosis/Necrosis
2.8.5. Study of Mechanism of Internalization
2.8.6. Western Blot Analysis
2.9. Statistical Analysis
3. Results
3.1. Preparation and Characterization of ABNs
3.2. Size and Surface Charge Characterization of ABNs
3.3. In Vitro Release of Dox from ABNs
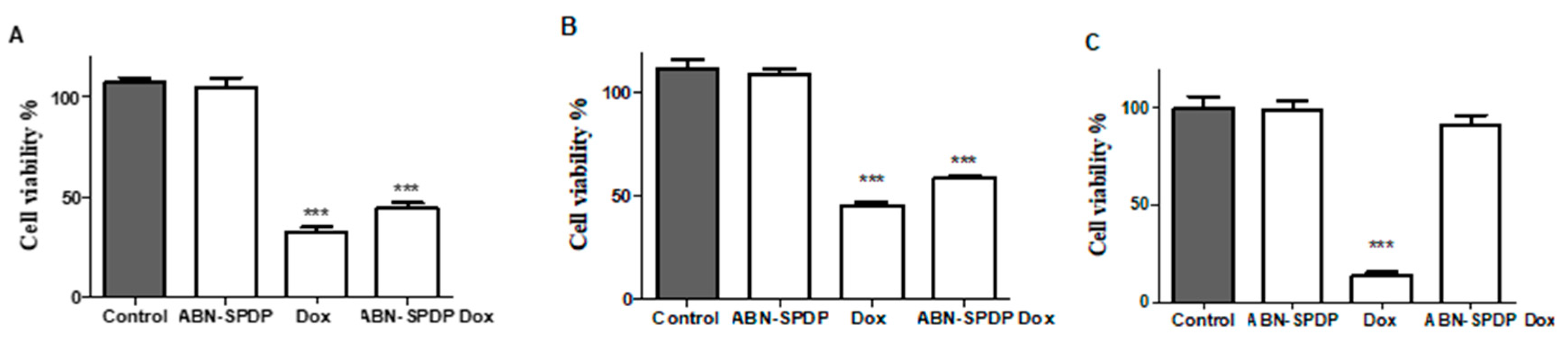
3.4. Cell Viability Studies
3.5. Stability Studies
3.6. Cell Studies
3.7. Effect on Apoptosis/Necrosis of the Cells
3.8. Determination of Dominant Cell Cycle Phase
3.9. Western Blot Analysis
3.10. Determination of the Mechanism of Internalization of NPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singal, P.K.; Iliskovic, N. Doxorubicin-induced cardiomyopathy. N. Engl. J. Med. 1998, 339, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Rivankar, S. An overview of doxorubicin formulations in cancer therapy. J. Cancer Res. Ther. 2014, 10, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; van der Meel, R.; Chen, X.; Lammers, T. The EPR effect and beyond: Strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics 2020, 10, 7921–7924. [Google Scholar] [CrossRef] [PubMed]
- Kalyane, D.; Raval, N.; Maheshwari, R.; Tambe, V.; Kalia, K.; Tekade, R.K. Employment of enhanced permeability and retention effect (EPR): Nanoparticle-based precision tools for targeting of therapeutic and diagnostic agent in cancer. Mater. Sci. Eng. C 2019, 98, 1252–1276. [Google Scholar] [CrossRef]
- Hoogenboezem, E.N.; Duvall, C.L. Harnessing albumin as a carrier for cancer therapies. Adv. Drug Deliv. Rev. 2018, 130, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.T.; Kuhlmann, M.; Hvam, M.L.; Howard, K.A. Albumin-based drug delivery: Harnessing nature to cure disease. Mol. Cell. Ther. 2016, 4, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curry, S.; Mandelkow, H.; Brick, P.; Franks, N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 1998, 5, 827–835. [Google Scholar] [CrossRef]
- Kragh-Hansen, U. Molecular Aspects of Ligand binding to serum albumin. Pharmacol. Rev. 1981, 33, 17–53. [Google Scholar]
- Patil, G.V. Biopolymer albumin for diagnosis and in drug delivery. Drug Dev. Res. 2003, 58, 219–247. [Google Scholar] [CrossRef]
- Evans, T.W. Review article: Albumin as a drug—biological effects of albumin unrelated to oncotic pressure. Aliment Pharmacol. Ther. 2002, 16, 6–11. [Google Scholar] [CrossRef]
- Kratz, F. Albumin, a versatile carrier in oncology. Int. J. Clin. Pharmacol. Ther. 2010, 48, 453–455. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Zhang, X. Strategies for Preparing Albumin-based Nanoparticles for Multifunctional Bioimaging and Drug Delivery. Theranostics 2017, 7, 3667–3689. [Google Scholar] [CrossRef] [PubMed]
- Onafuye, H.; Pieper, S.; Mulac, D.; Cinatl, J.; Wass, M.N.; Langer, K.; Michaelis, M. Doxorubicin-loaded human serum albumin nanoparticles overcome transporter-mediated drug resistance. Beilstein J. Nanotechnol. 2019, 10, 1707–1715. [Google Scholar] [CrossRef] [Green Version]
- Dreis, S.; Rothweiler, F.; Michaelis, M.; Cinatl, J.; Kreuter, J.; Langer, K. Preparation, characterisation and maintenance of drug efficacy of doxorubicin-loaded human serum albumin (HSA) nanoparticles. Int. J. Pharm. 2007, 341, 207–214. [Google Scholar] [CrossRef]
- Wang, W.; Huang, Y.; Zhao, S.; Shao, T.; Cheng, Y. Human serum albumin (HSA) nanoparticles stabilized with intermolecular disulfide bonds. Chem. Commun. 2013, 49, 2234–2236. [Google Scholar] [CrossRef] [PubMed]
- Amighi, F.; Emam-Djomeh, Z.; Labbafi-Mazraeh-Shahi, M. Effect of different cross-linking agents on the preparation of bovine serum albumin nanoparticles. J. Iran. Chem. Soc. 2020, 17, 1223–1235. [Google Scholar] [CrossRef]
- Niknejad, H.; Mahmoudzadeh, R. Comparison of different crosslinking methods for preparation of docetaxel-loaded albumin nanoparticles. Iran. J. Pharm. Res. 2015, 14, 385–394. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, W.; Huang, Y.; Fu, Y.; Cheng, Y. Paclitaxel loaded human serum albumin nanoparticles stabilized with intermolecular disulfide bonds. Medchemcomm 2014, 5, 1658–1663. [Google Scholar] [CrossRef]
- Silvestri, M.; Cristaudo, A.; Morrone, A.; Messina, C.; Bennardo, L.; Nisticò, S.P.; Mariano, M.; Cameli, N. Emerging Skin Toxicities in Patients with Breast Cancer Treated with New Cyclin-Dependent Kinase 4/6 Inhibitors: A Systematic Review. Drug Saf. 2021. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, G.; Lin, X.; Chatzinikolaidou, M.; Jennissen, H.P.; Laub, M. Polyethylenimine-coated Albumin Nanoparticles for BMP-2 Delivery. Biotechnol. Prog. 2008, 24, 945–956. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; ISBN 3-900051-07-0. [Google Scholar]
- Merodio, M.; Arnedo, A.; Renedo, M.J.; Irache, J.M. Ganciclovir-loaded albumin nanoparticles: Characterization and in vitro release properties. Eur. J. Pharm. Sci. 2001, 12, 251–259. [Google Scholar] [CrossRef]
- Jahanban-Esfahlan, A.; Dastmalchi, S.; Davaran, S. A simple improved desolvation method for the rapid preparation of albumin nanoparticles. Int. J. Biol. Macromol. 2016, 91, 703–709. [Google Scholar] [CrossRef]
- Jiang, J.; Oberdörster, G.; Biswas, P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J. Nanopart. Res. 2009, 11, 77–89. [Google Scholar] [CrossRef]
- Borm, P.J.A.; Robbins, D.; Haubold, S.; Kuhlbusch, T.; Fissan, H.; Donaldson, K.; Schins, R.; Stone, V.; Kreyling, W.; Lademann, J.; et al. The potential risks of nanomaterials: A review carried out for ECETOC. Part. Fibre Toxicol. 2006, 3, 1–35. [Google Scholar] [CrossRef] [Green Version]
- Renwick, L.C.; Donaldson, K.; Clouter, A. Impairment of alveolar macrophage phagocytosis by ultrafine particles. Toxicol. Appl. Pharmacol. 2001, 172, 119–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, A.; Fujioka, K.; Oku, T.; Suga, M.; Sasaki, Y.F.; Ohta, T.; Yasuhara, M.; Suzuki, K.; Yamamoto, K. Physicochemical properties and cellular toxicity of nanocrystal quantum dots depend on their surface modification. Nano Lett. 2004, 4, 2163–2169. [Google Scholar] [CrossRef]
- Qiu, Y.; Park, K. Environment-sensitive hydrogels for drug delivery. Adv. Drug Deliv. Rev. 2012, 64, 49–60. [Google Scholar] [CrossRef]
- Prajapati, R.; Gontsarik, M.; Yaghmur, A.; Salentinig, S. pH-responsive nano-self-assemblies of the anticancer drug 2-Hydroxyoleic acid. Langmuir 2019, 35, 7954–7961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tannock, I.F.; Rotin, D. Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 1989, 49, 4373–4384. [Google Scholar]
- Catanzaro, G.; Curcio, M.; Cirillo, G.; Spizzirri, U.G.; Besharat, Z.M.; Abballe, L.; Vacca, A.; Iemma, F.; Picci, N.; Ferretti, E. Albumin nanoparticles for glutathione-responsive release of cisplatin: New opportunities for medulloblastoma. Int. J. Pharm. 2017, 517, 168–174. [Google Scholar] [CrossRef]
- Kennedy, L.; Sandhu, J.K.; Harper, M.E.; Cuperlovic-culf, M. Role of glutathione in cancer: From mechanisms to therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef]
- Estrela, J.M.; Ortega, A.; Obrador, E. Glutathione in cancer biology and therapy. Crit. Rev. Clin. Lab. Sci. 2006, 43, 143–181. [Google Scholar] [CrossRef] [PubMed]
- Ballatori, N.; Krance, S.M.; Notenboom, S.; Shi, S.; Tieu, K.; Hammond, C.L. Glutathione dysregulation and the etiology and progression of human diseases. Biol. Chem. 2009, 390, 191–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curcio, M.; Blanco-Fernández, B.; Costoya, A.; Concheiro, A.; Puoci, F.; Alvarez-Lorenzo, C. Glucose cryoprotectant affects glutathione-responsive antitumor drug release from polysaccharide nanoparticles. Eur. J. Pharm. Biopharm. 2015, 93, 281–292. [Google Scholar] [CrossRef]
- KIPP, J. The role of solid nanoparticle technology in the parenteral delivery of poorly water-soluble drugs. Int. J. Pharm. 2004, 284, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Oncul, S.; Ercan, A. Discrimination of the effects of doxorubicin on two different breast cancer cell lines on account of multidrug resistance and apoptosis. Indian J. Pharm. Sci. 2017, 79, 599–607. [Google Scholar] [CrossRef]
- Mo, Y.; Barnett, M.E.; Takemoto, D.; Davidson, H.; Kompella, U.B. Human serum albumin nanoparticles for efficient delivery of Cu, Zn superoxide dismutase gene. Mol. Vis. 2007, 13, 746–757. [Google Scholar]
- Lv, L.; An, X.; Li, H.; Ma, L. Effect of miR-155 knockdown on the reversal of doxorubicin resistance in human lung cancer A549/dox cells. Oncol. Lett. 2016, 11, 1161–1166. [Google Scholar] [CrossRef] [Green Version]
- Wagh, J.; Patel, K.J.; Soni, P.; Desai, K.; Upadhyay, P.; Soni, H.P. Transfecting pDNA to E. coli DH5α using bovine serum albumin nanoparticles as a delivery vehicle. J. Biol. Chem. Lumin. 2015, 30, 583–591. [Google Scholar] [CrossRef] [PubMed]
- McGregor, D.; Bolt, H.; Cogliano, V.; Richter-Reichhelm, H.B. Formaldehyde and glutaraldehyde and nasal cytotoxicity: Case study within the context of the 2006 IPCS human framework for the analysis of a cancer mode of action for humans. Crit. Rev. Toxicol. 2006, 36, 821–835. [Google Scholar] [CrossRef] [PubMed]
- Albanese, A.; Tang, P.S.; Chan, W.C.W. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 2012, 14, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar-On, O.; Shapira, M.; Hershko, D.D. Differential effects of doxorubicin treatment on cell cycle arrest and Skp2 expression in breast cancer cells. Anticancer Drugs 2007, 18, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Troester, M.A.; Hoadley, K.A.; Sørlie, T.; Herbert, B.S.; Børresen-Dale, A.L.; Lønning, P.E.; Shay, J.W.; Kaufmann, W.K.; Perou, C.M. Cell-type-specific responses to chemotherapeutics in breast cancer. Cancer Res. 2004, 64, 4218–4226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elzoghby, A.O.; Samy, W.M.; Elgindy, N.A. Albumin-based nanoparticles as potential controlled release drug delivery systems. J. Control Release 2012, 157, 168–182. [Google Scholar] [CrossRef]




| MCF-7 | MDA-MB-231 | |||||
|---|---|---|---|---|---|---|
| Healthy Cells | Apoptotic Cells | Necrotic Cells | Healthy Cells | Apoptotic Cells | Necrotic Cells | |
| Control | 99.45 ± 0.45 | 0.15 ± 0.05 | 0.10 ± 0.10 | 99.75 ± 0.15 | 0.2 ± 0.10 | 0.05 ± 0.10 |
| ABNs | 95.15 ± 0.85 | 1.40 ± 0.10 | 4.00 ± 0.20 | 95.75 ± 3.15 | 1.15 ± 0.15 | 3.65 ± 0.25 |
| Dox alone 2 µM | 55.45 ± 0.85 | 3.70 ± 0.20 | 18.40 ± 0.10 | 72.00 ± 0.50 | 6.20 ± 0.10 | 12.95 ± 0.35 |
| ABN-SPDP Dox 1 µM | 65.70 ± 0.40 | 2.15 ± 0.15 | 17.45 ± 1.05 | 69.20 ± 0.51 | 5.65 ± 0.05 | 14.80 ± 0.10 |
| ABN-SPDP Dox 2 µM | 60.90 ± 0.40 | 2.70 ± 0.10 | 20.90 ± 3.80 | 63.80 ± 0.48 | 7.15 ± 0.25 | 21.00 ± 0.30 |
| ABN-SPDP Dox 4 µM | 50.95 ± 6.35 | 3.20 ± 1.00 | 24.05 ± 4.65 | 44.50 ± 2.48 | 7.30 ± 0.20 | 33.90 ± 0.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prajapati, R.; Garcia-Garrido, E.; Somoza, Á. Albumin-Based Nanoparticles for the Delivery of Doxorubicin in Breast Cancer. Cancers 2021, 13, 3011. https://doi.org/10.3390/cancers13123011
Prajapati R, Garcia-Garrido E, Somoza Á. Albumin-Based Nanoparticles for the Delivery of Doxorubicin in Breast Cancer. Cancers. 2021; 13(12):3011. https://doi.org/10.3390/cancers13123011
Chicago/Turabian StylePrajapati, Rama, Eduardo Garcia-Garrido, and Álvaro Somoza. 2021. "Albumin-Based Nanoparticles for the Delivery of Doxorubicin in Breast Cancer" Cancers 13, no. 12: 3011. https://doi.org/10.3390/cancers13123011
APA StylePrajapati, R., Garcia-Garrido, E., & Somoza, Á. (2021). Albumin-Based Nanoparticles for the Delivery of Doxorubicin in Breast Cancer. Cancers, 13(12), 3011. https://doi.org/10.3390/cancers13123011








