Circular RNAs: Emerging Regulators of the Major Signaling Pathways Involved in Cancer Progression
Abstract
Simple Summary
Abstract
1. Introduction
2. Biogenesis of circRNAs

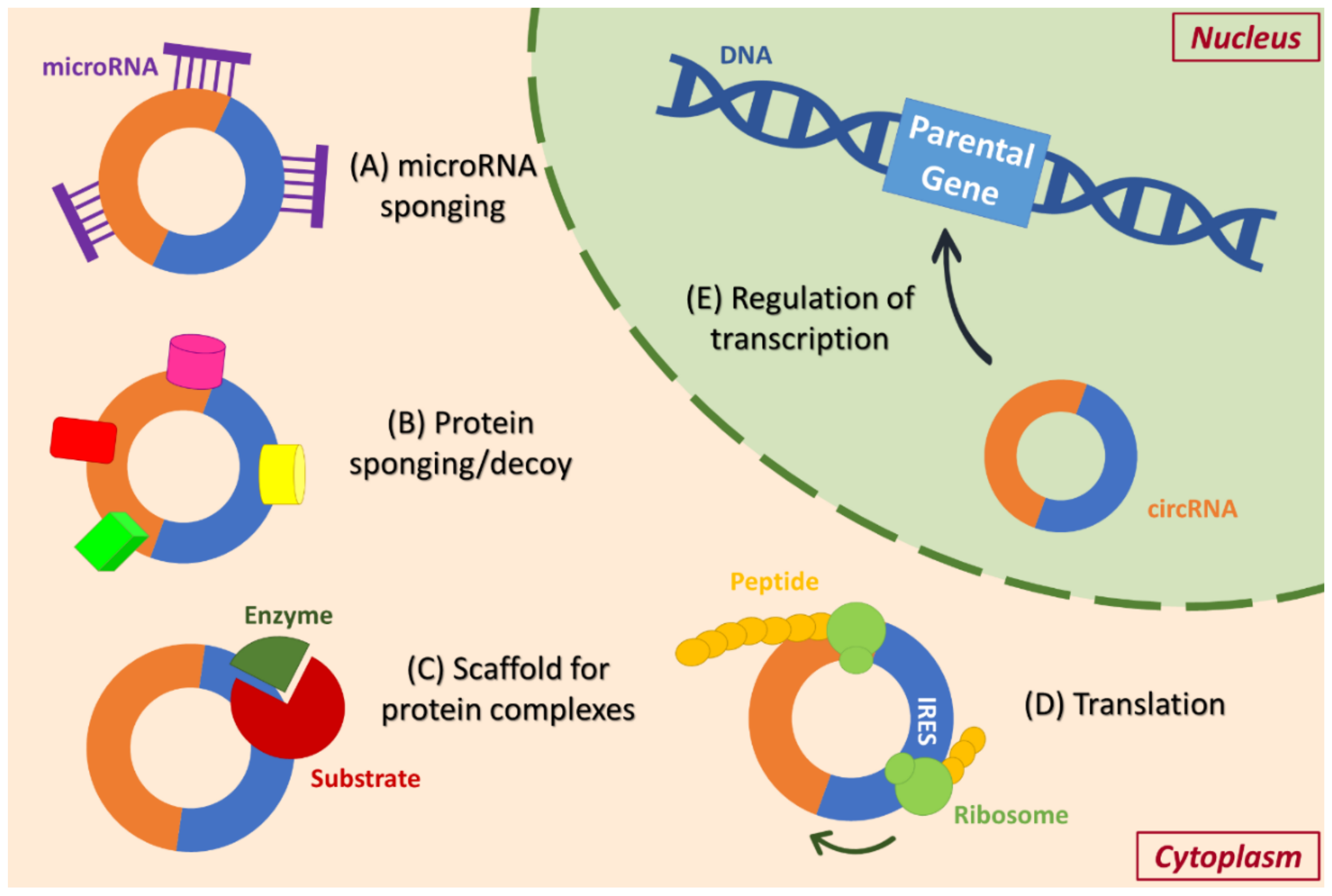
3. Functions of circRNAs and Their Involvement in Signal Transduction
4. CircRNA-Mediated Regulation of Major Signaling Pathways
4.1. VEGF
4.2. WNT/β-Catenin
4.3. MAPK
4.4. PI3K/AKT
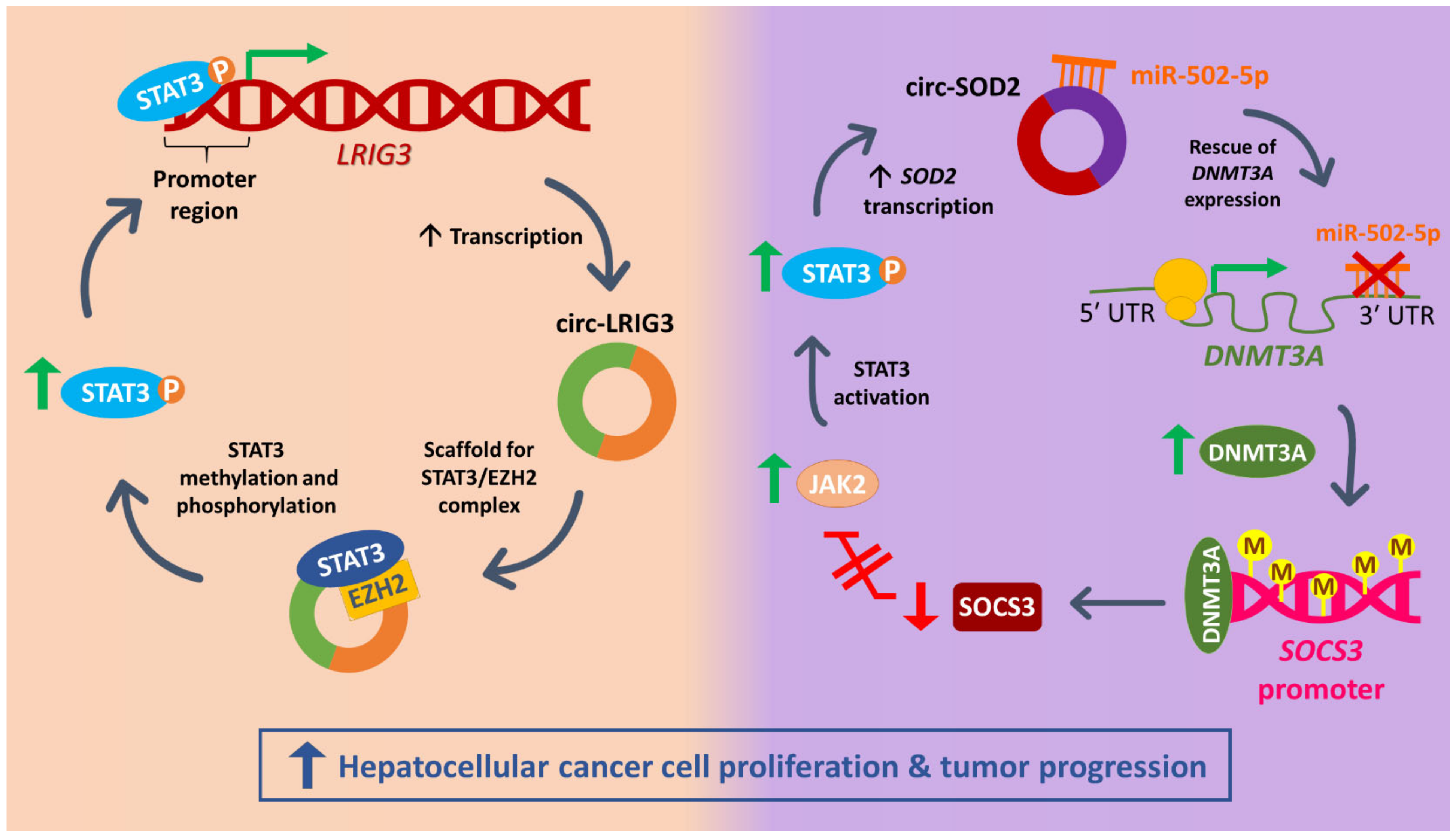
4.5. JAK/STAT
4.6. TGF-β/SMAD
4.7. Hippo/YAP
4.8. Notch
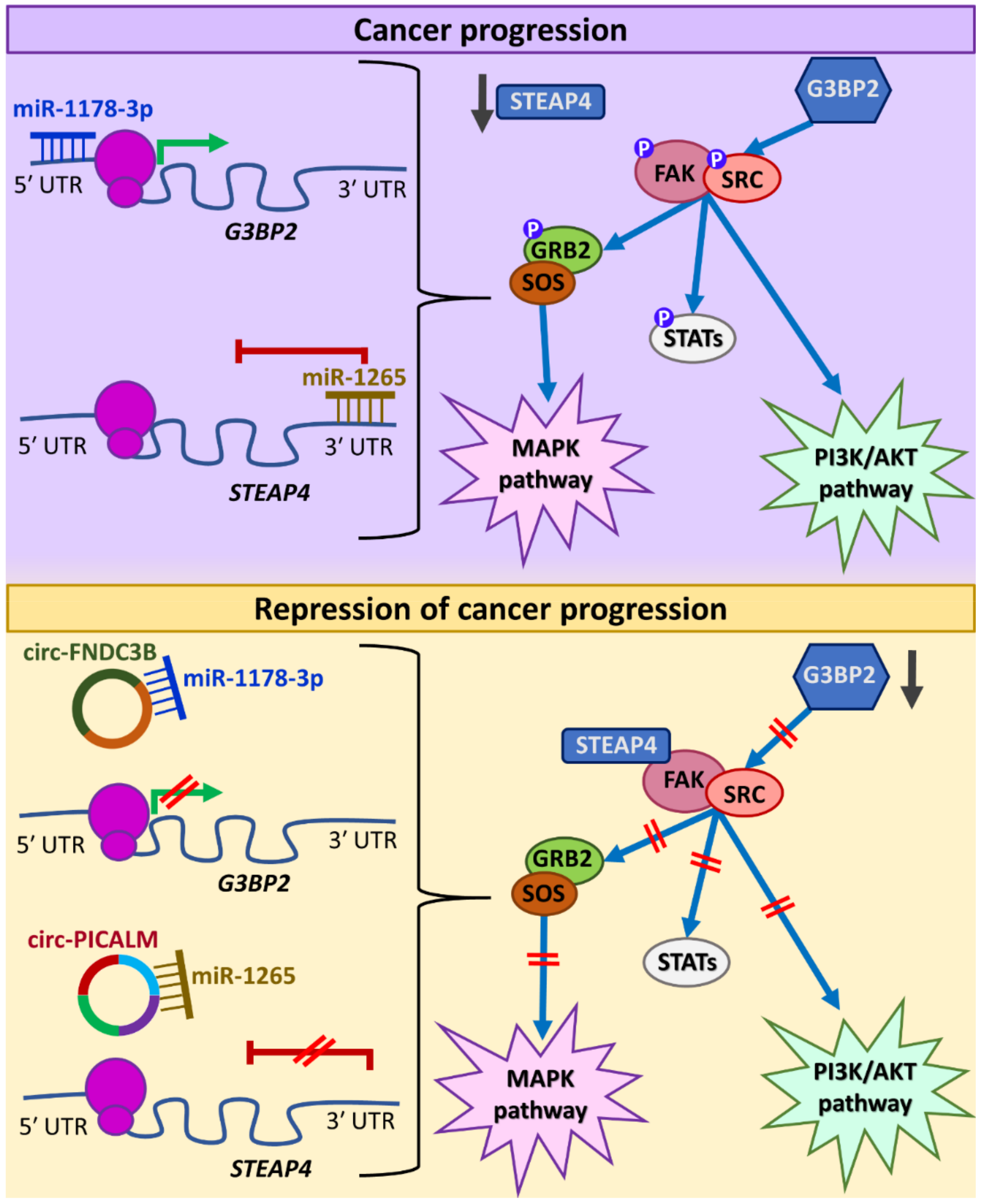
5. CircRNAs Affecting Multiple Signaling Pathways
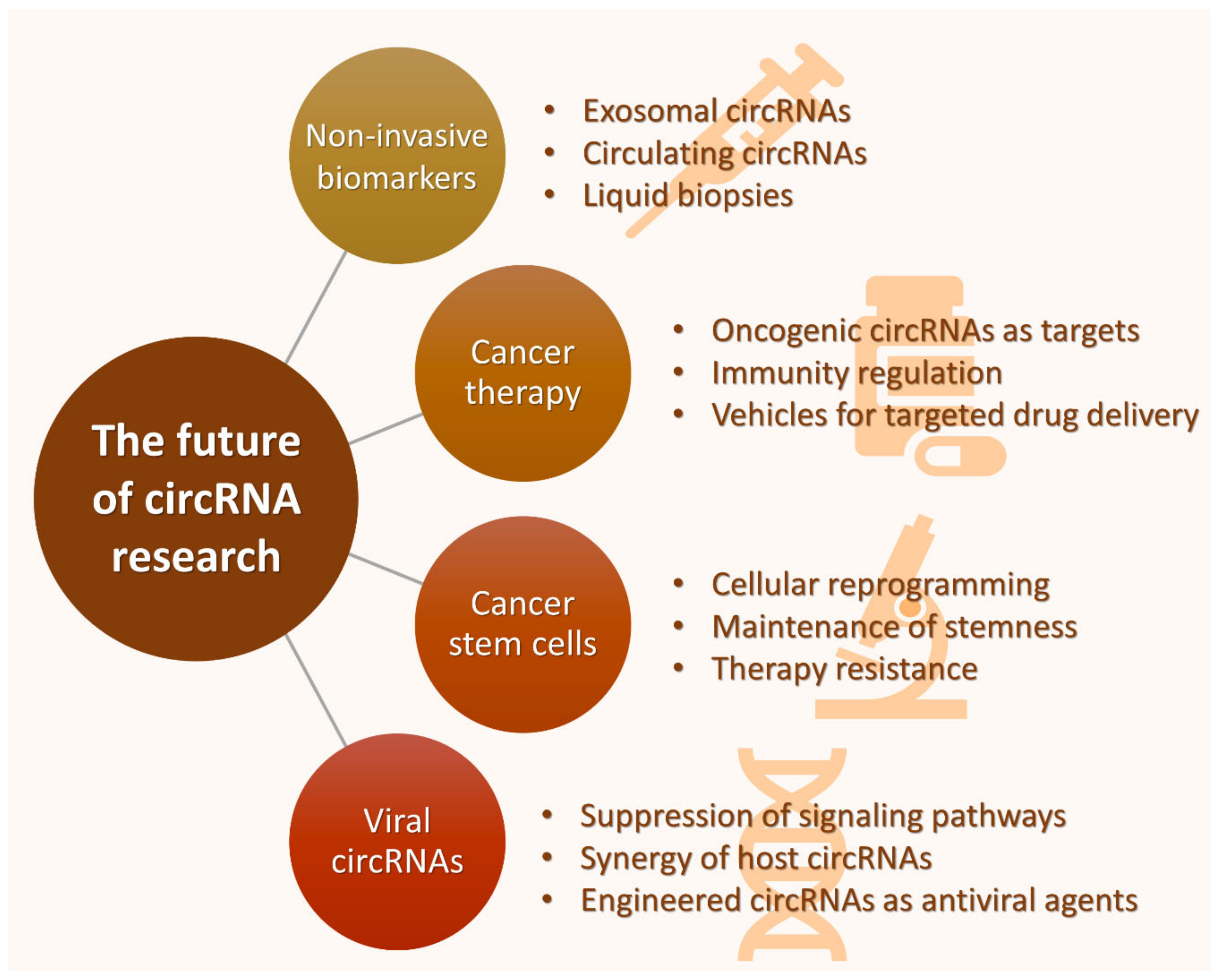
6. Future Perspectives
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baloghova, N.; Lidak, T.; Cermak, L. Ubiquitin Ligases Involved in the Regulation of Wnt, TGF-beta, and Notch Signaling Pathways and Their Roles in Mouse Development and Homeostasis. Genes 2019, 10, 815. [Google Scholar] [CrossRef] [PubMed]
- Sadoughi, F.; Hallajzadeh, J.; Asemi, Z.; Mansournia, M.A.; Alemi, F.; Yousefi, B. Signaling pathways involved in cell cycle arrest during the DNA breaks. DNA Repair 2021, 98, 103047. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.; Chory, J. Crosstalk in cellular signaling: Background noise or the real thing? Dev. Cell 2011, 21, 985–991. [Google Scholar] [CrossRef]
- Fairlie, W.D.; Tran, S.; Lee, E.F. Crosstalk between apoptosis and autophagy signaling pathways. Int. Rev. Cell Mol. Biol. 2020, 352, 115–158. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Vlahopoulos, S.A.; Cen, O.; Hengen, N.; Agan, J.; Moschovi, M.; Critselis, E.; Adamaki, M.; Bacopoulou, F.; Copland, J.A.; Boldogh, I.; et al. Dynamic aberrant NF-kappaB spurs tumorigenesis: A new model encompassing the microenvironment. Cytokine Growth Factor Rev. 2015, 26, 389–403. [Google Scholar] [CrossRef]
- Sever, R.; Brugge, J.S. Signal transduction in cancer. Cold Spring Harb. Perspect Med. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Mol. Biol. Rep. 2020, 47, 4587–4629. [Google Scholar] [CrossRef]
- Klein, C.A. Cancer progression and the invisible phase of metastatic colonization. Nat. Rev. Cancer 2020, 20, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Pulte, D.; Jansen, L.; Brenner, H. Changes in long term survival after diagnosis with common hematologic malignancies in the early 21st century. Blood Cancer J. 2020, 10, 56. [Google Scholar] [CrossRef]
- Yokota, J. Tumor progression and metastasis. Carcinogenesis 2000, 21, 497–503. [Google Scholar] [CrossRef]
- Misiorek, J.O.; Przybyszewska-Podstawka, A.; Kalafut, J.; Paziewska, B.; Rolle, K.; Rivero-Muller, A.; Nees, M. Context Matters: NOTCH Signatures and Pathway in Cancer Progression and Metastasis. Cells 2021, 10, 94. [Google Scholar] [CrossRef]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Artemaki, P.I.; Scorilas, A.; Kontos, C.K. Circular RNAs: A New Piece in the Colorectal Cancer Puzzle. Cancers 2020, 12, 2462. [Google Scholar] [CrossRef] [PubMed]
- Karousi, P.; Artemaki, P.I.; Sotiropoulou, C.D.; Christodoulou, S.; Scorilas, A.; Kontos, C.K. Identification of Two Novel Circular RNAs Deriving from BCL2L12 and Investigation of Their Potential Value as a Molecular Signature in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 8867. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, Y.; Wang, W.; Su, W.; Liu, Y.; Wang, Y.; Fan, C.; Li, X.; Li, G.; Li, Y.; et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018, 425, 134–142. [Google Scholar] [CrossRef]
- Shen, B.; Wang, Z.; Li, Z.; Song, H.; Ding, X. Circular RNAs: An emerging landscape in tumor metastasis. Am. J. Cancer Res. 2019, 9, 630–643. [Google Scholar]
- Liu, W.G.; Xu, Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/β-catenin pathway and indicates a poor prognosis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7905–7912. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Noto, J.J.; Schmidt, C.A.; Matera, A.G. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2017, 14, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Yang, Q.; Du, W.W.; Wu, N.; Yang, W.; Awan, F.M.; Fang, L.; Ma, J.; Li, X.; Zeng, Y.; Yang, Z.; et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017, 24, 1609–1620. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Du, W.W.; Wu, Y.; Yang, Z.; Awan, F.M.; Li, X.; Yang, W.; Zhang, C.; Yang, Q.; Yee, A.; et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics 2017, 7, 3842–3855. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Papatsirou, M.; Artemaki, P.I.; Scorilas, A.; Kontos, C.K. The role of circular RNAs in therapy resistance of patients with solid tumors. Pers. Med. 2020, 17, 469–490. [Google Scholar] [CrossRef]
- Chen, C.H.; Su, Y.J.; Ding, H.; Duan, J.; Wang, J. Circular RNA ZNF292 affects proliferation and apoptosis of hepatocellular carcinoma cells by regulating Wnt/beta-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12124–12130. [Google Scholar] [CrossRef]
- Wang, X.; Ji, C.; Hu, J.; Deng, X.; Zheng, W.; Yu, Y.; Hua, K.; Zhou, X.; Fang, L. Hsa_circ_0005273 facilitates breast cancer tumorigenesis by regulating YAP1-hippo signaling pathway. J. Exp. Clin. Cancer Res. 2021, 40, 29. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, S.; Zhang, M.; Yang, L.; Zhong, J.; Li, B.; Li, F.; Xia, X.; Li, X.; Zhou, H.; et al. A novel protein encoded by circular SMO RNA is essential for Hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol. 2021, 22, 33. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Zhou, Y.; Wang, Q.; Zheng, Z.; Xu, B.; Wu, C.; Zhou, Q.; Hu, W.; Wu, C.; et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 2019, 18, 47. [Google Scholar] [CrossRef]
- Veikkola, T.; Alitalo, K. VEGFs, receptors and angiogenesis. Semin Cancer Biol. 1999, 9, 211–220. [Google Scholar] [CrossRef]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef]
- Liu, H.; Bi, J.; Dong, W.; Yang, M.; Shi, J.; Jiang, N.; Lin, T.; Huang, J. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol. Cancer 2018, 17, 161. [Google Scholar] [CrossRef]
- Zhong, Z.; Huang, M.; Lv, M.; He, Y.; Duan, C.; Zhang, L.; Chen, J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017, 403, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, C.; Zou, Y.; Yu, J.; Gui, Y. Circular RNA MYLK promotes tumour growth and metastasis via modulating miR-513a-5p/VEGFC signalling in renal cell carcinoma. J. Cell. Mol. Med. 2020, 24, 6609–6621. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Li, Y.; Luo, Y.; Zhu, J.; Zheng, H.; Gao, B.; Guo, X.; Li, Z.; Chen, R.; Chen, C. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol. Cancer 2020, 19, 82. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chen, M.; Ai, G.; Mao, W.; Li, H.; Zhou, J. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 115, 108957. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Huang, Z.; Mo, X.; Song, Y.; Li, X.; Li, X.; Zhang, M. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J. Exp. Clin. Cancer Res. 2020, 39, 91. [Google Scholar] [CrossRef]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Zhang, J.; Yu, L. Circular RNAs Regulate Cancer Onset and Progression via Wnt/β-Catenin Signaling Pathway. Yonsei Med. J. 2019, 60, 1117–1128. [Google Scholar] [CrossRef]
- Wu, Z.; Zheng, M.; Zhang, Y.; Xie, M.; Tian, S.; Ding, T.; Li, L.; Guan, Q. Hsa_circ_0043278 functions as competitive endogenous RNA to enhance glioblastoma multiforme progression by sponging miR-638. Aging 2020, 12, 21114–21128. [Google Scholar] [CrossRef]
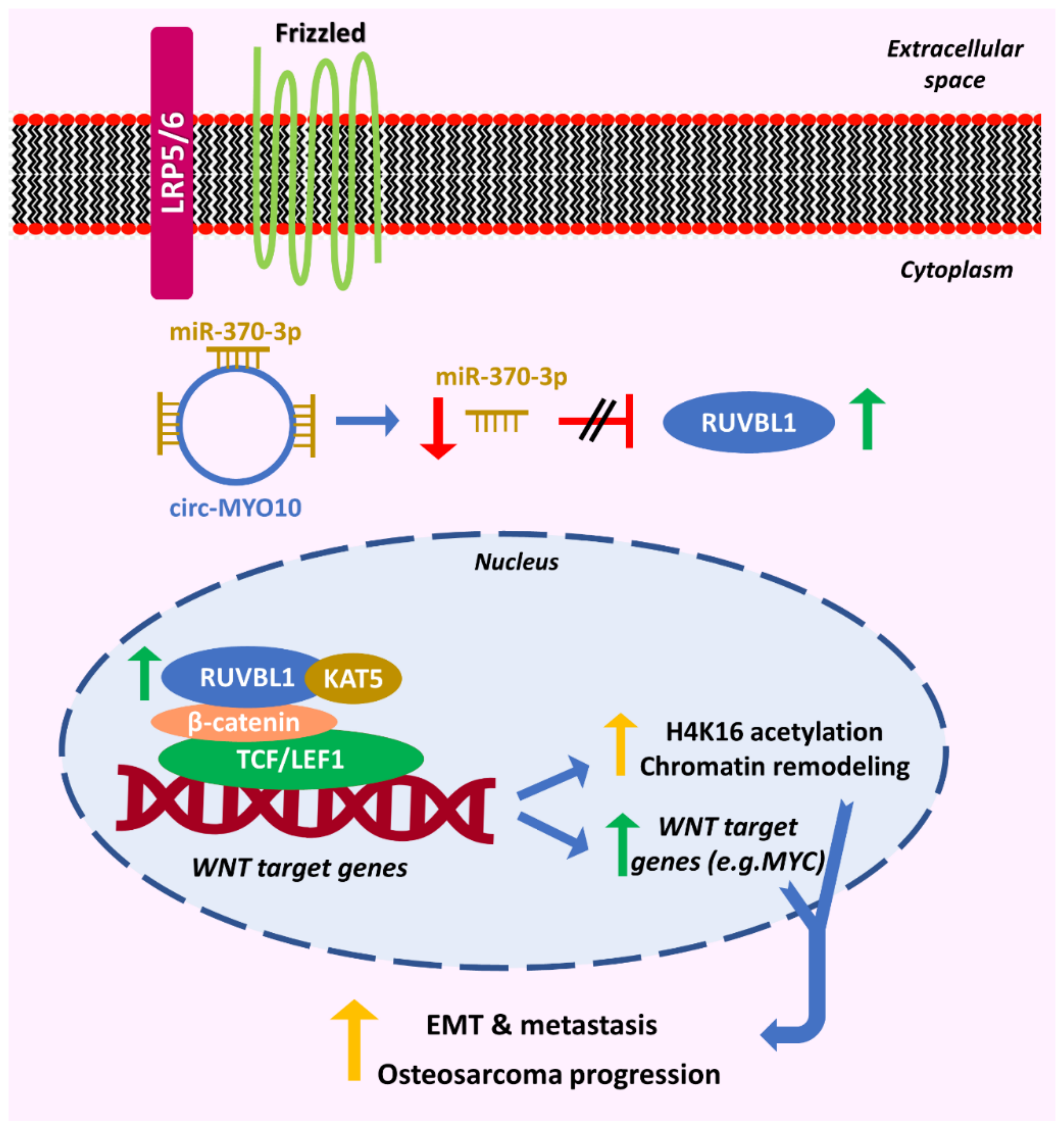
- Chen, J.; Liu, G.; Wu, Y.; Ma, J.; Wu, H.; Xie, Z.; Chen, S.; Yang, Y.; Wang, S.; Shen, P.; et al. CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of β-catenin/LEF1 complex via effects on chromatin remodeling. Mol. Cancer 2019, 18, 150. [Google Scholar] [CrossRef]
- Wu, Z.; Shi, W.; Jiang, C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis. Biochem. Biophys. Res. Commun. 2018, 502, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Huang, J.; Nie, C.; Liu, B.; He, G.; Han, J.; Pang, R.; Ding, Z.; Xu, J.; Zhang, J. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J. Exp. Clin. Cancer Res. 2018, 37, 275. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, B.; Ru, Z.; Cong, L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018, 504, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.T.; Liu, L.B.; Li, X.M.; Wang, Y.F.; Xie, P.J.; Li, Q.; Wang, R.; Wei, Q.; Kang, Y.H.; Meng, R.; et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/β-catenin pathway. Neoplasma 2019, 66, 232–239. [Google Scholar] [CrossRef]
- Shen, Q.; He, T.; Yuan, H. Hsa_circ_0002577 promotes endometrial carcinoma progression via regulating miR-197/CTNND1 axis and activating Wnt/β-catenin pathway. Cell Cycle 2019, 18, 1229–1240. [Google Scholar] [CrossRef]
- Hu, X.; Wu, D.; He, X.; Zhao, H.; He, Z.; Lin, J.; Wang, K.; Wang, W.; Pan, Z.; Lin, H.; et al. circGSK3β promotes metastasis in esophageal squamous cell carcinoma by augmenting β-catenin signaling. Mol. Cancer 2019, 18, 160. [Google Scholar] [CrossRef]
- Fang, G.; Ye, B.L.; Hu, B.R.; Ruan, X.J.; Shi, Y.X. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 2018, 504, 184–189. [Google Scholar] [CrossRef]
- Geng, Y.; Zheng, X.; Hu, W.; Wang, Q.; Xu, Y.; He, W.; Wu, C.; Zhu, D.; Wu, C.; Jiang, J. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin. Sci. 2019, 133, 1197–1213. [Google Scholar] [CrossRef]
- Cai, J.; Chen, Z.; Wang, J.; Wang, J.; Chen, X.; Liang, L.; Huang, M.; Zhang, Z.; Zuo, X. circHECTD1 facilitates glutaminolysis to promote gastric cancer progression by targeting miR-1256 and activating β-catenin/c-Myc signaling. Cell Death Dis. 2019, 10, 576. [Google Scholar] [CrossRef]
- Dai, X.; Liu, J.; Guo, X.; Cheng, A.; Deng, X.; Guo, L.; Wang, Z. Circular RNA circFGD4 suppresses gastric cancer progression via modulating miR-532-3p/APC/β-catenin signalling pathway. Clin. Sci. 2020, 134, 1821–1839. [Google Scholar] [CrossRef]
- Guo, X.; Dai, X.; Liu, J.; Cheng, A.; Qin, C.; Wang, Z. Circular RNA circREPS2 Acts as a Sponge of miR-558 to Suppress Gastric Cancer Progression by Regulating RUNX3/β-catenin Signaling. Mol. Ther. Nucleic Acids 2020, 21, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Li, Q.; Chen, L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Arch. Biochem. Biophys. 2019, 661, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Lu, G.; Luo, Z.; Gui, F.; Wu, J.; Zhang, D.; Ni, Y. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem. Biophys. Res. Commun. 2018, 497, 626–632. [Google Scholar] [CrossRef]
- Yao, Y.; Hua, Q.; Zhou, Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/β-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2019, 508, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Liu, Y.R.; Zhou, J.H.; Li, W.; Guo, H.H.; Ma, H.P. Enhanced expression of circular RNA hsa_circ_000984 promotes cells proliferation and metastasis in non-small cell lung cancer by modulating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3366–3374. [Google Scholar] [CrossRef]
- Fu, C.; Wang, S.; Jin, L.; Zhang, M.; Li, M. CircTET1 Inhibits Retinoblastoma Progression via Targeting miR-492 and miR-494-3p through Wnt/beta-catenin Signaling Pathway. Curr. Eye Res. 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Hao, A.; Zhang, Q.; Sui, G. The role of YY1 in oncogenesis and its potential as a drug target in cancer therapies. Curr Cancer Drug Targets 2015, 15, 145–157. [Google Scholar] [CrossRef]
- Yang, F.; Fang, E.; Mei, H.; Chen, Y.; Li, H.; Li, D.; Song, H.; Wang, J.; Hong, M.; Xiao, W.; et al. Cis-Acting circ-CTNNB1 Promotes β-Catenin Signaling and Cancer Progression via DDX3-Mediated Transactivation of YY1. Cancer Res. 2019, 79, 557–571. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef]
- Lee, S.; Rauch, J.; Kolch, W. Targeting MAPK Signaling in Cancer: Mechanisms of Drug Resistance and Sensitivity. Int. J. Mol. Sci 2020, 21, 1102. [Google Scholar] [CrossRef]
- Dhanasekaran, D.N.; Reddy, E.P. JNK-signaling: A multiplexing hub in programmed cell death. Genes Cancer 2017, 8, 682–694. [Google Scholar] [CrossRef]
- Zeke, A.; Misheva, M.; Remenyi, A.; Bogoyevitch, M.A. JNK Signaling: Regulation and Functions Based on Complex Protein-Protein Partnerships. Microbiol. Mol. Biol. Rev. 2016, 80, 793–835. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, B.; Liu, Y.; Yu, X.; Cheng, G. Dual effects of active ERK in cancer: A potential target for enhancing radiosensitivity. Oncol. Lett. 2020, 20, 993–1000. [Google Scholar] [CrossRef]
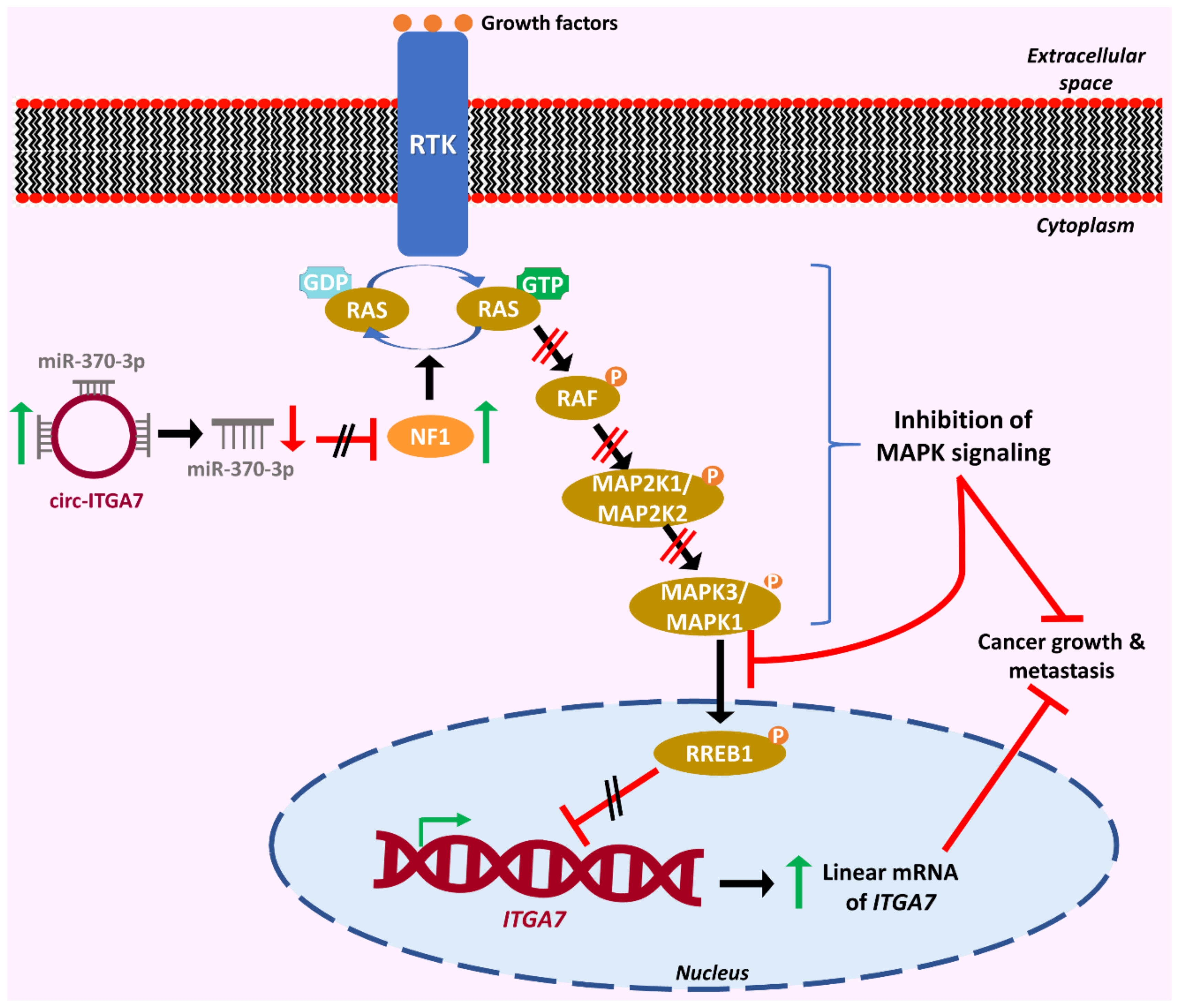
- Li, X.; Wang, J.; Zhang, C.; Lin, C.; Zhang, J.; Zhang, W.; Zhang, W.; Lu, Y.; Zheng, L.; Li, X. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J. Pathol. 2018, 246, 166–179. [Google Scholar] [CrossRef]
- Li, C.; Zhou, H. Circular RNA hsa_circRNA_102209 promotes the growth and metastasis of colorectal cancer through miR-761-mediated Ras and Rab interactor 1 signaling. Cancer Med. 2020, 9, 6710–6725. [Google Scholar] [CrossRef]
- Baxter, R.C. IGF binding proteins in cancer: Mechanistic and clinical insights. Nat. Rev. Cancer 2014, 14, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, X.; Chen, A.; Shi, W.; Wang, L.; Yi, R.; Qiu, J. circPIP5K1A serves as a competitive endogenous RNA contributing to ovarian cancer progression via regulation of miR-661/IGFBP5 signaling. J. Cell. Biochem. 2019, 120, 19406–19414. [Google Scholar] [CrossRef]
- Yang, D.; Qian, H.; Fang, Z.; Xu, A.; Zhao, S.; Liu, B.; Li, D. Silencing circular RNA VANGL1 inhibits progression of bladder cancer by regulating miR-1184/IGFBP2 axis. Cancer Med. 2020, 9, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhou, J. CircAGFG1 promotes cervical cancer progression via miR-370-3p/RAF1 signaling. BMC Cancer 2019, 19, 1067. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hou, L.; Liang, R.; Chen, X.; Zhang, R.; Chen, W.; Zhu, J. CircDLST promotes the tumorigenesis and metastasis of gastric cancer by sponging miR-502-5p and activating the NRAS/MEK1/ERK1/2 signaling. Mol. Cancer 2019, 18, 80. [Google Scholar] [CrossRef]
- Ouyang, Y.; Li, Y.; Huang, Y.; Li, X.; Zhu, Y.; Long, Y.; Wang, Y.; Guo, X.; Gong, K. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J. Cell. Physiol. 2019, 234, 10458–10469. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, R.; Zhang, D.; Lu, T.; Yu, W.; Wo, Y.; Liu, A.; Sui, T.; Cui, J.; Qin, Y.; et al. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl. Lung Cancer Res. 2019, 8, 862–875. [Google Scholar] [CrossRef]
- Bartel, C.A.; Parameswaran, N.; Cipriano, R.; Jackson, M.W. FAM83 proteins: Fostering new interactions to drive oncogenic signaling and therapeutic resistance. Oncotarget 2016, 7, 52597–52612. [Google Scholar] [CrossRef] [PubMed]
- Koveitypour, Z.; Panahi, F.; Vakilian, M.; Peymani, M.; Seyed Forootan, F.; Nasr Esfahani, M.H.; Ghaedi, K. Signaling pathways involved in colorectal cancer progression. Cell Biosci 2019, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Qi, X.; Zhang, X.; Fang, K.; Guo, Z.; Li, L. hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol. Carcinog. 2019, 58, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Liu, S.; Liu, F.; Sang, M.; Ju, Y.; Fan, X.; Gu, L.; Li, Z.; Geng, C.; Sang, M. ZEB1-Mediated Transcriptional Upregulation of circWWC3 Promotes Breast Cancer Progression through Activating Ras Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 22, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, L.; Chen, J.; Gao, H.; Zhao, W.; Huang, Y.; Jiang, T.; Zhou, J.; Chen, Y. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem. Biophys. Res. Commun. 2018, 505, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, H.; Li, P. Upregulation of hsa_circRNA_102958 Indicates Poor Prognosis and Promotes Ovarian Cancer Progression Through miR-1205/SH2D3A Axis. Cancer Manag. Res. 2020, 12, 4045–4053. [Google Scholar] [CrossRef]
- Huang, P.; Qi, B.; Yao, H.; Zhang, L.; Li, Y.; Li, Q. Circular RNA cSMARCA5 regulates the progression of cervical cancer by acting as a microRNA-432 sponge. Mol. Med. Rep. 2020, 21, 1217–1223. [Google Scholar] [CrossRef]
- Mo, J.; Zhao, Y.; Ao, Z.; Chen, L.; Lin, S.; Zeng, W.; Wu, H.; Liu, J. Circ-APBB1IP as a Prognostic Biomarker Promotes Clear Cell Renal Cell Carcinoma Progression Through The ERK1/2 Signaling Pathway. Int. J. Med. Sci. 2020, 17, 1177–1186. [Google Scholar] [CrossRef]
- Sun, M.; Zhao, W.; Chen, Z.; Li, M.; Li, S.; Wu, B.; Bu, R. Circular RNA CEP128 promotes bladder cancer progression by regulating Mir-145-5p/Myd88 via MAPK signaling pathway. Int. J. Cancer 2019, 145, 2170–2181. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, Y.; Chu, F.; Xu, L.; Wu, H. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int. 2020, 20, 74. [Google Scholar] [CrossRef]
- Hu, Z.Q.; Zhou, S.L.; Li, J.; Zhou, Z.J.; Wang, P.C.; Xin, H.Y.; Mao, L.; Luo, C.B.; Yu, S.Y.; Huang, X.W.; et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology 2020, 72, 906–922. [Google Scholar] [CrossRef]
- Liu, G.; Shi, H.; Deng, L.; Zheng, H.; Kong, W.; Wen, X.; Bi, H. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2019, 513, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Guo, H.; Niu, M.; Zheng, X.; Zhang, Y.; Xue, X.; Bo, Y.; Guan, X.; Li, Z.; Guo, Y.; et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Mol. Cancer 2020, 19, 166. [Google Scholar] [CrossRef] [PubMed]
- Bian, A.; Wang, Y.; Liu, J.; Wang, X.; Liu, D.; Jiang, J.; Ding, L.; Hui, X. Circular RNA Complement Factor H (CFH) Promotes Glioma Progression by Sponging miR-149 and Regulating AKT1. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 5704–5712. [Google Scholar] [CrossRef]
- Sun, D.; Liu, J.; Zhou, L. Upregulation of circular RNA circ-FAM53B predicts adverse prognosis and accelerates the progression of ovarian cancer via the miR-646/VAMP2 and miR-647/MDM2 signaling pathways. Oncol. Rep. 2019, 42, 2728–2737. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Li, L.; Zhu, T.; Liu, G. Circular RNA circ_0102049 promotes cell progression as ceRNA to target MDM2 via sponging miR-1304-5p in osteosarcoma. Pathol. Res. Pract. 2019, 215, 152688. [Google Scholar] [CrossRef]
- Yang, Y.; Ding, L.; Li, Y.; Xuan, C. Hsa_circ_0039411 promotes tumorigenesis and progression of papillary thyroid cancer by miR-1179/ABCA9 and miR-1205/MTA1 signaling pathways. J. Cell. Physiol. 2020, 235, 1321–1329. [Google Scholar] [CrossRef]
- Guo, X.; Zhou, Q.; Su, D.; Luo, Y.; Fu, Z.; Huang, L.; Li, Z.; Jiang, D.; Kong, Y.; Li, Z.; et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol. Cancer 2020, 19, 83. [Google Scholar] [CrossRef]
- Qiao, J.; Liu, M.; Tian, Q.; Liu, X. Microarray analysis of circRNAs expression profile in gliomas reveals that circ_0037655 could promote glioma progression by regulating miR-214/PI3K signaling. Life Sci. 2020, 245, 117363. [Google Scholar] [CrossRef]
- Shi, F.; Shi, Z.; Zhao, Y.; Tian, J. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem. Biophys. Res. Commun. 2019, 510, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.; Zhang, X.Y.; Sun, D.K.; Tian, L.Q.; Xu, P. Up-regulated circular RNA hsa_circ_0067934 contributes to glioblastoma progression through activating PI3K-AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3447–3454. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yan, X.; Zhang, H.; Zhan, X. CircRNA circ_0067934 Overexpression Correlates with Poor Prognosis and Promotes Thyroid Carcinoma Progression. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 1342–1349. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, X.; Liu, A.; Xu, J.; Lai, J.; Guan, H.; Ma, J. Circ_PSD3 promotes the progression of papillary thyroid carcinoma via the miR-637/HEMGN axis. Life Sci. 2020, 118622. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Wang, Q.; Song, H.; Shao, D.; Xue, J. circ_103809 promotes breast cancer progression by regulating the PI3K/AKT signaling pathway. Oncol. Lett. 2020, 19, 3725–3730. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.H.; Wang, Y.; Xu, D. Hsa_circ_001569 is an unfavorable prognostic factor and promotes cell proliferation and metastasis by modulating PI3K-AKT pathway in breast cancer. Cancer Biomark. Sect. A Dis. Markers 2019, 25, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Yalan, S.; Yanfang, L.; He, C.; Yujie, T. Circular RNA circRHOBTB3 inhibits ovarian cancer progression through PI3K/AKT signaling pathway. Panminerva Med. 2020. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, L.; Sun, X. CircRNA hsa_circ_0002577 accelerates endometrial cancer progression through activating IGF1R/PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 2020, 39, 169. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Liu, T.; Feng, H.; Yang, R.; Zhao, X.; Chen, W.; Jiang, B.; Qin, H.; Guo, X.; Liu, M.; et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol. Cancer 2019, 18, 111. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Mingyan, E.; Wang, C.; Liu, G.; Shi, M.; Liu, S. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int. J. Biol. Macromol. 2019, 125, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Cheng, X.; Liu, X.; Xia, C.; Zhang, H.; Pan, D.; Zhang, X.; Li, Y. Circ_0026344 restrains metastasis of human colorectal cancer cells via miR-183. Artif. CellsNanomed. Biotechnol. 2019, 47, 4038–4045. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Yang, Y.; Wang, C.; Yang, W.; Yan, Y.; Wang, Z.; Xu, J.; Jiang, K. Circular RNA 0047905 acts as a sponge for microRNA4516 and microRNA1227-5p, initiating gastric cancer progression. Cell Cycle 2019, 18, 1560–1572. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.K.; Pu, K.; Su, H.X.; Zhang, J.; Zheng, Y.; Ji, R.; Guo, Q.H.; Wang, Y.P.; Guan, Q.L.; Zhou, Y.N. Circular RNA hsa_circ_0010882 promotes the progression of gastric cancer via regulation of the PI3K/Akt/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef]
- Rong, D.; Lu, C.; Zhang, B.; Fu, K.; Zhao, S.; Tang, W.; Cao, H. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol. Cancer 2019, 18, 25. [Google Scholar] [CrossRef]
- Han, J.; Zhao, G.; Ma, X.; Dong, Q.; Zhang, H.; Wang, Y.; Cui, J. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem. Biophys. Res. Commun. 2018, 503, 2429–2435. [Google Scholar] [CrossRef]
- Hong, W.; Zhang, Y.; Ding, J.; Yang, Q.; Xie, H.; Gao, X. circHIPK3 Acts as Competing Endogenous RNA and Promotes Non-Small-Cell Lung Cancer Progression through the miR-107/BDNF Signaling Pathway. Biomed. Res. Int. 2020, 2020, 6075902. [Google Scholar] [CrossRef]
- Yao, J.; Xu, G.; Zhu, L.; Zheng, H. circGFRA1 Enhances NSCLC Progression by Sponging miR-188-3p. Oncotargets Ther. 2020, 13, 549–558. [Google Scholar] [CrossRef]
- Yin, D.; Wei, G.; Yang, F.; Sun, X. Circular RNA has circ 0001591 promoted cell proliferation and metastasis of human melanoma via ROCK1/PI3K/AKT by targeting miR-431-5p. Hum. Exp. Toxicol. 2020. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, Q.; Song, C.; Ma, R.; Li, X. Circ_0007841 promotes the progression of multiple myeloma through targeting miR-338-3p/BRD4 signaling cascade. Cancer Cell Int. 2020, 20, 383. [Google Scholar] [CrossRef]
- Chen, Z.L.; Li, X.N.; Ye, C.X.; Chen, H.Y.; Wang, Z.J. Elevated Levels of circRUNX1 in Colorectal Cancer Promote Cell Growth and Metastasis via miR-145-5p/IGF1 Signalling. Oncotargets Ther. 2020, 13, 4035–4048. [Google Scholar] [CrossRef] [PubMed]
- Aaronson, D.S.; Horvath, C.M. A road map for those who don’t know JAK-STAT. Science 2002, 296, 1653–1655. [Google Scholar] [CrossRef]
- Thomas, S.J.; Snowden, J.A.; Zeidler, M.P.; Danson, S.J. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 2015, 113, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Huang, Q.; Cheng, S.; Wu, S.; Sang, H.; Hou, J. Circ_ZNF124 promotes non-small cell lung cancer progression by abolishing miR-337-3p mediated downregulation of JAK2/STAT3 signaling pathway. Cancer Cell Int. 2019, 19, 291. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Gao, J.; Zhou, S.; Li, Y.; Wang, Y.; Jin, L.; Li, J.; Liu, B.; Zhang, B.; Han, S.; et al. A novel circular RNA circ-LRIG3 facilitates the malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J. Exp. Clin. Cancer Res. 2020, 39, 252. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Song, J.; Tang, B.; Fang, S.; Zhang, D.; Zheng, L.; Wu, F.; Gao, Y.; Chen, C.; Hu, X.; et al. CircSOD2 induced epigenetic alteration drives hepatocellular carcinoma progression through activating JAK2/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2020, 39, 259. [Google Scholar] [CrossRef]
- Wu, M.; Sun, T.; Xing, L. Circ_0004913 Inhibits Cell Growth, Metastasis, and Glycolysis by Absorbing miR-184 to Regulate HAMP in Hepatocellular Carcinoma. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef]
- Liu, Y.; Song, J.; Liu, Y.; Zhou, Z.; Wang, X. Transcription activation of circ-STAT3 induced by Gli2 promotes the progression of hepatoblastoma via acting as a sponge for miR-29a/b/c-3p to upregulate STAT3/Gli2. J. Exp. Clin. Cancer Res. 2020, 39, 101. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, Y. Circular RNA HIPK3 promotes glioma progression by binding to miR-124-3p. Gene 2019, 690, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.; Xu, M.; Yuan, X.; Xie, H.; Zhao, J. Circular RNA circCUL3 Accelerates the Warburg Effect Progression of Gastric Cancer through Regulating the STAT3/HK2 Axis. Mol. Ther. Nucleic Acids 2020, 22, 310–318. [Google Scholar] [CrossRef]
- Xu, Z.; Tie, X.; Li, N.; Yi, Z.; Shen, F.; Zhang, Y. Circular RNA hsa_circ_0000654 promotes esophageal squamous cell carcinoma progression by regulating the miR-149-5p/IL-6/STAT3 pathway. Iubmb Life 2020, 72, 426–439. [Google Scholar] [CrossRef]
- Liu, L.; Liu, F.B.; Huang, M.; Xie, K.; Xie, Q.S.; Liu, C.H.; Shen, M.J.; Huang, Q. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat. Dis. Int. Hbpd Int. 2019, 18, 580–586. [Google Scholar] [CrossRef]
- Yan, M.; Gao, H.; Lv, Z.; Liu, Y.; Zhao, S.; Gong, W.; Liu, W. Circular RNA PVT1 promotes metastasis via regulating of miR-526b/FOXC2 signals in OS cells. J. Cell. Mol. Med. 2020, 24, 5593–5604. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.; He, B.; Yang, B.B.; Xu, T.; Chen, X.; Xu, M.; Liu, X.; Sun, H.; Pan, Y.; Wang, S. The pro-metastasis effect of circANKS1B in breast cancer. Mol. Cancer 2018, 17, 160. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; You, J.; Xue, H.; Tan, X.; Chao, C. CircCTDP1 promotes nasopharyngeal carcinoma progression via a microRNA-320b/HOXA10/TGFβ2 pathway. Int. J. Mol. Med. 2020, 45, 836–846. [Google Scholar] [CrossRef]
- Xu, G.; Chen, Y.; Fu, M.; Zang, X.; Cang, M.; Niu, Y.; Zhang, W.; Zhang, Y.; Mao, Z.; Shao, M.; et al. Circular RNA CCDC66 promotes gastric cancer progression by regulating c-Myc and TGF-β signaling pathways. J. Cancer 2020, 11, 2759–2768. [Google Scholar] [CrossRef]
- Wang, L.; Tong, X.; Zhou, Z.; Wang, S.; Lei, Z.; Zhang, T.; Liu, Z.; Zeng, Y.; Li, C.; Zhao, J.; et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol. Cancer 2018, 17, 140. [Google Scholar] [CrossRef]
- Peng, Q.S.; Cheng, Y.N.; Zhang, W.B.; Fan, H.; Mao, Q.H.; Xu, P. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit Hippo signaling pathway. Cell Death Dis. 2020, 11, 112. [Google Scholar] [CrossRef]
- Chen, L.Y.; Zhi, Z.; Wang, L.; Zhao, Y.Y.; Deng, M.; Liu, Y.H.; Qin, Y.; Tian, M.M.; Liu, Y.; Shen, T.; et al. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J. Pathol. 2019, 248, 103–115. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, Y.; Qi, L.; Ding, L.; Jiang, H.; Yu, H. NFIX Circular RNA Promotes Glioma Progression by Regulating miR-34a-5p via Notch Signaling Pathway. Front. Mol. Neurosci. 2018, 11, 225. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Mishra, L.; Deng, C.-X. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci 2018, 14, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Shah, C.A.; Wang, H.; Bei, L.; Platanias, L.C.; Eklund, E.A. HoxA10 regulates transcription of the gene encoding transforming growth factor beta2 (TGFbeta2) in myeloid cells. J. Biol. Chem. 2011, 286, 3161–3176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.Y.; Liu, Q.H.; Wang, T.J.; Zhao, J.; Cheng, X.H.; Wang, J.S. CircZFR serves as a prognostic marker to promote bladder cancer progression by regulating miR-377/ZEB2 signaling. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Zhu, Y.; Casado, M.; Vaulont, S.; Sharma, K. Role of upstream stimulatory factors in regulation of renal transforming growth factor-beta1. Diabetes 2005, 54, 1976–1984. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bracken, C.P.; Smith, E.; Bert, A.G.; Wright, J.A.; Roslan, S.; Morris, M.; Wyatt, L.; Farshid, G.; Lim, Y.-Y.; et al. An autocrine TGF-β/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell 2011, 22, 1686–1698. [Google Scholar] [CrossRef]
- Fujita, H.; Kang, M.; Eren, M.; Gleaves, L.A.; Vaughan, D.E.; Kume, T. Foxc2 is a common mediator of insulin and transforming growth factor beta signaling to regulate plasminogen activator inhibitor type I gene expression. Circ Res. 2006, 98, 626–634. [Google Scholar] [CrossRef]
- Gumbiner, B.M.; Kim, N.-G. The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 2014, 127, 709–717. [Google Scholar] [CrossRef]
- Zygulska, A.L.; Krzemieniecki, K.; Pierzchalski, P. Hippo pathway—Brief overview of its relevance in cancer. J. Physiol Pharm. 2017, 68, 311–335. [Google Scholar]
- Allenspach, E.J.; Maillard, I.; Aster, J.C.; Pear, W.S. Notch signaling in cancer. Cancer Biol. Ther. 2002, 1, 466–476. [Google Scholar] [CrossRef]
- Chen, L.; Yang, X.; Zhao, J.; Xiong, M.; Almaraihah, R.; Chen, Z.; Hou, T. Circ_0008532 promotes bladder cancer progression by regulation of the miR-155-5p/miR-330-5p/MTGR1 axis. J. Exp. Clin. Cancer Res. 2020, 39, 94. [Google Scholar] [CrossRef]
- Parkin, A.; Man, J.; Timpson, P.; Pajic, M. Targeting the complexity of Src signalling in the tumour microenvironment of pancreatic cancer: From mechanism to therapy. FEBS J. 2019, 286, 3510–3539. [Google Scholar] [CrossRef] [PubMed]
- Worthmuller, J.; Ruegg, C. The Crosstalk between FAK and Wnt Signaling Pathways in Cancer and Its Therapeutic Implication. Int. J. Mol. Sci 2020, 21. [Google Scholar] [CrossRef]
- Yan, D.; Dong, W.; He, Q.; Yang, M.; Huang, L.; Kong, J.; Qin, H.; Lin, T.; Huang, J. Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation. EBioMedicine 2019, 48, 316–331. [Google Scholar] [CrossRef]
- Deng, L.; Liu, G.; Zheng, C.; Zhang, L.; Kang, Y.; Yang, F. Circ-LAMP1 promotes T-cell lymphoblastic lymphoma progression via acting as a ceRNA for miR-615-5p to regulate DDR2 expression. Gene 2019, 701, 146–151. [Google Scholar] [CrossRef]
- Shi, Y.; Fang, N.; Li, Y.; Guo, Z.; Jiang, W.; He, Y.; Ma, Z.; Chen, Y. Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020, 111, 2824–2836. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Zhao, W.; Liu, G.; Yang, Q. Circ-PGAM1 promotes malignant progression of epithelial ovarian cancer through regulation of the miR-542-3p/CDC5L/PEAK1 pathway. Cancer Med. 2020, 9, 3500–3521. [Google Scholar] [CrossRef]
- Li, W.; Xu, Y.; Wang, X.; Cao, G.; Bu, W.; Wang, X.; Fang, Z.; Xu, Y.; Dong, M.; Tao, Q. circCCT3 Modulates Vascular Endothelial Growth Factor A and Wnt Signaling to Enhance Colorectal Cancer Metastasis Through Sponging miR-613. DNA Cell Biol. 2020, 39, 118–125. [Google Scholar] [CrossRef]
- Wang, L.; Li, H.; Qiao, Q.; Ge, Y.; Ma, L.; Wang, Q. Circular RNA circSEMA5A promotes bladder cancer progression by upregulating ENO1 and SEMA5A expression. Aging 2020, 12, 21674–21686. [Google Scholar] [CrossRef]
- Sun, H.; Wang, Q.; Yuan, G.; Quan, J.; Dong, D.; Lun, Y.; Sun, B. Hsa_circ_0001649 restrains gastric carcinoma growth and metastasis by downregulation of miR-20a. J. Clin. Lab. Anal. 2020, 34, e23235. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, W.; Zhou, Q.; Chen, C.; Yuan, W.; Liu, J.; Li, X.; Sun, Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer 2020, 19, 14. [Google Scholar] [CrossRef]
- Tan, K.E.; Lim, Y.Y. Viruses join the circular RNA world. FEBS J. 2020. [Google Scholar] [CrossRef]
| Pathway | Cancer | circRNA | Gene of Origin | Interactions | Effect | Reference |
|---|---|---|---|---|---|---|
| VEGF | Bladder cancer | circ-MYLK | MYLK | miR-29a-3p/VEGFA | Promotes cancer progression | [43] |
| Cervical cancer | circ_0023404 | RNF121 | miR-5047/VEGFA | Promotes cancer metastasis and chemoresistance | [46] | |
| Colorectal cancer | circ_001971 (circ_0001060) 1 | UXS1 | miR-29c-3p/VEGFA | Promotes cancer growth, angiogenesis, and metastasis | [47] | |
| Renal cell carcinoma | circ-MYLK | MYLK | miR-513a-5p/VEGFC | Promotes cancer growth and metastasis | [44] | |
| Pancreatic cancer | circ-NFIB1 (circ_0086375) 1 | NFIB | miR-486-5p/ PIK3R1/VEGFC | Inhibits lymphangiogenesis and lymphatic metastasis, suppresses cancer progression | [45] | |
| WNT/β-catenin | Glioblastoma multiforme | circ_0043278 | TADA2A | miR-638 | Promotes cancer progression | [50] |
| Osteosarcoma | circ-MYO10 | MYO10 | miR-370-3p/RUVBL1 | Regulates chromatin remodeling; promotes cancer progression | [51] | |
| circ_0002052 | PAPPA | miR-1205/APC2 | Suppresses cancer progression | [52] | ||
| Papillary thyroid cancer | circ_102171 | SMURF2 | CTNNBIP1 | Promotes cancer progression | [53] | |
| circ-ITCH | ITCH | miR-22-3p/CBL | Suppresses cancer progression | [54] | ||
| Breast cancer | circ-ITCH | ITCH | miR-214-3p/ITCH, miR-17-5p/ITCH | Suppresses cancer progression | [55] | |
| Endometrial | circ_0002577 | WDR26 | miR-197-3p/CTNND1 | Promotes cancer progression | [56] | |
| Esophageal squamous cell carcinoma | circ-GSK3B | GSK3B | - | Promotes metastasis | [57] | |
| Colorectal cancer | circ_100290 | - | miR-516b-5p/FZD4 | Promotes cancer progression | [58] | |
| circ_0009361 | GNB1 | miR-582-3p/APC2 | Suppresses cancer progression | [59] | ||
| Gastric cancer | circ-HECTD1 | HECTD1 | miR-1256 | Promotes glutaminolysis and cancer progression | [60] | |
| circ-HIPK3 | HIPK3 | WNT1, TCF4, β-catenin | Promotes cancer progression | [20] | ||
| circ-FGD4 | FGD4 | miR-532-3p/APC | Suppresses cancer progression | [61] | ||
| circ-REPS2 | REPS2 | miR-558/RUNX3 | [62] | |||
| Hepatocellular carcinoma | circ-ZFR | ZFR | miR-3619-5p/β-catenin | Promotes cancer progression | [63] | |
| circ_0067934 | PRKCI | miR-1324/FZD5 | Promotes tumor growth and metastasis | [64] | ||
| Lung cancer | circ_0006427 | BCAR3 | miR-6783-3p/DKK1 | Suppresses cancer progression | [65] | |
| Non-small cell lung cancer | circ_000984 (circ_0001724) 1 | CDK6 | - | Promotes cell proliferation and metastasis | [66] | |
| Retinoblastoma | circ-TET1 | TET1 | miR-492, miR-494-3p | Suppresses cancer progression | [67] |
| Cancer | CircRNA | Gene of Origin | Interactions | Effect | Reference |
|---|---|---|---|---|---|
| Breast cancer | circ_0006528 | PRELID2 | miR-7-5p | Promotes cancer progression | [86] |
| circ-WWC3 | WWC3 | miR-26b-3p, miR-660-3p | [87] | ||
| Ovarian | circ-ITCH | ITCH | miR-145-5p/RASA1 | Suppresses cancer progression | [88] |
| circ-PIP5K1A | PIP5K1A | miR-661/IGFBP5 | Promotes cancer progression | [78] | |
| circ_102958 | - | miR-1205/SH2D3A | [89] | ||
| Cervical cancer | circ-SMARCA5 | SMARCA5 | miR-432-5p | Promotes cell proliferation | [90] |
| circ-AGFG1 | AGFG1 | miR-370-3p/RAF1 | Promotes cancer progression | [80] | |
| Clear Cell Renal Cell Carcinoma | circ-APBB1IP | APBB1IP | - | Promotes cancer progression | [91] |
| Bladder cancer | circ-CEP128 | CEP128 | miR-145-5p/MYD88 | Promotes cancer progression | [92] |
| circ-VANGL1 | VANGL1 | miR-1184/IGFBP2 | [79] | ||
| Colorectal cancer | circ_102209 | - | miR-761/RIN1 | Promotes cell growth and metastasis | [76] |
| circITGA7 | ITGA7 | miR-370-3p/NF1/ RREB1/ITGA7 | Suppresses tumor growth and metastasis | [75] | |
| Gastric cancer | circ_0032821 | CEP128 | - | Promotes cell proliferation and metastasis | [93] |
| circ-PDSS1 | PDSS1 | miR-186-5p/NEK2 | Promotes cancer progression | [82] | |
| circ-DLST | DLST | miR-502-5p/ NRAS/MAP2K1/MAPK1,miR-502-5p/ NRAS/MAP2K1/MAPK3 | [81] | ||
| Hepatocellular carcinoma | circ-ASAP1 | ASAP1 | miR-326/MAPK1, miR-532-5p/MAPK1 | Promotes cancer progression | [94] |
| Non-small cell lung cancer | circ-FOXM1 | FOXM1 | miR-1304-5p/PPDPF, miR-1304-5p/MACC1 | Promotes cancer progression and metastasis | [95] |
| circ-ZKSCAN1 | ZKSCAN1 | miR-330-5p/FAM83A | Promotes cancer progression | [83] |
| Cancer | circRNA | Gene of Origin | Interactions | Effect | Reference |
|---|---|---|---|---|---|
| Glioma | circ_0015758 | CFH | miR-149-3p/AKT1 | Promotes cancer progression | [97] |
| circ_0037655 | CREBBP | miR-214-3p/PI3K | [102] | ||
| circ_0014359 | NUP210L | miR-153-3p | [103] | ||
| Glioblastoma | circ_0067934 | PRKCI | - | Promotes cancer progression | [104] |
| Osteosarcoma | circ_0102049 | ATL1 | miR-1304-5p/MDM2 | Promotes cancer progression | [99] |
| Laryngeal squamous cell carcinoma | circ-PARD3 | PARD3 | miR-145-5p/ PRKCI/AKT1/MTOR | Promotes cancer progression | [96] |
| Thyroid Carcinoma | circ_0067934 | PRKCI | - | Promotes cancer progression | [105] |
| Papillary thyroid carcinoma | circ-PSD3 | PSD3 | miR-637/HEMGN | Promotes cancer progression | [106] |
| circ_0039411 | MMP2 | miR-1179/ABCA9, miR-1205/MTA1 | Promotes cancer progression | [100] | |
| Breast cancer | circ_103809 | - | Promotes cancer progression | [107] | |
| circ_001569 (circ_0000677) 1 | ABCC1 | - | [108] | ||
| Ovarian cancer | circ-FAM53B | FAM53B | miR-646/VAMP2, miR-647/MDM2 | Promotes cancer progression | [98] |
| circ-RHOBTB3 | RHOBTB3 | - | Suppresses cancer progression | [109] | |
| Endometrial cancer | circ_0002577 | WDR26 | IGF1R/PI3K/AKT | Promotes cancer progression | [110] |
| Bladder cancer | circ-SLC8A1 | SLC8A1 | miR-130b-3p, miR-494-3p | Suppresses cancer progression | [111] |
| Esophageal squamous cell carcinoma | circ-VRK1 | VRK1 | miR-624-3p/ PTEN/PI3K/AKT | Suppresses cancer progression | [112] |
| Colorectal cancer | circ-RUNX1 | RUNX1 | miR-145-5p/IGF1 | Promotes cancer progression | [45] |
| circ_0026344 | ACVRL1 | miR-183-5p | Suppresses cancer progression | [113] | |
| Gastric cancer | circ_0047905 | SERPINB5 | miR-4516, miR-1227-5p | Promotes cancer progression | [114] |
| circ_0010882 | RPL11 | - | [115] | ||
| circ-NRIP1 | NRIP1 | miR-149-5p | [116] | ||
| circ-PSMC3 | PSMC3 | miR-296-5p | Suppresses cancer progression | [117] | |
| Pancreatic cancer | circ-BFAR | BFAR | miR-34b-5p/ MET/AKT1 | Promotes cancer progression | [101] |
| Lung cancer | circ-BANP | BANP | miR-503-3p/LARP1 | Promotes cancer progression | [118] |
| Non-Small Cell Lung Cancer | circ-HIPK3 | HIPK3 | miR-107/BDNF | Promotes cancer progression | [119] |
| circ-GFRA1 | GFRA1 | miR-188-3p | [120] | ||
| Melanoma | circ_0001591 (circ_001436) 1 | HIST1H2AG | miR-431-5p/ ROCK1/PI3K/AKT | Promotes cell proliferation and cancer progression | [121] |
| Multiple myeloma | circ_0007841 | SEC61A1 | miR-338-3p/BRD4 | Promotes cancer progression and cell proliferation | [122] |
| Pathway | Cancer | CircRNA | Gene of Origin | Interactions | Effect | Reference |
|---|---|---|---|---|---|---|
| JAK/STAT | Glioma | circ-HIPK3 (circ_0000284) 1 | HIPK3 | miR-124-3p/STAT3 | Promotes cancer progression | [131] |
| Gastric cancer | circ-CUL3 (circ_0008309) 1 | CUL3 | miR-515-5p/ STAT3/HK2 | Promotes Warburg effect progression | [132] | |
| Hepatocellular cancer | circ-LRIG3 (circ_0027345) 1 | LRIG3 | EZH2/STAT3 | Promotes cancer progression | [127] | |
| circ-SOD2 (circ_0004662) 1 | SOD2 | miR-502-5p/DNMT3A/ SOCS3/JAK2/STAT3 | Induces epigenetic alterations, promotes cancer progression | [128] | ||
| circ_0004913 | TEX2 | miR-184/HAMP/ JAK2/STAT3 | Suppresses cancer progression | [129] | ||
| Hepatoblastoma | circ-STAT3(circ_0043800) 1 | STAT3 | miR-29-3p family/ STAT3, miR-29-3p family/GLI2 | Promotes cancer progression | [130] | |
| Non-small cell lung cancer | circ-ZNF124 | ZNF124 | miR-337-3p/ JAK2/STAT3 | Promotes cancer progression | [81] | |
| Esophageal squamous cell carcinoma | circ_0000654 (circ_000608) 1 | CHO2 | miR-149-5p/ IL6/STAT3 | Promotes cancer progression | [133] | |
| Pancreatic cancer | CDR1as (ciRS-7, circ_0001946) 1 | CDR1 | miR-7-5p/EGFR/STAT3 | Promotes cancer progression | [134] | |
| TGF-β/SMAD | Osteosarcoma | circ-PVT1 | PVT1 | miR-526b-5p/ FOXC2 | Promotes metastasis | [135] |
| Breast cancer | circ_0007294 | ANKS1B | miR-148a-3p/USF1, miR-152-3p/USF1 | Promotes metastasis | [136] | |
| Nasopharyngeal carcinoma | circ-CTDP1 | CTDP1 | miR-320b/HOXA10 | Promotes metastasis | [137] | |
| Bladder cancer | circ_0072088 | ZFR | miR-377-3p/ZEB2 | Promotes metastasis | [83] | |
| Gastric cancer | circ- CCDC66 | CCDC66 | - | Promotes cell growth and metastasis | [138] | |
| Non-small cell lung cancer | circ_0008305 | PTK2 (FAK) | miR-200b-3p/TRIM33, miR-429/TRIM33 | Suppresses metastasis | [139] | |
| Hippo/ YAP | Colon cancer | circ-PPP1R12A | PPP1R12A | - | Promotes migration, invasion, and proliferation | [39] |
| Oral squamous cell carcinoma | circ_0000140 (circ_002059) 1 | KIAA0907 | miR-31-5p/LATS2 | Suppresses cell growth and metastasis | [140] | |
| Notch | Colon cancer | circ-NSD2 | NSD2 | miR-199b-5p/Jag1 | Promotes metastasis (in mice) | [141] |
| Bladder cancer | circ_0008532 | CBFA2T2 | miR-155-5p/CBFA2T2, miR-330-5p/CBFA2T2 | Promotes migration and invasion | [47] | |
| Glioma | circ-NFIX | NFIX | miR-34a-5p/NOTCH1 | Promotes cancer progression | [142] |
| Cancer | circRNA | Gene of Origin | Interactions | Effect | Reference |
|---|---|---|---|---|---|
| Ovarian cancer | circ-PGAM1 | PGAM1 | miR-542-3p/PEA K1/MAPK1, miR-542-3p/PEAK1/MAPK3, miR-542-3p/PEAK1/JAK2 | Promotes cancer progression | [159] |
| Esophageal cancer | circ-LPAR3 | LPAR3 | miR-198/MET/MAP2K7, miR-198/MET/AKT1 | Promotes migration and invasion | [158] |
| Bladder cancer | circ-SEMA5A | SEMA5A | miR-330-5p/ENO1 | Promotes cancer progression | [161] |
| circ-FNDC3B | FNDC3B | miR-1178-3p/G3BP2/ SRC/FAK | Suppresses tumor invasion and metastasis | [42] | |
| circ-PICALM | PICALM | miR-1265/STEAP4/FAK | Suppresses tumor invasion and metastasis | [156] | |
| Colon cancer | circ-CCT3 | CCT3 | miR-613/WNT3, miR-613/VEGFA | Promotes cancer metastasis | [160] |
| Gastric cancer | circ_0001649 (circ_001599) 1 | SHPRH | miR-20a-5p | Suppresses cancer progression | [162] |
| T-cell lymphoblastic lymphoma | circ-LAMP1 | LAMP1 | miR-615-5p/DDR2 | Promotes cancer progression | [157] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papatsirou, M.; Artemaki, P.I.; Karousi, P.; Scorilas, A.; Kontos, C.K. Circular RNAs: Emerging Regulators of the Major Signaling Pathways Involved in Cancer Progression. Cancers 2021, 13, 2744. https://doi.org/10.3390/cancers13112744
Papatsirou M, Artemaki PI, Karousi P, Scorilas A, Kontos CK. Circular RNAs: Emerging Regulators of the Major Signaling Pathways Involved in Cancer Progression. Cancers. 2021; 13(11):2744. https://doi.org/10.3390/cancers13112744
Chicago/Turabian StylePapatsirou, Maria, Pinelopi I. Artemaki, Paraskevi Karousi, Andreas Scorilas, and Christos K. Kontos. 2021. "Circular RNAs: Emerging Regulators of the Major Signaling Pathways Involved in Cancer Progression" Cancers 13, no. 11: 2744. https://doi.org/10.3390/cancers13112744
APA StylePapatsirou, M., Artemaki, P. I., Karousi, P., Scorilas, A., & Kontos, C. K. (2021). Circular RNAs: Emerging Regulators of the Major Signaling Pathways Involved in Cancer Progression. Cancers, 13(11), 2744. https://doi.org/10.3390/cancers13112744











