Mapping of Metabolic Heterogeneity of Glioma Using MR-Spectroscopy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Ethical Approval
2.2. Patient Enrolment
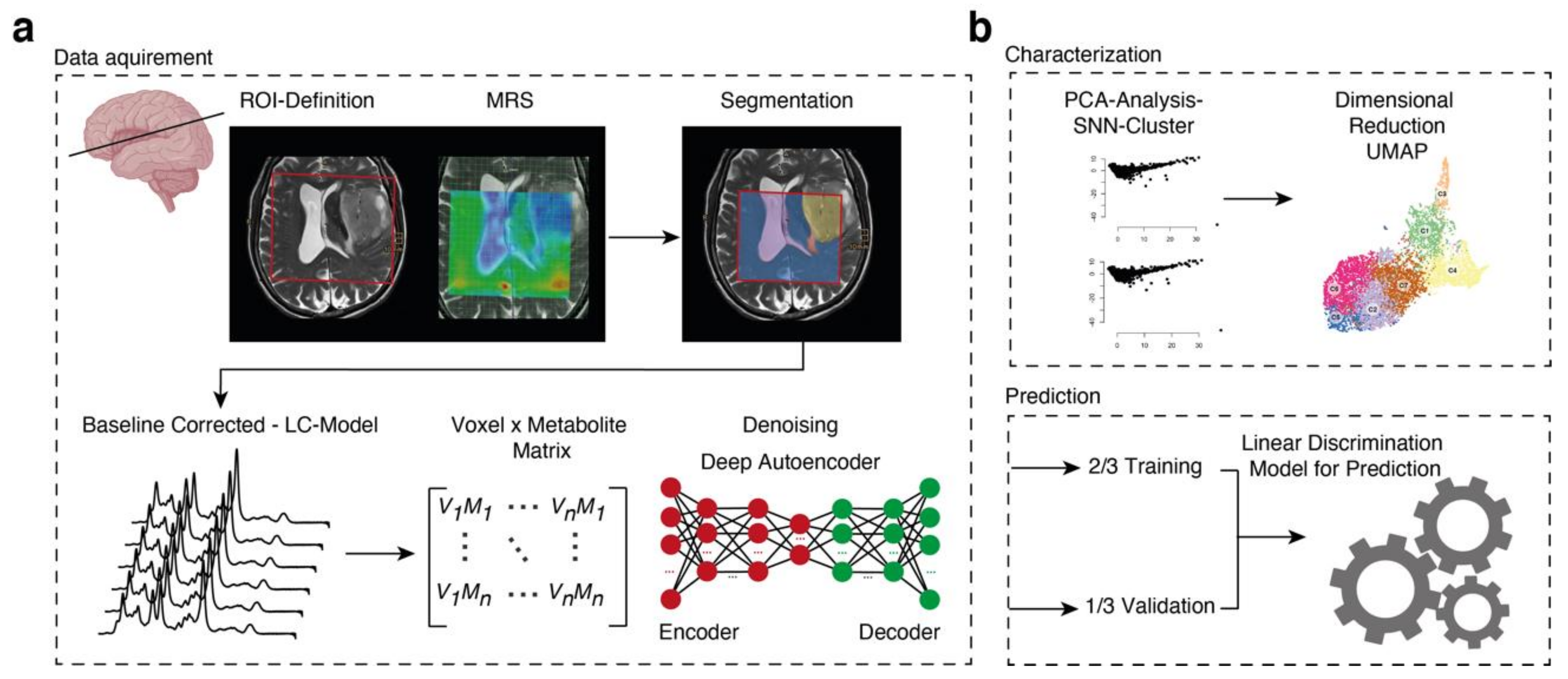
2.3. Imaging Acquisition and Data
2.4. Data Processing
2.5. Segmentation
2.6. Tumor Tissue Sampling
2.7. Deep Autoencoder for Denoising
2.8. Hyperparameter Search
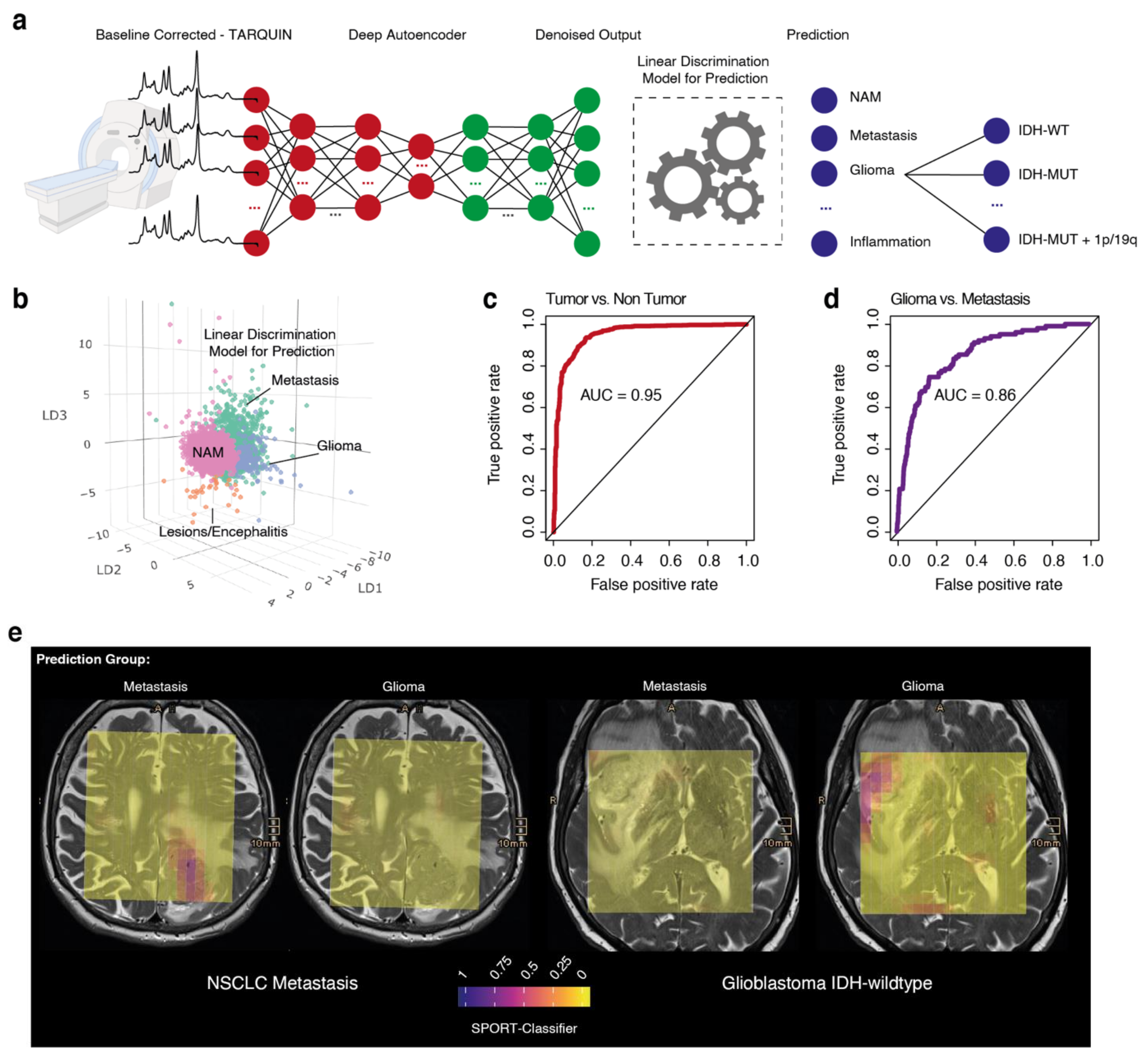
2.9. Prediction Model
2.10. Dimensional Reduction and Clustering
2.11. Spatial Data Analysis
2.12. Mean Spectra
2.13. Data Availability
3. Results
3.1. Patient Demographics
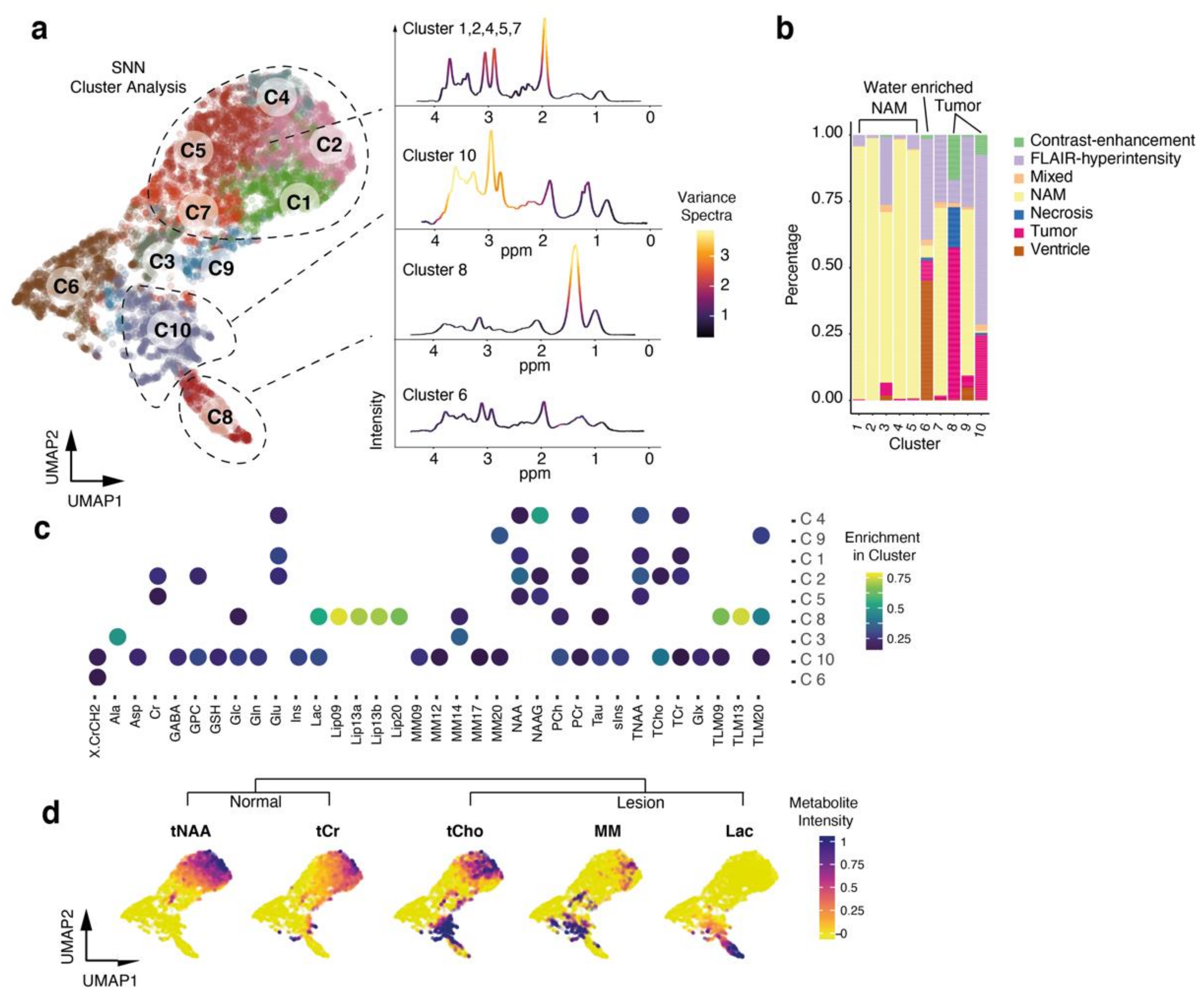
3.2. Unsupervised Analysis of Metabolomic Heterogeneity and Diversity in MRS Data
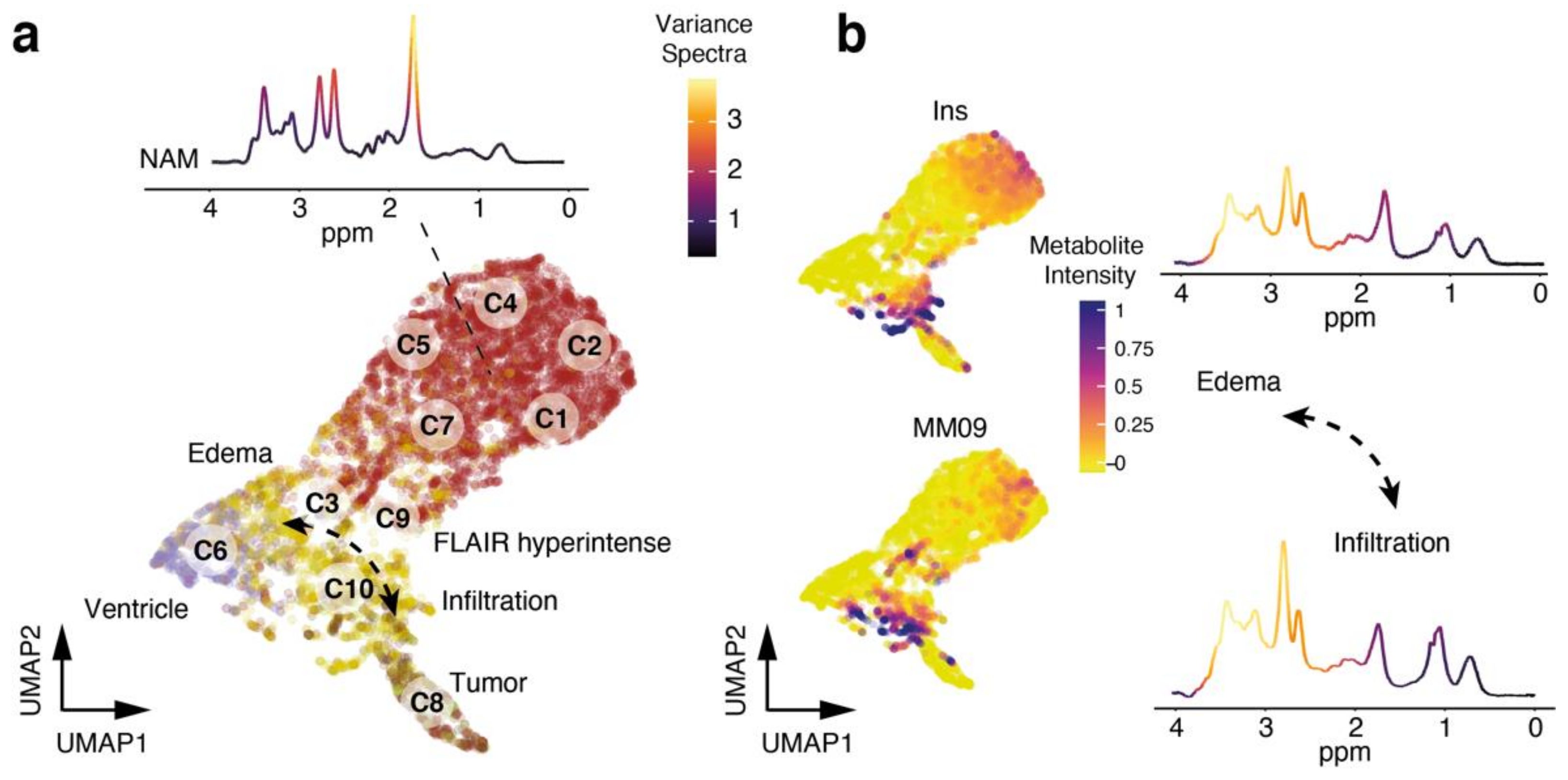
3.3. Cluster Analysis Reflects Regional Differences and Pathological Spectra
3.4. Prediction of Tumor Regions
3.5. Exploration of Metabolic Diversity in Pathological Lesions
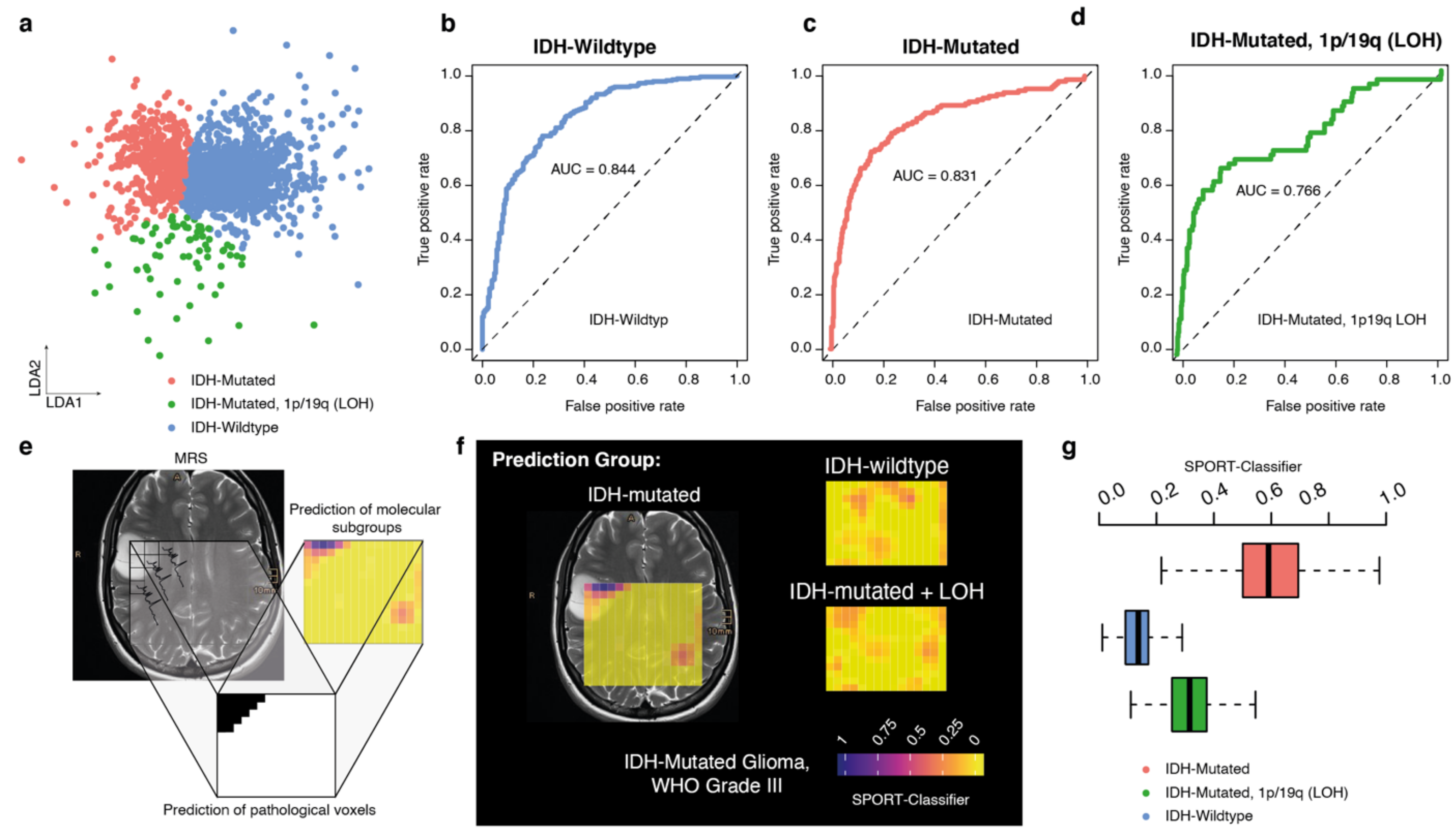
3.6. Prediction Model for Molecular Tumor Subgroups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, C.; Ganji, S.K.; DeBerardinis, R.J.; Hatanpaa, K.J.; Rakheja, D.; Kovacs, Z.; Yang, X.-L.; Mashimo, T.; Raisanen, J.M.; Marin-Valencia, I.; et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med. 2012, 18, 624–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diamandis, E.; Gabriel, C.P.S.; Würtemberger, U.; Guggenberger, K.; Urbach, H.; Staszewski, O.; Lassmann, S.; Schnell, O.; Grauvogel, J.; Mader, I.; et al. MR-spectroscopic imaging of glial tumors in the spotlight of the 2016 WHO classification. J. Neurooncol. 2018, 139, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Di Ieva, A.; Magnussen, J.S.; McIntosh, J.; Mulcahy, M.J.; Pardey, M.; Choi, C. Magnetic Resonance Spectroscopic Assessment of Isocitrate Dehydrogenase Status in Gliomas: The New Frontiers of Spectrobiopsy in Neurodiagnostics. World Neurosurg. 2020, 133, e421–e427. [Google Scholar] [CrossRef]
- Heiland, D.H.; Mader, I.; Schlosser, P.; Pfeifer, D.; Carro, M.S.; Lange, T.; Schwarzwald, R.; Vasilikos, I.; Urbach, H.; Weyerbrock, A. Integrative Network-based Analysis of Magnetic Resonance Spectroscopy and Genome Wide Expression in Glioblastoma multiforme. Sci. Rep. 2016, 6, 29052. [Google Scholar] [CrossRef]
- Zarinabad, N.; Abernethy, L.J.; Avula, S.; Davies, N.P.; Rodriguez Gutierrez, D.; Jaspan, T.; MacPherson, L.; Mitra, D.; Rose, H.E.L.; Wilson, M.; et al. Application of pattern recognition techniques for classification of pediatric brain tumors by in vivo 3T 1 H-MR spectroscopy—A multi-center study. Magn. Reson. Med. 2018, 79, 2359–2366. [Google Scholar] [CrossRef] [Green Version]
- Manias, K.A.; Harris, L.M.; Davies, N.P.; Natarajan, K.; MacPherson, L.; Foster, K.; Brundler, M.-A.; Hargrave, D.R.; Payne, G.S.; Leach, M.O.; et al. Prospective multicentre evaluation and refinement of an analysis tool for magnetic resonance spectroscopy of childhood cerebellar tumours. Pediatr. Radiol. 2018, 48, 1630–1641. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M.; Cummins, C.L.; Macpherson, L.; Sun, Y.; Natarajan, K.; Grundy, R.G.; Arvanitis, T.N.; Kauppinen, R.A.; Peet, A.C. Magnetic resonance spectroscopy metabolite profiles predict survival in paediatric brain tumours. Eur. J. Cancer 2013, 49, 457–464. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M. Adaptive Baseline Fitting for 1H MR Spectroscopy Analysis. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Eraslan, G.; Simon, L.M.; Mircea, M.; Mueller, N.S.; Theis, F.J. Single-cell RNA-seq denoising using a deep count autoencoder. Nat. Commun. 2019, 10, 390. [Google Scholar] [CrossRef] [Green Version]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.-A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018, 37, 38–44. [Google Scholar] [CrossRef]
- Kueckelhaus, J.; von Ehr, J.; Ravi, V.M.; Will, P.; Joseph, K.M.; Beck, J.; Hofmann, U.G.; Delev, D.; Schnell, O.; Heiland, H.D. Inferring spatially transient gene expression pattern from spatial transcriptomic studies. BioRxiv 2020. [Google Scholar] [CrossRef]
- Peet, A.C.; Arvanitis, T.N.; Leach, M.O.; Waldman, A.D. Functional imaging in adult and paediatric brain tumours. Nat. Rev. Clin. Oncol. 2012, 9, 700–711. [Google Scholar] [CrossRef]
- Perreault, S.; Ramaswamy, V.; Achrol, A.S.; Chao, K.; Liu, T.T.; Shih, D.; Remke, M.; Schubert, S.; Bouffet, E.; Fisher, P.G.; et al. MRI surrogates for molecular subgroups of medulloblastoma. Am. J. Neuroradiol. 2014, 35, 1263–1269. [Google Scholar] [CrossRef] [Green Version]
- Blüml, S.; Margol, A.S.; Sposto, R.; Kennedy, R.J.; Robison, N.J.; Vali, M.; Hung, L.T.; Muthugounder, S.; Finlay, J.L.; Erdreich-Epstein, A.; et al. Molecular subgroups of medulloblastoma identification using noninvasive magnetic resonance spectroscopy. Neuro-Oncology 2016, 18, 126–131. [Google Scholar] [CrossRef] [Green Version]
- Harris, L.M.; Davies, N.; Macpherson, L.; Foster, K.; Lateef, S.; Natarajan, K.; Sgouros, S.; Brundler, M.-A.; Arvanitis, T.N.; Grundy, R.G.; et al. The use of short-echo-time 1H MRS for childhood cerebellar tumours prior to histopathological diagnosis. Pediatr. Radiol. 2007, 37, 1101–1109. [Google Scholar] [CrossRef]
- Demerath, T.; Simon-Gabriel, C.P.; Kellner, E.; Schwarzwald, R.; Lange, T.; Heiland, D.H.; Reinacher, P.; Staszewski, O.; Mast, H.; Kiselev, V.G.; et al. Mesoscopic imaging of glioblastomas: Are diffusion, perfusion and spectroscopic measures influenced by the radiogenetic phenotype? Neuroradiol. J. 2017, 30, 36–47. [Google Scholar] [CrossRef]
- Bertholdo, D.; Watcharakorn, A.; Castillo, M. Brain proton magnetic resonance spectroscopy: Introduction and overview. Neuroimaging Clin. N. Am. 2013, 23, 359–380. [Google Scholar] [CrossRef]
- Lin, A.; Ross, B.D.; Harris, K.; Wong, W. Efficacy of proton magnetic resonance spectroscopy in neurological diagnosis and neurotherapeutic decision making. NeuroRx 2005, 2, 197–214. [Google Scholar] [CrossRef]
- Hajek, M.; Dezortova, M. Introduction to clinical in vivo MR spectroscopy. Eur. J. Radiol. 2008, 67, 185–193. [Google Scholar] [CrossRef]
- Chaumeil, M.M.; Lupo, J.M.; Ronen, S.M. Magnetic resonance (MR) metabolic imaging in glioma. Brain Pathol. 2015, 25, 769–780. [Google Scholar] [CrossRef]
- Kickingereder, P.; Götz, M.; Muschelli, J.; Wick, A.; Neuberger, U.; Shinohara, R.T.; Sill, M.; Nowosielski, M.; Schlemmer, H.-P.; Radbruch, A.; et al. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clin. Cancer Res. 2016, 22, 5765–5771. [Google Scholar] [CrossRef] [Green Version]
- Dowling, C.; Bollen, A.W.; Noworolski, S.M.; McDermott, M.W.; Barbaro, N.M.; Day, M.R.; Henry, R.G.; Chang, S.M.; Dillon, W.P.; Nelson, S.J.; et al. Preoperative proton MR spectroscopic imaging of brain tumors: Correlation with histopathologic analysis of resection specimens. Am. J. Neuroradiol. 2001, 22, 604–612. [Google Scholar]
- Bulik, M.; Jancalek, R.; Vanicek, J.; Skoch, A.; Mechl, M. Potential of MR spectroscopy for assessment of glioma grading. Clin. Neurol. Neurosurg. 2013, 115, 146–153. [Google Scholar] [CrossRef]
- Shimizu, H.; Kumabe, T.; Shirane, R.; Yoshimoto, T. Correlation between choline level measured by proton MR spectroscopy and Ki-67 labeling index in gliomas. Am. J. Neuroradiol. 2000, 21, 659–665. [Google Scholar] [PubMed]
- Heiland, D.H.; Gaebelein, A.; Börries, M.; Wörner, J.; Pompe, N.; Franco, P.; Heynckes, S.; Bartholomae, M.; Hailín, D.Ó.; Carro, M.S.; et al. Microenvironment-Derived Regulation of HIF Signaling Drives Transcriptional Heterogeneity in Glioblastoma Multiforme. Mol. Cancer Res. 2018, 16, 655–668. [Google Scholar] [CrossRef] [Green Version]
- Mader, I.; Rauer, S.; Gall, P.; Klose, U. (1)H MR spectroscopy of inflammation, infection and ischemia of the brain. Eur. J. Radiol. 2008, 67, 250–257. [Google Scholar] [CrossRef]
- Durmo, F.; Rydelius, A.; Cuellar Baena, S.; Askaner, K.; Lätt, J.; Bengzon, J.; Englund, E.; Chenevert, T.L.; Björkman-Burtscher, I.M.; Sundgren, P.C. Multivoxel 1H-MR Spectroscopy Biometrics for Preoprerative Differentiation Between Brain Tumors. Tomography 2018, 4, 172–181. [Google Scholar] [CrossRef]
- Majós, C.; Aguilera, C.; Alonso, J.; Julià-Sapé, M.; Castañer, S.; Sánchez, J.J.; Samitier, A.; León, A.; Rovira, A.; Arús, C. Proton MR spectroscopy improves discrimination between tumor and pseudotumoral lesion in solid brain masses. Am. J. Neuroradiol. 2009, 30, 544–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haris, M.; Cai, K.; Singh, A.; Hariharan, H.; Reddy, R. In vivo mapping of brain myo-inositol. Neuroimage 2011, 54, 2079–2085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manias, K.; Gill, S.K.; Zarinabad, N.; Davies, P.; English, M.; Ford, D.; MacPherson, L.; Nicklaus-Wollenteit, I.; Oates, A.; Solanki, G.; et al. Evaluation of the added value of 1H-magnetic resonance spectroscopy for the diagnosis of pediatric brain lesions in clinical practice. Neurooncol. Pract. 2018, 5, 18–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellström, J.; Romanos Zapata, R.; Libard, S.; Wikström, J.; Ortiz-Nieto, F.; Alafuzoff, I.; Raininko, R. The value of magnetic resonance spectroscopy as a supplement to MRI of the brain in a clinical setting. PLoS ONE 2018, 13, e0207336. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco, P.; Huebschle, I.; Simon-Gabriel, C.P.; Dacca, K.; Schnell, O.; Beck, J.; Mast, H.; Urbach, H.; Wuertemberger, U.; Prinz, M.; et al. Mapping of Metabolic Heterogeneity of Glioma Using MR-Spectroscopy. Cancers 2021, 13, 2417. https://doi.org/10.3390/cancers13102417
Franco P, Huebschle I, Simon-Gabriel CP, Dacca K, Schnell O, Beck J, Mast H, Urbach H, Wuertemberger U, Prinz M, et al. Mapping of Metabolic Heterogeneity of Glioma Using MR-Spectroscopy. Cancers. 2021; 13(10):2417. https://doi.org/10.3390/cancers13102417
Chicago/Turabian StyleFranco, Pamela, Irene Huebschle, Carl Philipp Simon-Gabriel, Karam Dacca, Oliver Schnell, Juergen Beck, Hansjoerg Mast, Horst Urbach, Urs Wuertemberger, Marco Prinz, and et al. 2021. "Mapping of Metabolic Heterogeneity of Glioma Using MR-Spectroscopy" Cancers 13, no. 10: 2417. https://doi.org/10.3390/cancers13102417





