Therapeutic Silencing of BCL-2 Using NK Cell-Derived Exosomes as a Novel Therapeutic Approach in Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Primary Cells
2.2. Lentiviral Vector Packaging, Titration, and Cell Transduction
2.3. Flow Cytometry
2.4. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.5. Analysis of NK Cell Cytotoxicity
2.6. Exosomes Isolation and Identification
2.7. Exosome Uptake
2.8. Live Cell Analysis
2.9. Caspase-9 Activation
2.10. Statistical Analysis
3. Results
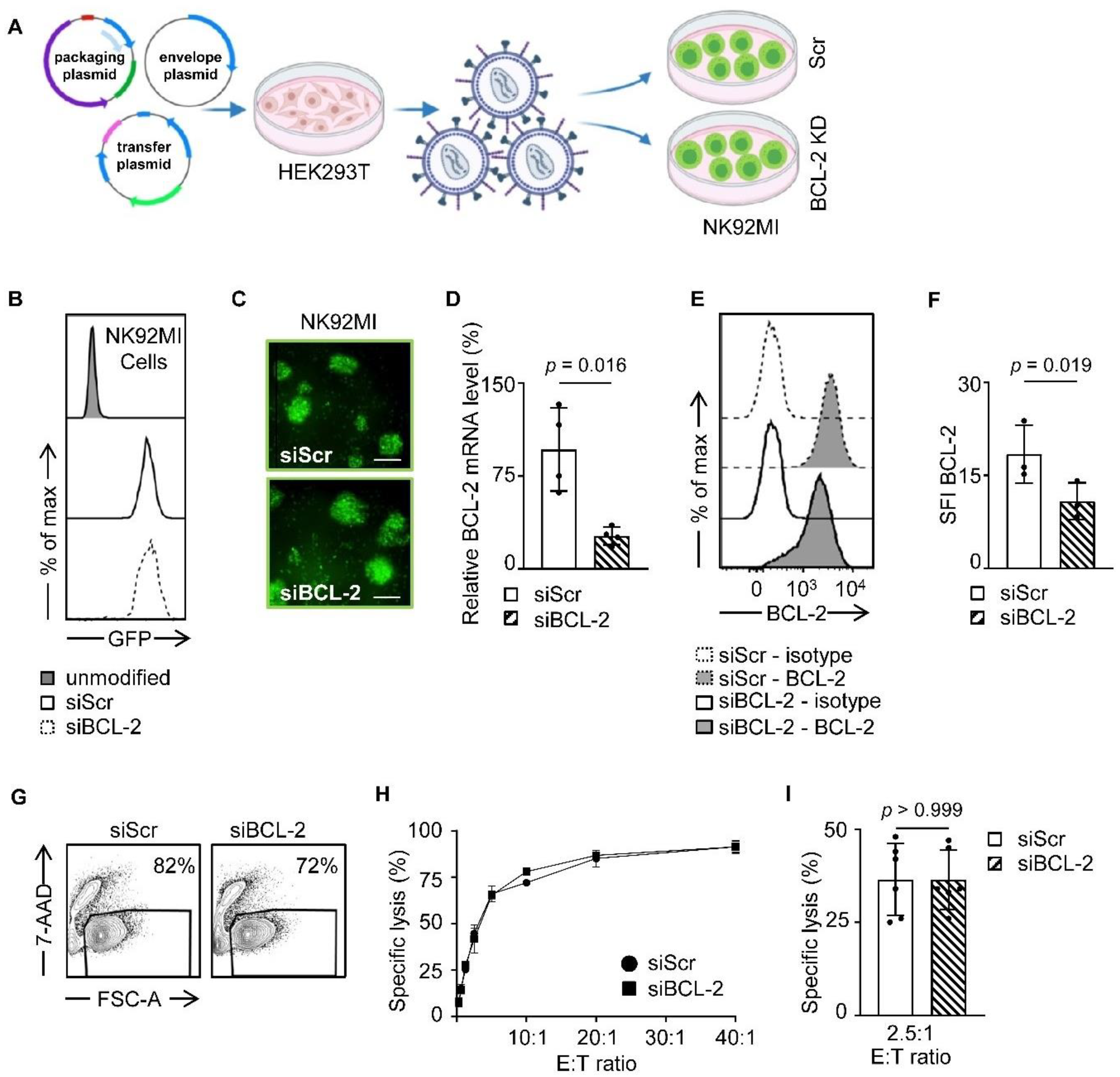
3.1. Generation of BCL-2 shRNA Producing NK92MI Cells
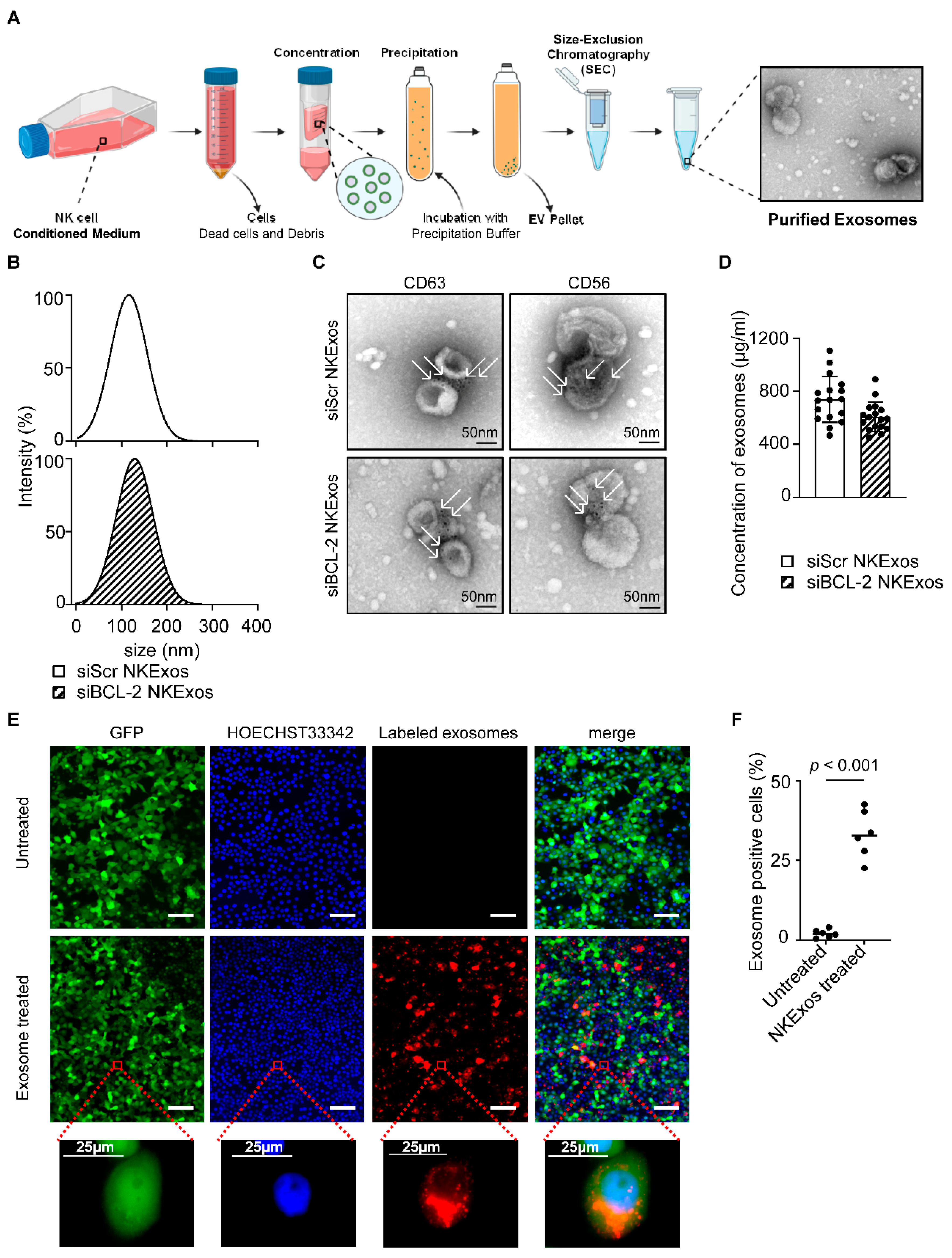
3.2. Characterization of NK Cell-Derived Exosomes (NKExos)
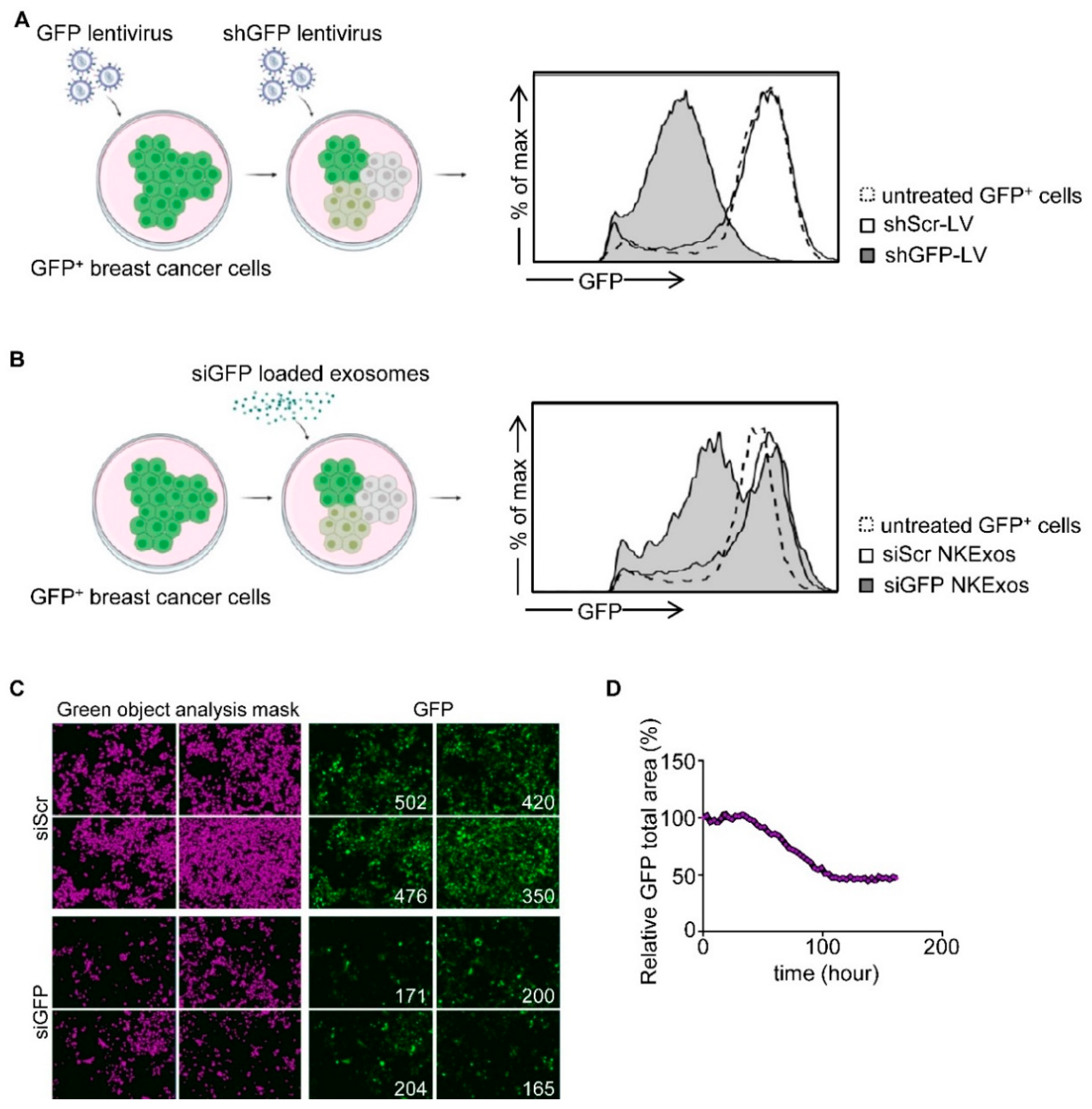
3.3. Silencing of GFP by siGFP Loaded NKExos
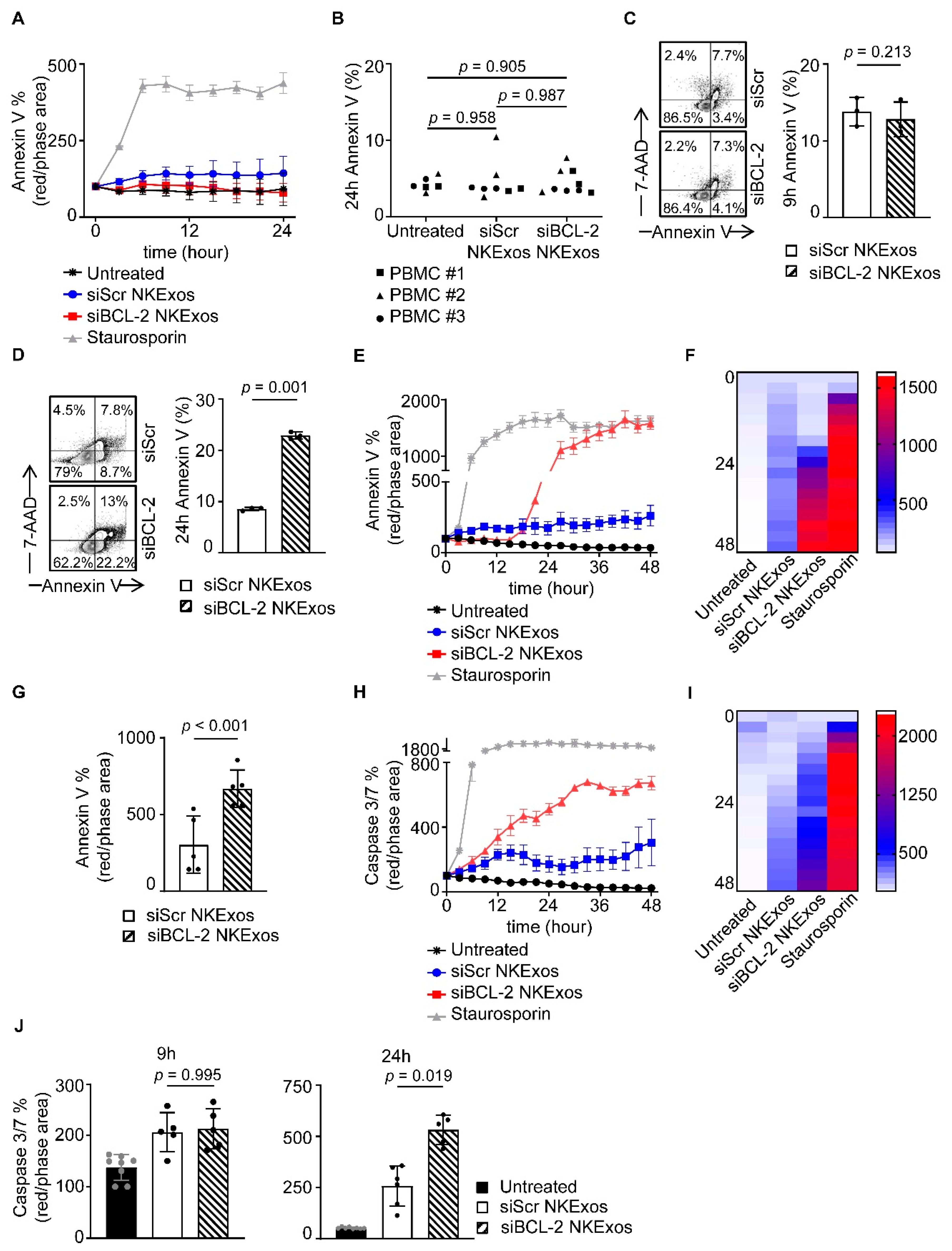
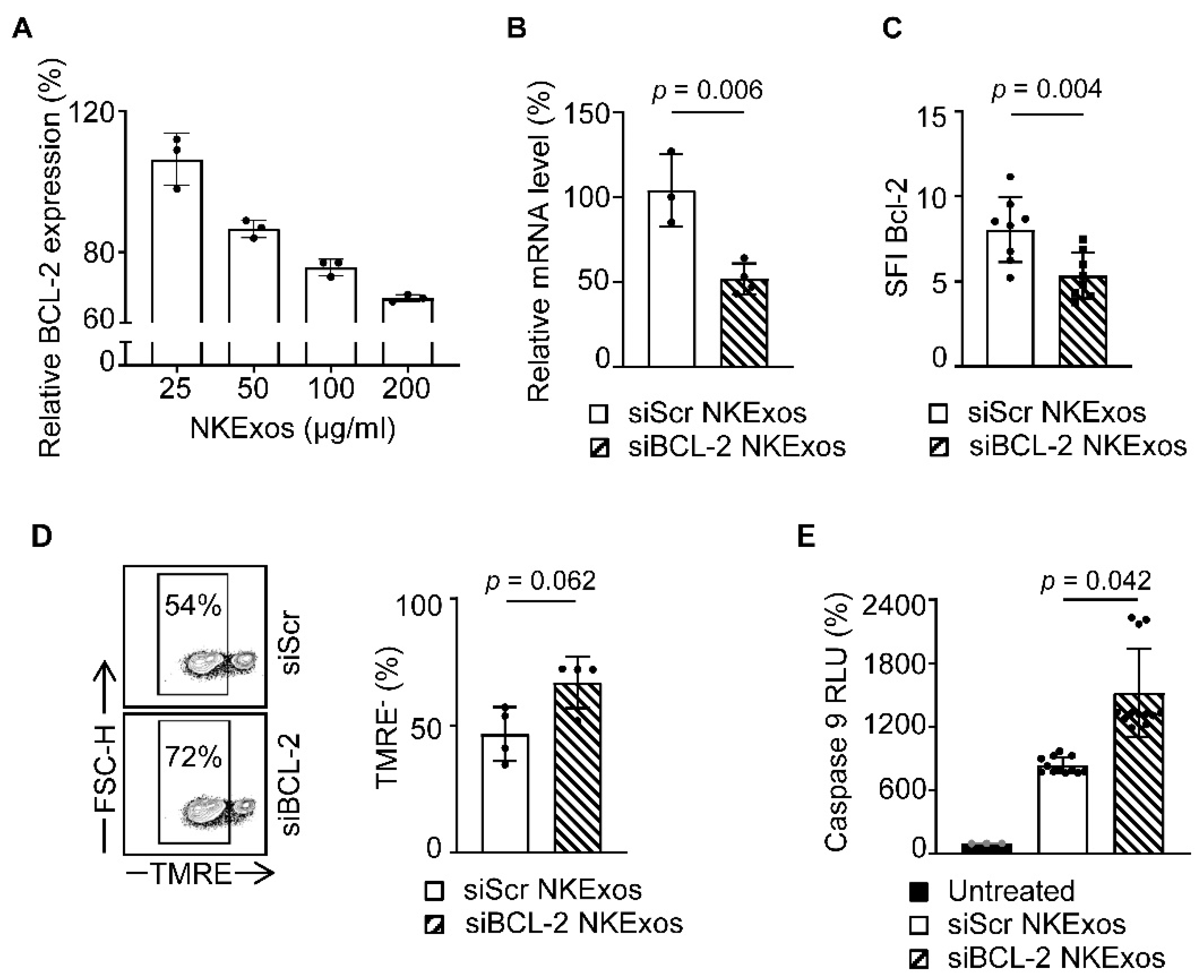
3.4. siBCL-2 NKExos Promote Apoptosis of Breast Cancer Cells
3.5. siBCL-2 NKExos Enhance the Intrinsic Apoptosis Pathway of Breast Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Merino, D.; Lok, S.W.; Visvader, J.E.; Lindeman, G.J. Targeting BCL-2 to enhance vulnerability to therapy in estrogen receptor-positive breast cancer. Oncogene 2016, 35, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.; et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef]
- Jain, V.; Kumar, H.; Anod, H.V.; Chand, P.; Gupta, N.V.; Dey, S.; Kesharwani, S.S. A review of nanotechnology-based approaches for breast cancer and triple-negative breast cancer. J. Control Release 2020, 326, 628–647. [Google Scholar] [CrossRef]
- Peng, L.; Jiang, J.; Tang, B.; Nice, E.C.; Zhang, Y.Y.; Xie, N. Managing therapeutic resistance in breast cancer: From the lncRNAs perspective. Theranostics 2020, 10, 10360–10377. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Shim, M.K.; Yang, S.; Moon, Y.; Song, S.; Choi, J.; Kim, J.; Kim, K. Combination of cancer-specific prodrug nanoparticle with Bcl-2 inhibitor to overcome acquired drug resistance. J. Control Release 2020. [Google Scholar] [CrossRef]
- Persidis, A. Cancer multidrug resistance. Nat. Biotechnol. 1999, 17, 94–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Seebacher, N.; Shi, H.; Kan, Q.; Duan, Z. Novel strategies to prevent the development of multidrug resistance (MDR) in cancer. Oncotarget 2017, 8, 84559–84571. [Google Scholar] [CrossRef] [PubMed]
- Vaillant, F.; Merino, D.; Lee, L.; Breslin, K.; Pal, B.; Ritchie, M.E.; Smyth, G.K.; Christie, M.; Phillipson, L.J.; Burns, C.J.; et al. Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell 2013, 24, 120–129. [Google Scholar] [CrossRef]
- Parton, M.; Dowsett, M.; Smith, I. Studies of apoptosis in breast cancer. BMJ 2001, 322, 1528–1532. [Google Scholar] [CrossRef]
- Akar, U.; Chaves-Reyez, A.; Barria, M.; Tari, A.; Sanguino, A.; Kondo, Y.; Kondo, S.; Arun, B.; Lopez-Berestein, G.; Ozpolat, B. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy 2008, 4, 669–679. [Google Scholar] [CrossRef]
- Teixeira, C.; Reed, J.C.; Pratt, M.A. Estrogen promotes chemotherapeutic drug resistance by a mechanism involving Bcl-2 proto-oncogene expression in human breast cancer cells. Cancer Res. 1995, 55, 3902–3907. [Google Scholar]
- Delbridge, A.R.; Grabow, S.; Strasser, A.; Vaux, D.L. Thirty years of BCL-2: Translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer 2016, 16, 99–109. [Google Scholar] [CrossRef]
- Juin, P.; Geneste, O.; Gautier, F.; Depil, S.; Campone, M. Decoding and unlocking the BCL-2 dependency of cancer cells. Nat. Rev. Cancer 2013, 13, 455–465. [Google Scholar] [CrossRef]
- Korycka-Wolowiec, A.; Wolowiec, D.; Kubiak-Mlonka, A.; Robak, T. Venetoclax in the treatment of chronic lymphocytic leukemia. Expert Opin. Drug Metab. Toxicol. 2019, 15, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Thol, F.; Ganser, A. Treatment of Relapsed Acute Myeloid Leukemia. Curr. Treat. Options Oncol. 2020, 21, 66. [Google Scholar] [CrossRef]
- Tekade, R.K.; Tekade, M.; Kesharwani, P.; D’Emanuele, A. RNAi-combined nano-chemotherapeutics to tackle resistant tumors. Drug Discov. Today 2016, 21, 1761–1774. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Farooqi, A.A.; Desai, N.N.; Qureshi, M.Z.; Librelotto, D.R.N.; Gasparri, M.L.; Bishayee, A.; Nabavi, S.M.; Curti, V.; Daglia, M. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 2018, 36, 328–334. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Andaloussi, S.E.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Batrakova, E.V.; Kim, M.S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control Release 2015, 219, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Y.; Jin, X.; Hu, D.; Xia, C.; Xu, H.; Hu, J. NK cell-derived exosomes carry miR-207 and alleviate depression-like symptoms in mice. J. Neuroinflamm. 2020, 17, 126. [Google Scholar] [CrossRef] [PubMed]
- Neviani, P.; Wise, P.M.; Murtadha, M.; Liu, C.W.; Wu, C.H.; Jong, A.Y.; Seeger, R.C.; Fabbri, M. Natural Killer-Derived Exosomal miR-186 Inhibits Neuroblastoma Growth and Immune Escape Mechanisms. Cancer Res. 2019, 79, 1151–1164. [Google Scholar] [CrossRef]
- Di Pace, A.L.; Tumino, N.; Besi, F.; Alicata, C.; Conti, L.A.; Munari, E.; Maggi, E.; Vacca, P.; Moretta, L. Characterization of Human NK Cell-Derived Exosomes: Role of DNAM1 Receptor In Exosome-Mediated Cytotoxicity Against Tumor. Cancers 2020, 12, 661. [Google Scholar] [CrossRef]
- Fais, S. NK cell-released exosomes: Natural nanobullets against tumors. Oncoimmunology 2013, 2, e22337. [Google Scholar] [CrossRef]
- Jong, A.Y.; Wu, C.H.; Li, J.; Sun, J.; Fabbri, M.; Wayne, A.S.; Seeger, R.C. Large-scale isolation and cytotoxicity of extracellular vesicles derived from activated human natural killer cells. J. Extracell. Vesicles 2017, 6, 1294368. [Google Scholar] [CrossRef]
- Wen, C.; Seeger, R.C.; Fabbri, M.; Wang, L.; Wayne, A.S.; Jong, A.Y. Biological roles and potential applications of immune cell-derived extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1400370. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Kalimuthu, S.; Gangadaran, P.; Oh, J.M.; Lee, H.W.; Baek, S.H.; Jeong, S.Y.; Lee, S.W.; Lee, J.; Ahn, B.C. Exosomes Derived From Natural Killer Cells Exert Therapeutic Effect in Melanoma. Theranostics 2017, 7, 2732–2745. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.A.; Dykxhoorn, D.M.; Palliser, D.; Mizuno, H.; Yu, E.Y.; An, D.S.; Sabatini, D.M.; Chen, I.S.; Hahn, W.C.; Sharp, P.A.; et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 2003, 9, 493–501. [Google Scholar] [CrossRef]
- Schmied, B.J.; Riegg, F.; Zekri, L.; Grosse-Hovest, L.; Buhring, H.J.; Jung, G.; Salih, H.R. An Fc-Optimized CD133 Antibody for Induction of Natural Killer Cell Reactivity against Colorectal Cancer. Cancers 2019, 11, 789. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, L.; Wang, S.; Han, Q.; Zhao, R.C. Exosomes secreted by mesenchymal stem cells promote endothelial cell angiogenesis by transferring miR-125a. J. Cell Sci. 2016, 129, 2182–2189. [Google Scholar] [CrossRef]
- Lugini, L.; Cecchetti, S.; Huber, V.; Luciani, F.; Macchia, G.; Spadaro, F.; Paris, L.; Abalsamo, L.; Colone, M.; Molinari, A.; et al. Immune surveillance properties of human NK cell-derived exosomes. J. Immunol. 2012, 189, 2833–2842. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Huang, W.; Fan, M. Emerging therapies for breast cancer. J. Hematol. Oncol. 2017, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Radosa, J.C.; Stotz, L.; Muller, C.; Kaya, A.C.; Solomayer, E.F.; Radosa, M.P. Clinical Data on Immunotherapy in Breast Cancer. Breast Care 2020, 15, 450–469. [Google Scholar] [CrossRef] [PubMed]
- Polk, A.; Svane, I.M.; Andersson, M.; Nielsen, D. Checkpoint inhibitors in breast cancer—Current status. Cancer Treat. Rev. 2018, 63, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, A.; Cui, H.; Caligiuri, M.A.; Yu, J. Chimeric antigen receptor-engineered natural killer cells for cancer immunotherapy. J. Hematol. Oncol. 2020, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, A.; Jimeno, A. Bispecific antibodies for cancer therapy: A review. Pharmacol. Ther. 2018, 185, 122–134. [Google Scholar] [CrossRef]
- Zekri, L.; Vogt, F.; Osburg, L.; Muller, S.; Kauer, J.; Manz, T.; Pflugler, M.; Maurer, A.; Heitmann, J.S.; Hagelstein, I.; et al. An IgG-based bispecific antibody for improved dual targeting in PSMA-positive cancer. EMBO Mol. Med. 2021, 13, e11902. [Google Scholar] [CrossRef] [PubMed]
- Arnould, L.; Gelly, M.; Penault-Llorca, F.; Benoit, L.; Bonnetain, F.; Migeon, C.; Cabaret, V.; Fermeaux, V.; Bertheau, P.; Garnier, J.; et al. Trastuzumab-based treatment of HER2-positive breast cancer: An antibody-dependent cellular cytotoxicity mechanism? Br. J. Cancer 2006, 94, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Vadlamudi, R.K.; Adam, L. Apoptosis in mammary gland and cancer. Endocr. Relat. Cancer 2000, 7, 257–269. [Google Scholar] [CrossRef]
- Du, C.; Zhang, X.; Yao, M.; Lv, K.; Wang, J.; Chen, L.; Chen, Y.; Wang, S.; Fu, P. Bcl-2 promotes metastasis through the epithelial-to-mesenchymal transition in the BCap37 medullary breast cancer cell line. Oncol. Lett. 2018, 15, 8991–8998. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Wickman, E.; DeRenzo, C.; Gottschalk, S. CAR T Cell Therapy for Solid Tumors: Bright Future or Dark Reality? Mol. Ther. 2020, 28, 2320–2339. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.J.; Kim, O.Y.; Gho, Y.S. Extracellular vesicles as emerging intercellular communicasomes. BMB Rep. 2014, 47, 531–539. [Google Scholar] [CrossRef]
- Ngoi, N.Y.L.; Choong, C.; Lee, J.; Bellot, G.; Wong, A.L.A.; Goh, B.C.; Pervaiz, S. Targeting Mitochondrial Apoptosis to Overcome Treatment Resistance in Cancer. Cancers 2020, 12, 574. [Google Scholar] [CrossRef]
- Mato, A.R.; Thompson, M.; Allan, J.N.; Brander, D.M.; Pagel, J.M.; Ujjani, C.S.; Hill, B.T.; Lamanna, N.; Lansigan, F.; Jacobs, R.; et al. Real-world outcomes and management strategies for venetoclax-treated chronic lymphocytic leukemia patients in the United States. Haematologica 2018, 103, 1511–1517. [Google Scholar] [CrossRef] [PubMed]
- Tekedereli, I.; Alpay, S.N.; Akar, U.; Yuca, E.; Ayugo-Rodriguez, C.; Han, H.D.; Sood, A.K.; Lopez-Berestein, G.; Ozpolat, B. Therapeutic Silencing of Bcl-2 by Systemically Administered siRNA Nanotherapeutics Inhibits Tumor Growth by Autophagy and Apoptosis and Enhances the Efficacy of Chemotherapy in Orthotopic Xenograft Models of ER (−) and ER (+) Breast Cancer. Mol. Ther. Nucleic Acids 2013, 2, e121. [Google Scholar] [CrossRef]
- Sun, W.; Liu, X.Y.; Ma, L.L.; Lu, Z.L. Tumor Targeting Gene Vector for Visual Tracking of Bcl-2 siRNA Transfection and Anti-Tumor Therapy. ACS Appl. Mater. Interfaces 2020, 12, 10193–10201. [Google Scholar] [CrossRef]
- Jiang, M.; Milner, J. Bcl-2 constitutively suppresses p53-dependent apoptosis in colorectal cancer cells. Genes Dev. 2003, 17, 832–837. [Google Scholar] [CrossRef]
- Ocker, M.; Neureiter, D.; Lueders, M.; Zopf, S.; Ganslmayer, M.; Hahn, E.G.; Herold, C.; Schuppan, D. Variants of bcl-2 specific siRNA for silencing antiapoptotic bcl-2 in pancreatic cancer. Gut 2005, 54, 1298–1308. [Google Scholar] [CrossRef]
- Wu, X.; Zheng, Y.; Yang, D.; Chen, T.; Feng, B.; Weng, J.; Wang, J.; Zhang, K.; Zhang, X. A strategy using mesoporous polymer nanospheres as nanocarriers of Bcl-2 siRNA towards breast cancer therapy. J. Mater. Chem. B 2019, 7, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Xue, S.; Sun, J.; Zhang, W.; Hu, P.; Ji, L.; Mao, Z. Effective combination treatment of lung cancer cells by single vehicular delivery of siRNA and different anticancer drugs. Int. J. Nanomed. 2016, 11, 4609–4624. [Google Scholar] [CrossRef] [PubMed]
- Reddy, T.L.; Garikapati, K.R.; Reddy, S.G.; Reddy, B.V.; Yadav, J.S.; Bhadra, U.; Bhadra, M.P. Simultaneous delivery of Paclitaxel and Bcl-2 siRNA via pH-Sensitive liposomal nanocarrier for the synergistic treatment of melanoma. Sci. Rep. 2016, 6, 35223. [Google Scholar] [CrossRef] [PubMed]
- Rossi, J.J.; Rossi, D.J. siRNA Drugs: Here to Stay. Mol. Ther. 2021, 29, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Kaban, K.; Salva, E.; Akbuga, J. The effects of chitosan/miR-200c nanoplexes on different stages of cancers in breast cancer cell lines. Eur. J. Pharm. Sci. 2016, 95, 103–110. [Google Scholar] [CrossRef]
- Kaban, K.; Salva, E.; Akbuga, J. In Vitro Dose Studies on Chitosan Nanoplexes for microRNA Delivery in Breast Cancer Cells. Nucleic Acid Ther. 2017, 27, 45–55. [Google Scholar] [CrossRef]
- Kaban, K.; Salva, E.; Akbuga, J. Modulation of the dual-faced effects of miR-141 with chitosan/miR-141 nanoplexes in breast cancer cells. J. Gene Med. 2019, 21, e3116. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Liew, M.W.J.; Venkatraman, S. Recent Advances in Chitosan-Based Carriers for Gene Delivery. Mar. Drugs 2019, 17, 381. [Google Scholar] [CrossRef]
- Setten, R.L.; Rossi, J.J.; Han, S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Bahal, R.; Rasmussen, T.P.; Manautou, J.E.; Zhong, X.B. The growth of siRNA-based therapeutics: Updated clinical studies. Biochem. Pharmacol. 2021, 114432. [Google Scholar] [CrossRef] [PubMed]
- El Andaloussi, S.; Lakhal, S.; Mager, I.; Wood, M.J. Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug Deliv. Rev. 2013, 65, 391–397. [Google Scholar] [CrossRef]
- Chen, S.; Tang, Y.; Liu, Y.; Zhang, P.; Lv, L.; Zhang, X.; Jia, L.; Zhou, Y. Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif. 2019, 52, e12669. [Google Scholar] [CrossRef]
- Morishita, M.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Pharmacokinetics of Exosomes-An Important Factor for Elucidating the Biological Roles of Exosomes and for the Development of Exosome-Based Therapeutics. J. Pharm. Sci. 2017, 106, 2265–2269. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Lin, Z.X.; Jiang, X.Y.; Yu, X.Y. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 2018, 39, 542–551. [Google Scholar] [CrossRef]
- Sancho-Albero, M.; Navascues, N.; Mendoza, G.; Sebastian, V.; Arruebo, M.; Martin-Duque, P.; Santamaria, J. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J. Nanobiotechnol. 2019, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Shoae-Hassani, A.; Hamidieh, A.A.; Behfar, M.; Mohseni, R.; Mortazavi-Tabatabaei, S.A.; Asgharzadeh, S. NK Cell-derived Exosomes From NK Cells Previously Exposed to Neuroblastoma Cells Augment the Antitumor Activity of Cytokine-activated NK Cells. J. Immunother. 2017, 40, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Hu, W.; Chen, H.; Shou, X.; Ye, T.; Xu, Y. Cocktail Strategy Based on NK Cell-Derived Exosomes and Their Biomimetic Nanoparticles for Dual Tumor Therapy. Cancers 2019, 11, 1560. [Google Scholar] [CrossRef] [PubMed]
| shRNA | Reporter | Vector ID | Target Sequences (5′-3′) |
| shScrambled | None | Addgene #1864 [33] | CCTAAGGTTAAGTCGCCCTCG |
| shGFP | None | Addgene #30323 [34] | GCAAGCTGACCCTGAAGTTCAT |
| shScrambled | eGFP | VB190131-1072gtc | CCTAAGGTTAAGTCGCCCTCG |
| shBCL-2 | eGFP | VB190131-1073stf | CGGGAGATAGTGATGAAGTACATCCATTA |
| Expression Vectors | Reporter | Vector ID | |
| Stuffer | eGFP | VB181209-1075efx | |
| pCMV-dR8.2 dvpr | - | Addgene #8455 [35] | |
| pCMV-VSV-G | - | Addgene #8454 [35] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaban, K.; Hinterleitner, C.; Zhou, Y.; Salva, E.; Kantarci, A.G.; Salih, H.R.; Märklin, M. Therapeutic Silencing of BCL-2 Using NK Cell-Derived Exosomes as a Novel Therapeutic Approach in Breast Cancer. Cancers 2021, 13, 2397. https://doi.org/10.3390/cancers13102397
Kaban K, Hinterleitner C, Zhou Y, Salva E, Kantarci AG, Salih HR, Märklin M. Therapeutic Silencing of BCL-2 Using NK Cell-Derived Exosomes as a Novel Therapeutic Approach in Breast Cancer. Cancers. 2021; 13(10):2397. https://doi.org/10.3390/cancers13102397
Chicago/Turabian StyleKaban, Kübra, Clemens Hinterleitner, Yanjun Zhou, Emine Salva, Ayse Gülten Kantarci, Helmut R. Salih, and Melanie Märklin. 2021. "Therapeutic Silencing of BCL-2 Using NK Cell-Derived Exosomes as a Novel Therapeutic Approach in Breast Cancer" Cancers 13, no. 10: 2397. https://doi.org/10.3390/cancers13102397
APA StyleKaban, K., Hinterleitner, C., Zhou, Y., Salva, E., Kantarci, A. G., Salih, H. R., & Märklin, M. (2021). Therapeutic Silencing of BCL-2 Using NK Cell-Derived Exosomes as a Novel Therapeutic Approach in Breast Cancer. Cancers, 13(10), 2397. https://doi.org/10.3390/cancers13102397






