BRAF/EZH2 Signaling Represses miR-129-5p Inhibition of SOX4 Thereby Modulating BRAFi Resistance in Melanoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sets
2.2. Cell Culture
2.3. miRNA Detection by Quantitative Real-Time PCR
2.4. Transcriptional Analysis by qRT-PCR
2.5. DNA Constructs and siRNA
2.6. Transfections
2.7. Immunoblot Snalyses
2.8. Cell Viability Assay
2.9. Cell Growth Assay
2.10. Cell Cycle Analysis
2.11. 3D Spheroid Growth Assay
2.12. Luciferase Reporter Assay
2.13. Statistical Analyses
3. Results
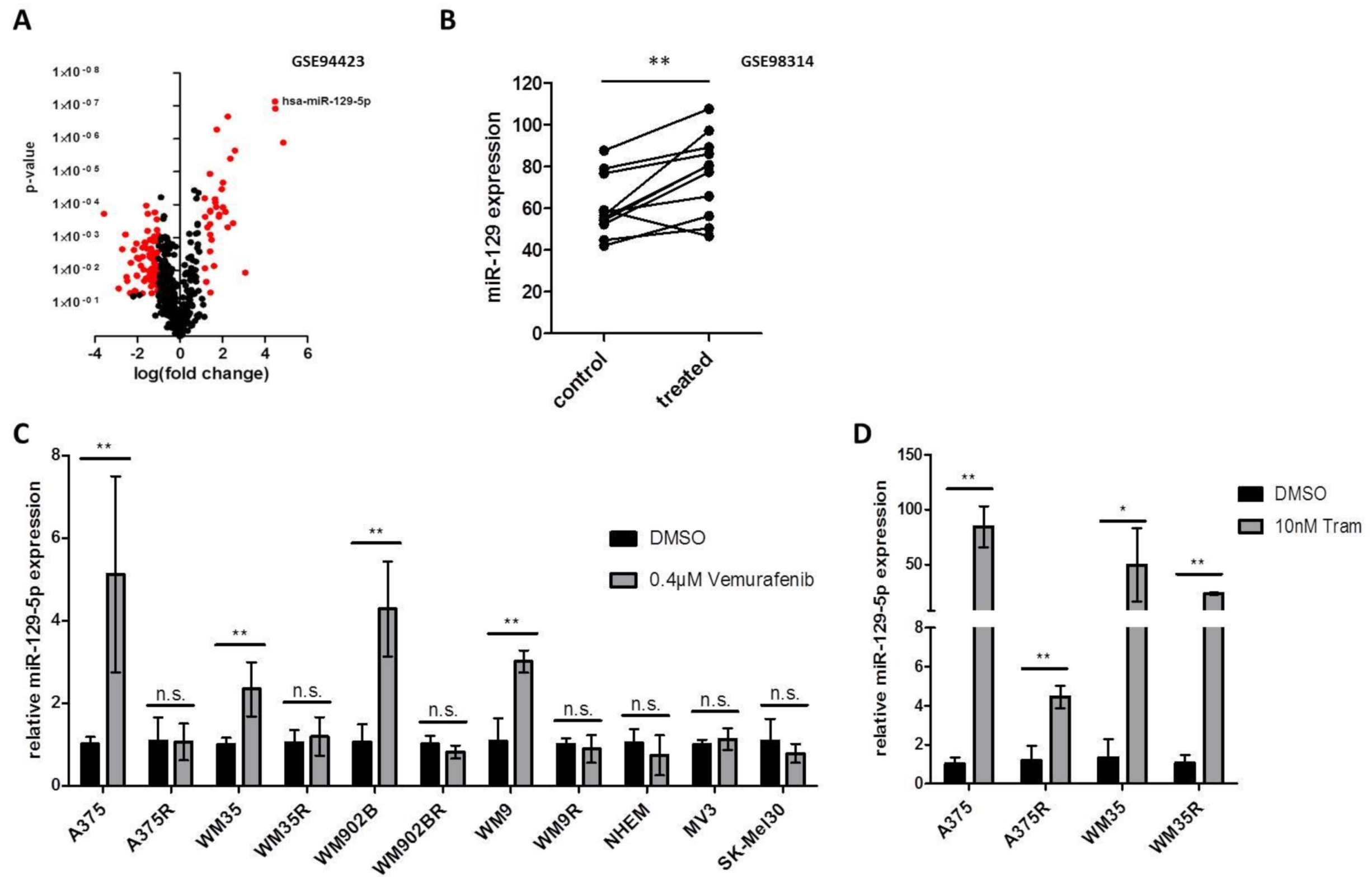
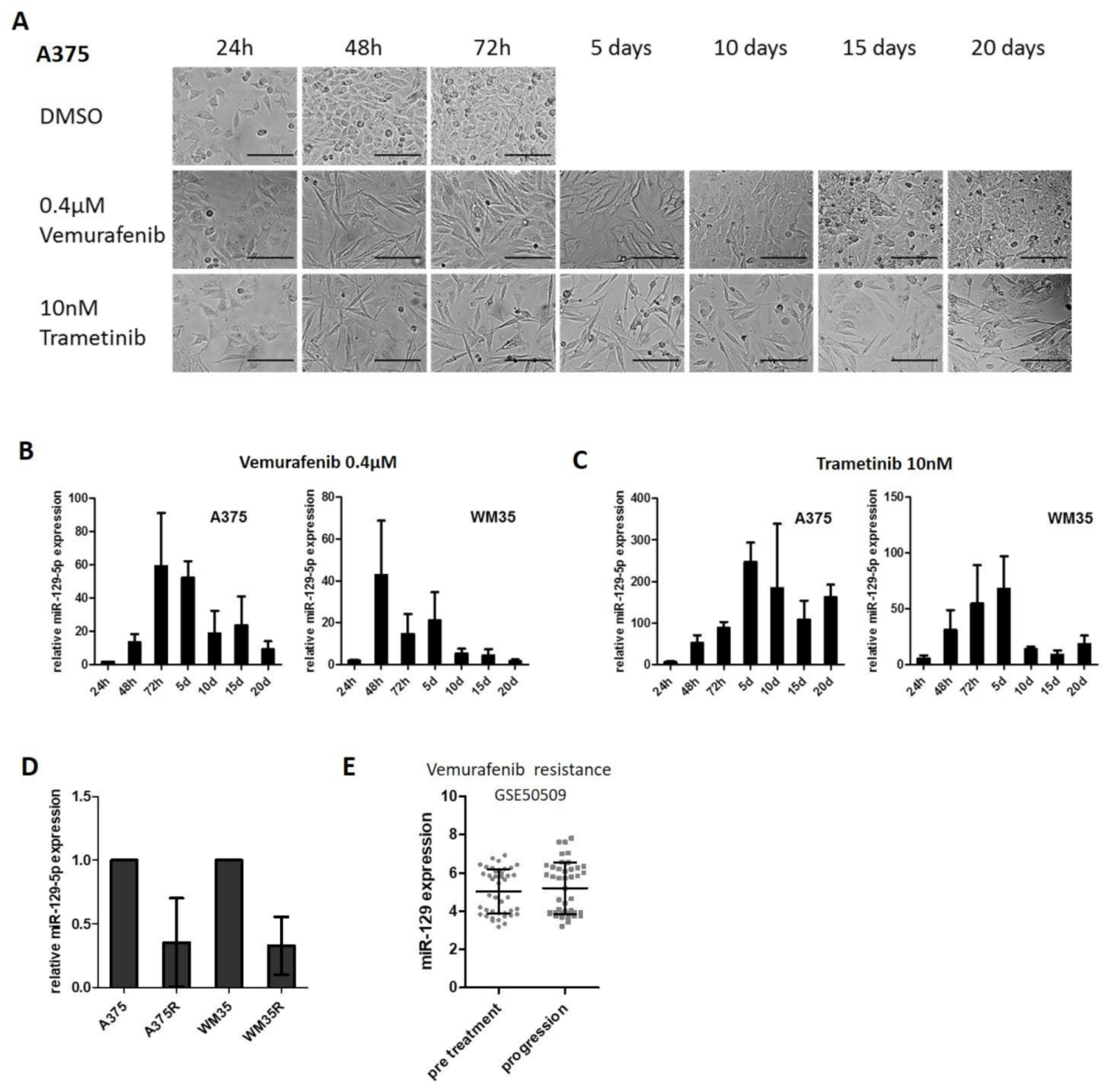
3.1. Expression of miR-129-5p Increases during BRAFi and MEKi Treatment
3.2. miR-129-5p Expression Decreases During Emergence of Resistance to BRAFi
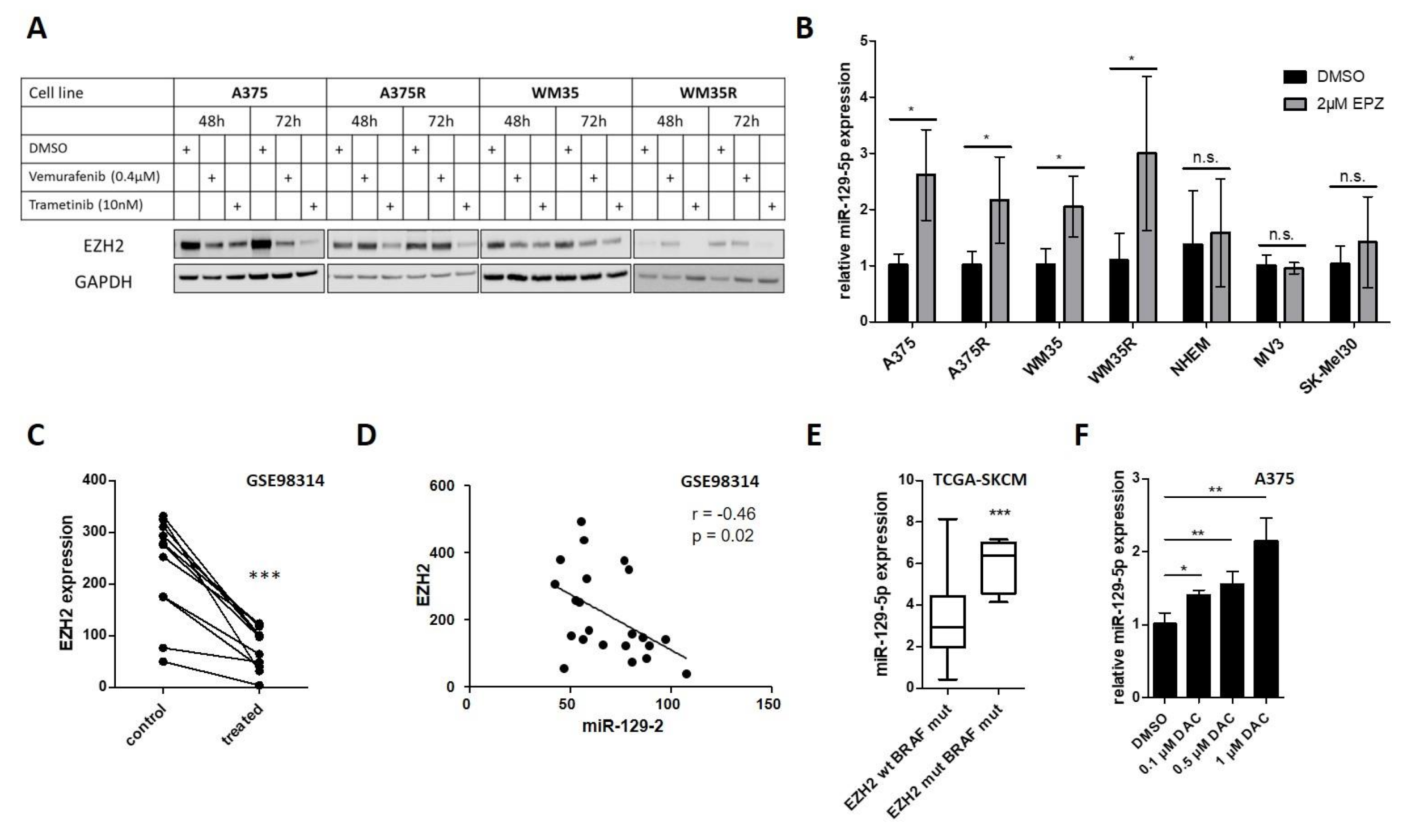
3.3. EZH2 Suppresses miR-129-5p Expression Downstream of Constitutive Active BRAF Signaling
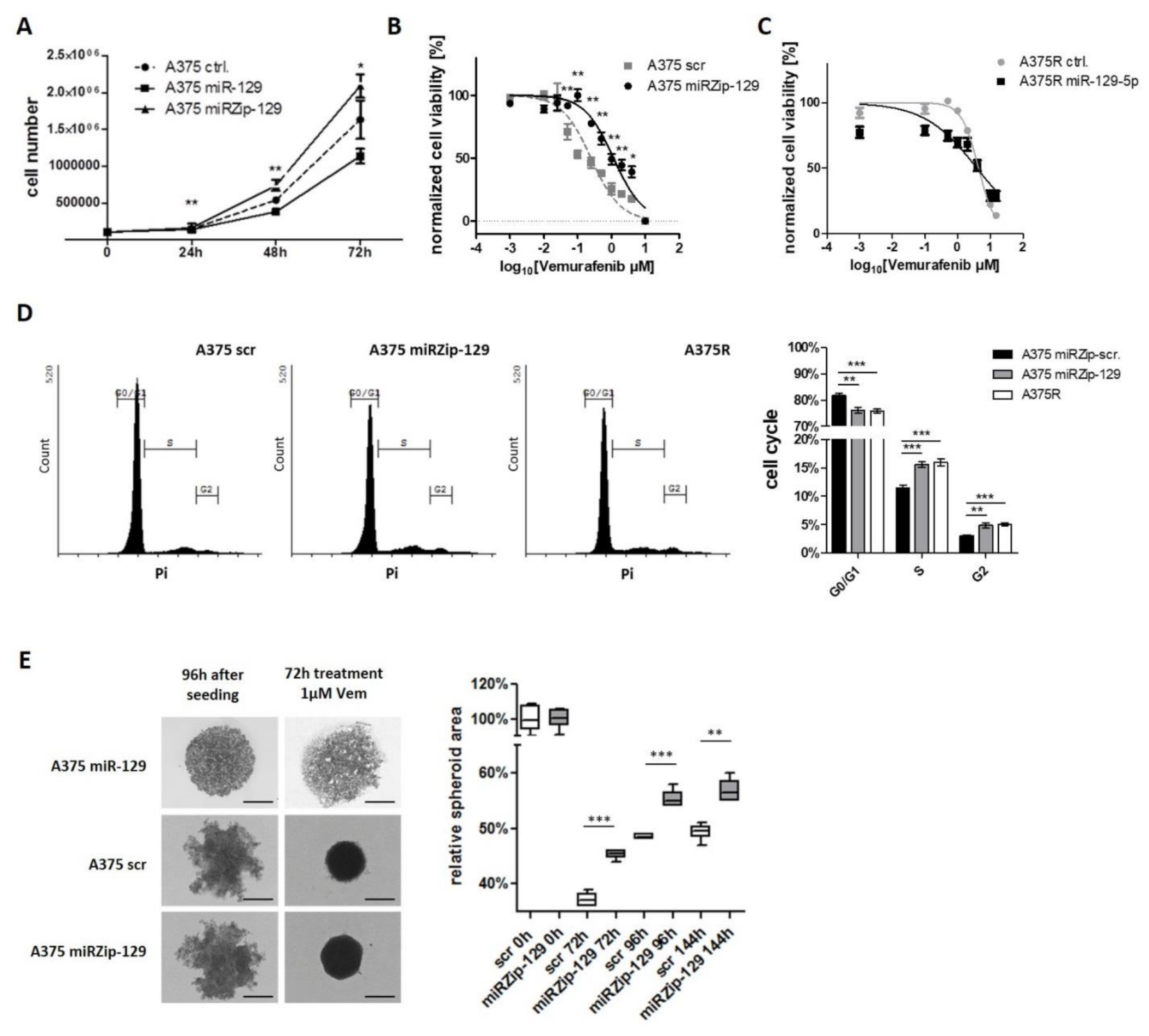
3.4. miR-129-5p Acts as Tumor Suppressor In Vitro and in a 3D Spheroid Model
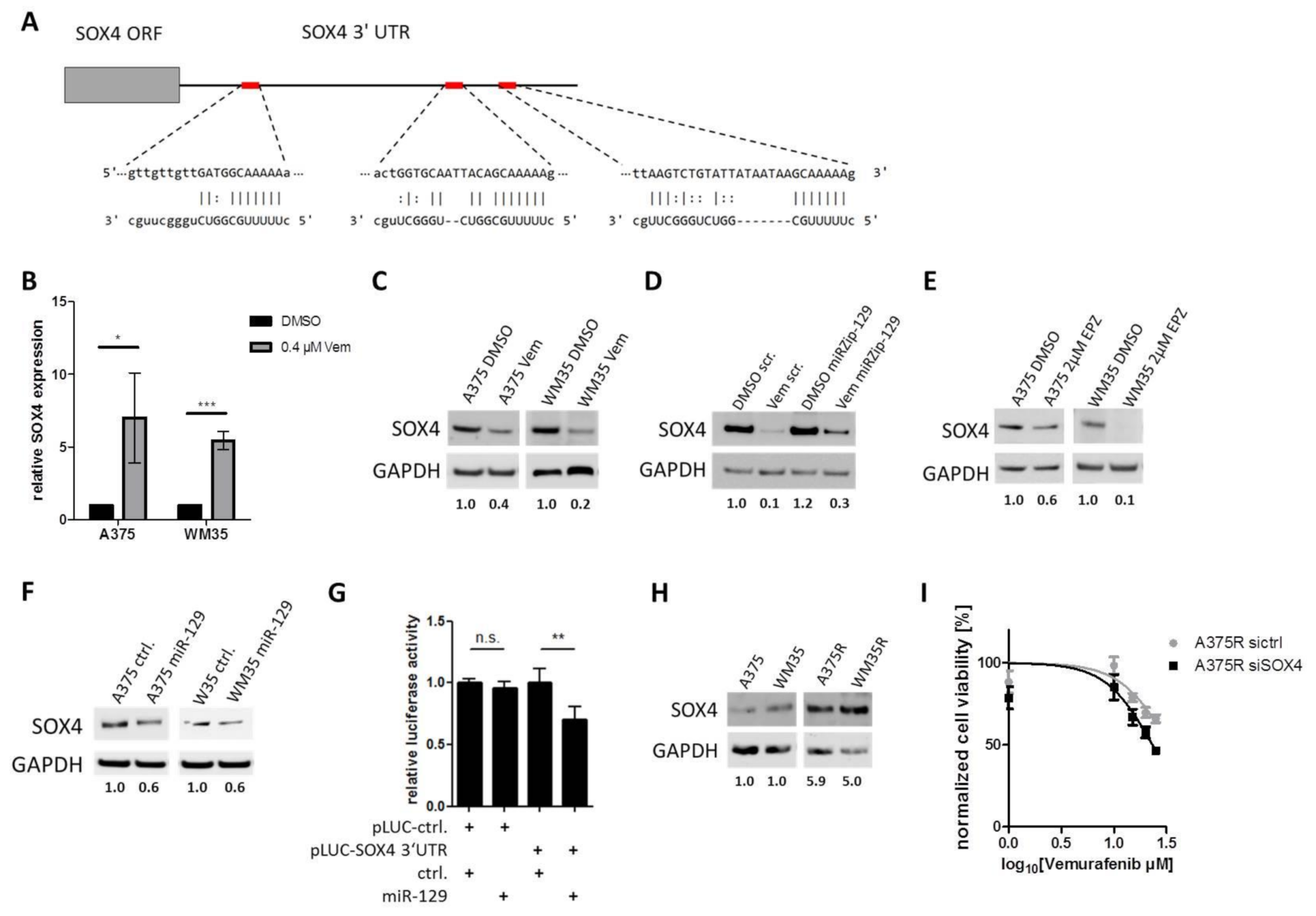
3.5. SOX4 is a Targeted by miR-129-5p During BRAFi Response
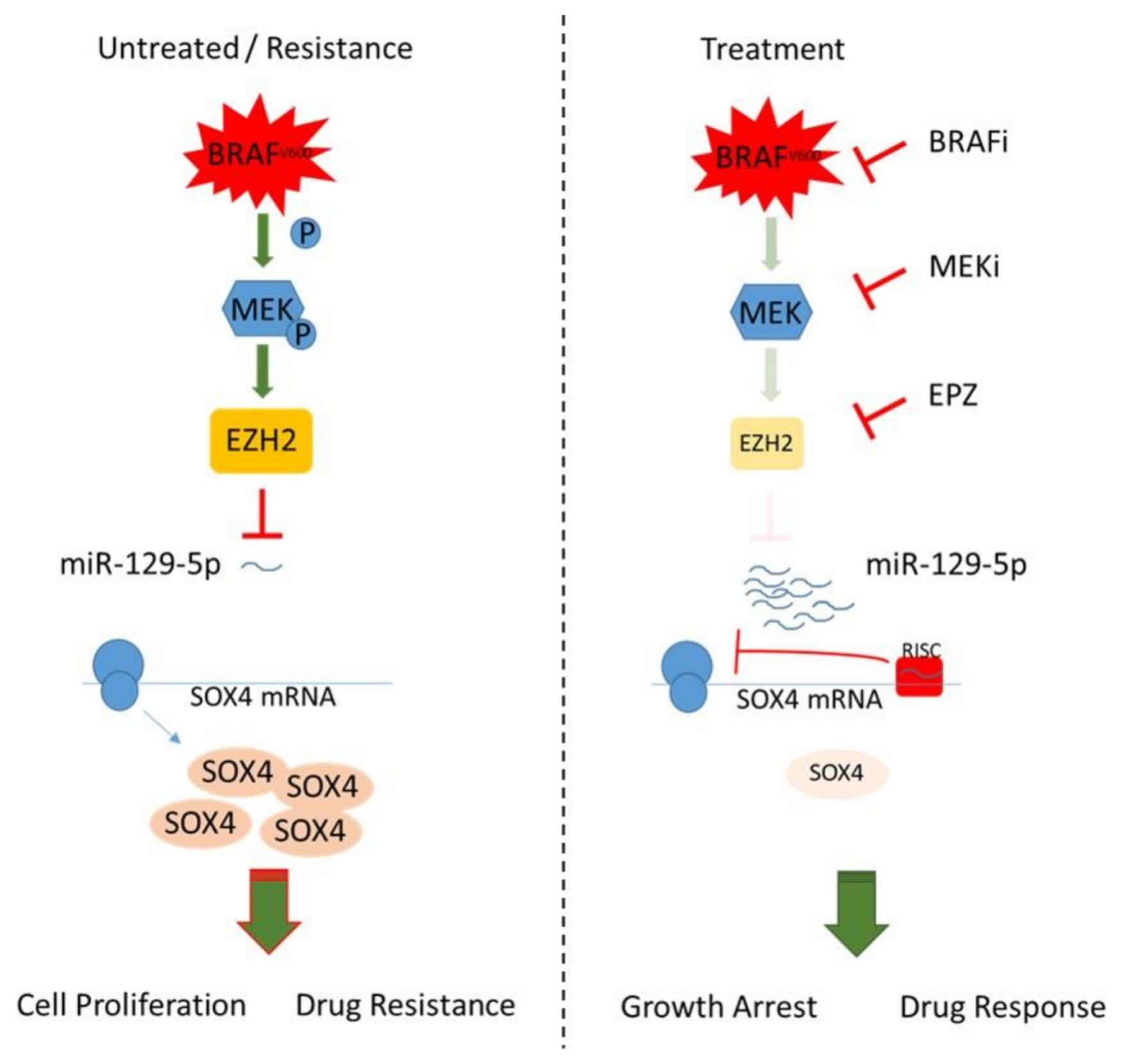
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Markovic, S.N.; Erickson, L.A.; Flotte, T.J.; Kottschade, L.A.; McWilliams, R.R.; Jakub, J.W.; Farley, D.R.; Tran, N.V.; Schild, S.E.; Olivier, K.R.; et al. Metastatic malignant melanoma. Gancer Ital. Derm. Venereol. 2009, 144, 1–26. [Google Scholar]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Sini, M.C.; Doneddu, V.; Paliogiannis, P.; Casula, M.; Colombino, M.; Manca, A.; Botti, G.; Ascierto, P.A.; Lissia, A.; Cossu, A.; et al. Genetic alterations in main candidate genes during melanoma progression. Oncotarget 2018, 9, 8531–8541. [Google Scholar] [CrossRef]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017, 545, 175–180. [Google Scholar] [CrossRef]
- Rabbie, R.; Ferguson, P.; Molina-Aguilar, C.; Adams, D.J.; Robles-Espinoza, C.D. Melanoma subtypes: Genomic profiles, prognostic molecular markers and therapeutic possibilities. J. Pathol. 2019, 247, 539–551. [Google Scholar] [CrossRef]
- Garman, B.; Anastopoulos, I.N.; Krepler, C.; Brafford, P.; Sproesser, K.; Jiang, Y.; Wubbenhorst, B.; Amaravadi, R.; Bennett, J.; Beqiri, M.; et al. Genetic and genomic characterization of 462 melanoma patient-derived xenografts, tumor biopsies, and cell lines. Cell Rep. 2017, 21, 1936–1952. [Google Scholar] [CrossRef]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A landscape of driver mutations in melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef]
- Sullivan, R.J.; Flaherty, K.T. Resistance to braf-targeted therapy in melanoma. Eur. J. Cancer 2013, 49, 1297–1304. [Google Scholar] [CrossRef]
- Larkin, J.; Ascierto, P.A.; Dreno, B.; Atkinson, V.; Liszkay, G.; Maio, M.; Mandala, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Combined vemurafenib and cobimetinib in braf-mutated melanoma. N. Engl. J. Med. 2014, 371, 1867–1876. [Google Scholar] [CrossRef]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef]
- Long, G.V.; Weber, J.S.; Infante, J.R.; Kim, K.B.; Daud, A.; Gonzalez, R.; Sosman, J.A.; Hamid, O.; Schuchter, L.; Cebon, J.; et al. Overall survival and durable responses in patients with braf v600 mutant metastatic melanoma receiving dabrafenib combined with trametinib (vol 34, pg 871, 2016). J. Clin. Oncol. 2019, 37, 355-355. [Google Scholar]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with braf v600e mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef]
- Nazarian, R.; Shi, H.B.; Wang, Q.; Kong, X.J.; Koya, R.C.; Lee, H.; Chen, Z.G.; Lee, M.K.; Attar, N.; Sazegar, H.; et al. Melanomas acquire resistance tob-raf(v600e) inhibition by rtk or n-ras upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Persaud, Y.; Janakiraman, M.; Kong, X.J.; Ng, C.; Moriceau, G.; Shi, H.B.; Atefi, M.; Titz, B.; Gabay, M.T.; et al. Raf inhibitor resistance is mediated by dimerization of aberrantly spliced braf(v600e). Nature 2011, 480, 387–390. [Google Scholar] [CrossRef]
- Shi, H.; Moriceau, G.; Kong, X.; Lee, M.K.; Lee, H.; Koya, R.C.; Ng, C.; Chodon, T.; Scolyer, R.A.; Dahlman, K.B.; et al. Melanoma whole-exome sequencing identifies (v600e)b-raf amplification-mediated acquired b-raf inhibitor resistance. Nat. Commun. 2012, 3, 724. [Google Scholar] [CrossRef]
- Johannessen, C.M.; Boehm, J.S.; Kim, S.Y.; Thomas, S.R.; Wardwell, L.; Johnson, L.A.; Emery, C.M.; Stransky, N.; Cogdill, A.P.; Barretina, J.; et al. Cot drives resistance to raf inhibition through map kinase pathway reactivation. Nature 2010, 468, 968–972. [Google Scholar] [CrossRef]
- Emery, C.M.; Vijayendran, K.G.; Zipser, M.C.; Sawyer, A.M.; Niu, L.; Kim, J.J.; Hatton, C.; Chopra, R.; Oberholzer, P.A.; Karpova, M.B.; et al. Mek1 mutations confer resistance to mek and b-raf inhibition. Proc. Natl. Acad. Sci. USA 2009, 106, 20411–20416. [Google Scholar] [CrossRef]
- Lito, P.; Rosen, N.; Solit, D.B. Tumor adaptation and resistance to raf inhibitors. Nat. Med. 2013, 19, 1401–1409. [Google Scholar] [CrossRef]
- Kun, E.; Tsang, Y.T.M.; Ng, C.W.; Gershenson, D.M.; Wong, K.K. Mek inhibitor resistance mechanisms and recent developments in combination trials. Cancer Treat. Rev. 2021, 92, 102137. [Google Scholar] [CrossRef]
- Johnson, D.B.; Menzies, A.M.; Zimmer, L.; Eroglu, Z.; Ye, F.; Zhao, S.; Rizos, H.; Sucker, A.; Scolyer, R.A.; Gutzmer, R.; et al. Acquired braf inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur. J. Cancer 2015, 51, 2792–2799. [Google Scholar] [CrossRef] [PubMed]
- Rizos, H.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Fung, C.; Hyman, J.; Haydu, L.E.; Mijatov, B.; Becker, T.M.; Boyd, S.C.; et al. Braf inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin. Cancer Res. 2014, 20, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Varrone, F.; Caputo, E. The mirnas role in melanoma and in its resistance to therapy. Int. J. Mol. Sci. 2020, 21, 878. [Google Scholar] [CrossRef]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal micrornas. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Krakowsky, R.H.E.; Wurm, A.A.; Gerloff, D.; Katzerke, C.; Brauer-Hartmann, D.; Hartmann, J.U.; Wilke, F.; Thiede, C.; Muller-Tidow, C.; Niederwieser, D.; et al. Mir-451a abrogates treatment resistance in flt3-itd-positive acute myeloid leukemia. Blood Cancer J. 2018, 8, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.W.; Zhao, W.J.; Zhu, Y.G.; Zhao, X.D. Mir-185 inhibits tumor growth and enhances chemo-resistance via targeting sry-related high mobility group box transcription factor 13 in non-small-cell carcinoma. Am. J. Transl. Res. 2018, 10, 2600. [Google Scholar]
- Gao, Y.W.; Liu, Z.Y.; Ding, Z.H.; Hou, S.C.; Li, J.; Jiang, K.H. Microrna-155 increases colon cancer chemoresistance to cisplatin by targeting forkhead box o3. Oncol. Lett. 2018, 15, 4781–4788. [Google Scholar] [CrossRef]
- Wurm, A.A.; Zjablovskaja, P.; Kardosova, M.; Gerloff, D.; Brauer-Hartmann, D.; Katzerke, C.; Hartmann, J.U.; Benoukraf, T.; Fricke, S.; Hilger, N.; et al. Disruption of the c/ebpalpha-mir-182 balance impairs granulocytic differentiation. Nat. Commun. 2017, 8, 46. [Google Scholar] [CrossRef]
- Gerloff, D.; Grundler, R.; Wurm, A.A.; Brauer-Hartmann, D.; Katzerke, C.; Hartmann, J.U.; Madan, V.; Muller-Tidow, C.; Duyster, J.; Tenen, D.G.; et al. Nf-kappab/stat5/mir-155 network targets pu.1 in flt3-itd-driven acute myeloid leukemia. Leukemia 2015, 29, 535–547. [Google Scholar] [CrossRef]
- Medarova, Z.; Pantazopoulos, P.; Yoo, B. Screening of potential mirna therapeutics for the prevention of multi-drug resistance in cancer cells. Sci. Rep. 2020, 10, 1970. [Google Scholar] [CrossRef]
- Linck-Paulus, L.; Hellerbrand, C.; Bosserhoff, A.K.; Dietrich, P. Dissimilar appearances are deceptive-common micrornas and therapeutic strategies in liver cancer and melanoma. Cells 2020, 9, 114. [Google Scholar] [CrossRef]
- Gerloff, D.; Sunderkotter, C.; Wohlrab, J. Importance of micrornas in skin oncogenesis and their suitability as agents and targets for topical therapy. Ski. Pharm. Physiol. 2020, 33, 270–279. [Google Scholar] [CrossRef]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of hcv infection by targeting microrna. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef]
- Sun, X.; Li, J.; Sun, Y.; Zhang, Y.; Dong, L.; Shen, C.; Yang, L.; Yang, M.; Li, Y.; Shen, G.; et al. Mir-7 reverses the resistance to brafi in melanoma by targeting egfr/igf-1r/craf and inhibiting the mapk and pi3k/akt signaling pathways. Oncotarget 2016, 7, 53558–53570. [Google Scholar] [CrossRef]
- Caporali, S.; Amaro, A.; Levati, L.; Alvino, E.; Lacal, P.M.; Mastroeni, S.; Ruffini, F.; Bonmassar, L.; Antonini Cappellini, G.C.; Felli, N.; et al. Mir-126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating adam9 and vegf-a. J. Exp. Clin. Cancer Res. Cr. 2019, 38, 272. [Google Scholar] [CrossRef]
- Fattore, L.; Mancini, R.; Acunzo, M.; Romano, G.; Lagana, A.; Pisanu, M.E.; Malpicci, D.; Madonna, G.; Mallardo, D.; Capone, M.; et al. Mir-579-3p controls melanoma progression and resistance to target therapy. Proc. Natl. Acad. Sci. USA 2016, 113, E5005–E5013. [Google Scholar] [CrossRef]
- Vergani, E.; Di Guardo, L.; Dugo, M.; Rigoletto, S.; Tragni, G.; Ruggeri, R.; Perrone, F.; Tamborini, E.; Gloghini, A.; Arienti, F.; et al. Overcoming melanoma resistance to vemurafenib by targeting ccl2-induced mir-34a, mir-100 and mir-125b. Oncotarget 2016, 7, 4428–4441. [Google Scholar] [CrossRef]
- Diaz-Martinez, M.; Benito-Jardon, L.; Alonso, L.; Koetz-Ploch, L.; Hernando, E.; Teixido, J. Mir-204-5p and mir-211-5p contribute to braf inhibitor resistance in melanoma. Cancer Res. 2018, 78, 1017–1030. [Google Scholar] [CrossRef]
- Lee, B.; Sahoo, A.; Sawada, J.; Marchica, J.; Sahoo, S.; Layng, F.; Finlay, D.; Mazar, J.; Joshi, P.; Komatsu, M.; et al. Microrna-211 modulates the dusp6-erk5 signaling axis to promote braf(v600e)-driven melanoma growth in vivo and braf/mek inhibitor resistance. J. Investig. Derm. 2020, 141, 385–394. [Google Scholar] [CrossRef]
- Vitiello, M.; Tuccoli, A.; D’Aurizio, R.; Sarti, S.; Giannecchini, L.; Lubrano, S.; Marranci, A.; Evangelista, M.; Peppicelli, S.; Ippolito, C.; et al. Context-dependent mir-204 and mir-211 affect the biological properties of amelanotic and melanotic melanoma cells. Oncotarget 2017, 8, 25395–25417. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.W.; Liu, J.C.; Deatherage, D.E.; Luo, J.Q.; Mutch, D.G.; Goodfellow, P.J.; Miller, D.S.; Huang, T.H.M. Epigenetic repression of microrna-129-2 leads to overexpression of sox4 oncogene in endometrial cancer. Cancer Res. 2009, 69, 9038–9046. [Google Scholar] [CrossRef] [PubMed]
- Luan, Q.X.; Zhang, B.G.; Li, X.J.; Guo, M.Y. Mir-129-5p is downregulated in breast cancer cells partly due to promoter h3k27m3 modification and regulates epithelial-mesenchymal transition and multi-drug resistance. Eur. Rev. Med. Pharm. 2016, 20, 4257–4265. [Google Scholar]
- Wu, Q.; Yang, Z.; Xia, L.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. Methylation of mir-129-5p cpg island modulates multi-drug resistance in gastric cancer by targeting abc transporters. Oncotarget 2014, 5, 11552–11563. [Google Scholar] [CrossRef] [PubMed]
- Vire, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.; Morey, L.; Van Eynde, A.; Bernard, D.; Vanderwinden, J.M.; et al. The polycomb group protein ezh2 directly controls DNA methylation. Nature 2006, 439, 871–874. [Google Scholar] [CrossRef]
- Mahmoud, F.; Shields, B.; Makhoul, I.; Hutchins, L.F.; Shalin, S.C.; Tackett, A.J. Role of ezh2 histone methyltrasferase in melanoma progression and metastasis. Cancer Biol. 2016, 17, 579–591. [Google Scholar] [CrossRef]
- Dai, W.; Xu, X.; Li, S.; Ma, J.; Shi, Q.; Guo, S.; Liu, L.; Guo, W.; Xu, P.; He, Y.; et al. Sox4 promotes proliferative signals by regulating glycolysis through akt activation in melanoma cells. J. Investig. Derm. 2017, 137, 2407–2416. [Google Scholar] [CrossRef]
- Liu, X.; Mi, J.; Qin, H.-H.; He, S.; Li, Z.; Chai, J.-X.; Li, M.; Xu, J.-H.; Wu, J.-F. Sox4 mediates braf inhibitor resistance in melanoma through regulation of igf-1r signaling: In vitro study. Int. J. Dermatol. Venereol. 2020, 3, 156–165. [Google Scholar] [CrossRef]
- Kozar, I.; Cesi, G.; Margue, C.; Philippidou, D.; Kreis, S. Impact of braf kinase inhibitors on the mirnomes and transcriptomes of melanoma cells. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2980–2992. [Google Scholar] [CrossRef]
- Grzywa, T.M.; Klicka, K.; Paskal, W.; Dudkiewicz, J.; Wejman, J.; Pyzlak, M.; Wlodarski, P.K. Mir-410-3p is induced by vemurafenib via er stress and contributes to resistance to braf inhibitor in melanoma. PLoS ONE 2020, 15, e0234707. [Google Scholar] [CrossRef]
- Fattore, L.; Ruggiero, C.F.; Pisanu, M.E.; Liguoro, D.; Cerri, A.; Costantini, S.; Capone, F.; Acunzo, M.; Romano, G.; Nigita, G.; et al. Reprogramming mirnas global expression orchestrates development of drug resistance in braf mutated melanoma. Cell Death Differ. 2019, 26, 1267–1282. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Menggen, Q.; Wuren, Q.; Shi, Q.; Pi, X. Long noncoding rna taurine-upregulated gene1 (tug1) promotes tumor growth and metastasis through tug1/mir-129-5p/astrocyte-elevated gene-1 (aeg-1) axis in malignant melanoma. Med. Sci. Monit. 2018, 24, 1547–1559. [Google Scholar] [CrossRef]
- Gao, Y.; Feng, B.; Han, S.; Lu, L.; Chen, Y.; Chu, X.; Wang, R.; Chen, L. Microrna-129 in human cancers: From tumorigenesis to clinical treatment. Cell Physiol. Biochem. 2016, 39, 2186–2202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, J.; Song, Y.; Wang, X. Mir-129-5p inhibits proliferation and invasion of chondrosarcoma cells by regulating sox4/wnt/beta-catenin signaling pathway. Cell Physiol. Biochem. 2017, 42, 242–253. [Google Scholar] [CrossRef]
- Wan, P.; Bai, X.; Yang, C.; He, T.; Luo, L.; Wang, Y.; Fan, M.; Wang, Z.; Lu, L.; Yin, Y.; et al. Mir-129-5p inhibits proliferation, migration, and invasion in rectal adenocarcinoma cells through targeting e2f7. J. Cell Physiol. 2020, 235, 5689–5701. [Google Scholar] [CrossRef]
- Gao, G.; Xiu, D.; Yang, B.; Sun, D.; Wei, X.; Ding, Y.; Ma, Y.; Wang, Z. Mir-129-5p inhibits prostate cancer proliferation via targeting etv1. Onco Targets 2019, 12, 3531–3544. [Google Scholar] [CrossRef]
- Feng, J.; Guo, J.; Wang, J.P.; Chai, B.F. Mir-129-5p inhibits proliferation of gastric cancer cells through targeted inhibition on hmgb1 expression. Eur. Rev. Med. Pharm. Sci. 2020, 24, 3665–3673. [Google Scholar]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. Ezh2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef]
- Tiffen, J.; Gallagher, S.J.; Filipp, F.; Gunatilake, D.; Emran, A.A.; Cullinane, C.; Dutton-Register, K.; Aoude, L.; Hayward, N.; Chatterjee, A.; et al. Ezh2 cooperates with DNA methylation to downregulate key tumor suppressors and ifn gene signatures in melanoma. J. Investig. Derm. 2020, 140, 2442–2454 e2445. [Google Scholar] [CrossRef]
- Zingg, D.; Debbache, J.; Schaefer, S.M.; Tuncer, E.; Frommel, S.C.; Cheng, P.; Arenas-Ramirez, N.; Haeusel, J.; Zhang, Y.; Bonalli, M.; et al. The epigenetic modifier ezh2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 2015, 6, 6051. [Google Scholar] [CrossRef]
- Tiffen, J.C.; Gallagher, S.J.; Tseng, H.Y.; Filipp, F.V.; Fazekas de St. Groth, B.; Hersey, P. Ezh2 as a mediator of treatment resistance in melanoma. Pigment. Cell Melanoma Res. 2016, 29, 500–507. [Google Scholar] [CrossRef]
- Grigore, F.; Yang, H.N.; Hanson, N.D.; VanBrocklin, M.W.; Sarver, A.L.; Robinson, J.P. Braf inhibition in melanoma is associated with the dysregulation of histone methylation and histone methyltransferases. Neoplasia 2020, 22, 376–389. [Google Scholar] [CrossRef]
- Qu, Y.; Yang, Q.; Liu, J.; Shi, B.; Ji, M.; Li, G.; Hou, P. C-myc is required for braf(v600e)-induced epigenetic silencing by h3k27me3 in tumorigenesis. Theranostics 2017, 7, 2092–2107. [Google Scholar] [CrossRef]
- Han, H.; Li, W.; Shen, H.; Zhang, J.; Zhu, Y.; Li, Y. Microrna-129-5p, a c-myc negative target, affects hepatocellular carcinoma progression by blocking the warburg effect. J. Mol. Cell Biol. 2016, 8, 400–410. [Google Scholar] [CrossRef]
- Xiao, Y.; Li, X.; Wang, H.; Wen, R.; He, J.; Tang, J. Epigenetic regulation of mir-129-2 and its effects on the proliferation and invasion in lung cancer cells. J. Cell Mol. Med. 2015, 19, 2172–2180. [Google Scholar] [CrossRef]
- Fesler, A.; Zhai, H.; Ju, J. Mir-129 as a novel therapeutic target and biomarker in gastrointestinal cancer. Onco Targets 2014, 7, 1481–1485. [Google Scholar]
- Yuan, C.; Yang, L. Long non-coding rna pitpna-as1 accelerates the progression of colorectal cancer through mir-129-5p/hmgb1 axis. Cancer Manag. Res. 2020, 12, 12497–12507. [Google Scholar] [CrossRef]
- Wang, H.; Li, H.; Yu, Y.; Jiang, Q.; Zhang, R.; Sun, H.; Xing, W.; Li, Y. Long non-coding rna xist promotes the progression of esophageal squamous cell carcinoma through sponging mir-129-5p and upregulating ccnd1 expression. Cell Cycle 2020, 20, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Li, Y.; Liu, W.; Wang, R.; Tang, A.; Hao, H.; Liu, Z.; Ou, H. Mir-129-2 suppresses proliferation and migration of esophageal carcinoma cells through downregulation of sox4 expression. Int. J. Mol. Med. 2013, 32, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Qu, S.; Li, X.; Zhong, J.; Chen, X.; Qu, Z.; Wu, D. Mir-129 suppresses tumor cell growth and invasion by targeting pak5 in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2015, 464, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhu, H.; Ma, H.; Yuan, L.; Hu, Q.; Yang, L. Linc01305 inhibits malignant progression of cervical cancer via mir-129-5p/sox4 axis. Am. J. Transl. Res. 2020, 12, 7581–7592. [Google Scholar]
- Tang, X.; Tang, J.; Liu, X.; Zeng, L.; Cheng, C.; Luo, Y.; Li, L.; Qin, S.L.; Sang, Y.; Deng, L.M.; et al. Downregulation of mir-129-2 by promoter hypermethylation regulates breast cancer cell proliferation and apoptosis. Oncol. Rep. 2016, 35, 2963–2969. [Google Scholar] [CrossRef]
- Zeng, A.; Yin, J.; Li, Y.; Li, R.; Wang, Z.; Zhou, X.; Jin, X.; Shen, F.; Yan, W.; You, Y. Mir-129-5p targets wnt5a to block pkc/erk/nf-kappab and jnk pathways in glioblastoma. Cell Death Dis. 2018, 9, 394. [Google Scholar] [CrossRef]
- Zeng, H.; Wang, L.; Wang, J.; Chen, T.; Li, H.; Zhang, K.; Chen, J.; Zhen, S.; Tuluhong, D.; Li, J.; et al. Microrna-129-5p suppresses adriamycin resistance in breast cancer by targeting sox2. Arch. Biochem. Biophys. 2018, 651, 52–60. [Google Scholar] [CrossRef]
- Lu, X.; Ma, J.; Chu, J.; Shao, Q.; Zhang, Y.; Lu, G.; Li, J.; Huang, X.; Li, W.; Li, Y.; et al. Mir-129-5p sensitizes the response of her-2 positive breast cancer to trastuzumab by reducing rps6. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 44, 2346–2356. [Google Scholar] [CrossRef]
- Wang, J.; Ye, C.; Liu, J.; Hu, Y. Uca1 confers paclitaxel resistance to ovarian cancer through mir-129/abcb1 axis. Biochem. Biophys. Res. Commun. 2018, 501, 1034–1040. [Google Scholar] [CrossRef]
- Cao, J.; Wang, Q.; Wu, G.; Li, S.; Wang, Q. Mir-129-5p inhibits gemcitabine resistance and promotes cell apoptosis of bladder cancer cells by targeting wnt5a. Int. Urol. Nephrol. 2018, 50, 1811–1819. [Google Scholar] [CrossRef]
- Giricz, O.; Mo, Y.; Dahlman, K.B.; Cotto-Rios, X.M.; Vardabasso, C.; Nguyen, H.; Matusow, B.; Bartenstein, M.; Polishchuk, V.; Johnson, D.B.; et al. The runx1/il-34/csf-1r axis is an autocrinally regulated modulator of resistance to braf-v600e inhibition in melanoma. JCI Insight 2018, 3, 14. [Google Scholar] [CrossRef]
- Tiwari, N.; Tiwari, V.K.; Waldmeier, L.; Balwierz, P.J.; Arnold, P.; Pachkov, M.; Meyer-Schaller, N.; Schubeler, D.; van Nimwegen, E.; Christofori, G. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling ezh2 expression and epigenetic reprogramming. Cancer Cell 2013, 23, 768–783. [Google Scholar] [CrossRef]
- Koumangoye, R.B.; Andl, T.; Taubenslag, K.J.; Zilberman, S.T.; Taylor, C.J.; Loomans, H.A.; Andl, C.D. Sox4 interacts with ezh2 and hdac3 to suppress microrna-31 in invasive esophageal cancer cells. Mol. Cancer 2015, 14, 24. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gebhardt, K.; Edemir, B.; Groß, E.; Nemetschke, L.; Kewitz-Hempel, S.; Moritz, R.K.C.; Sunderkötter, C.; Gerloff, D. BRAF/EZH2 Signaling Represses miR-129-5p Inhibition of SOX4 Thereby Modulating BRAFi Resistance in Melanoma. Cancers 2021, 13, 2393. https://doi.org/10.3390/cancers13102393
Gebhardt K, Edemir B, Groß E, Nemetschke L, Kewitz-Hempel S, Moritz RKC, Sunderkötter C, Gerloff D. BRAF/EZH2 Signaling Represses miR-129-5p Inhibition of SOX4 Thereby Modulating BRAFi Resistance in Melanoma. Cancers. 2021; 13(10):2393. https://doi.org/10.3390/cancers13102393
Chicago/Turabian StyleGebhardt, Kathleen, Bayram Edemir, Elisabeth Groß, Linda Nemetschke, Stefanie Kewitz-Hempel, Rose K. C. Moritz, Cord Sunderkötter, and Dennis Gerloff. 2021. "BRAF/EZH2 Signaling Represses miR-129-5p Inhibition of SOX4 Thereby Modulating BRAFi Resistance in Melanoma" Cancers 13, no. 10: 2393. https://doi.org/10.3390/cancers13102393
APA StyleGebhardt, K., Edemir, B., Groß, E., Nemetschke, L., Kewitz-Hempel, S., Moritz, R. K. C., Sunderkötter, C., & Gerloff, D. (2021). BRAF/EZH2 Signaling Represses miR-129-5p Inhibition of SOX4 Thereby Modulating BRAFi Resistance in Melanoma. Cancers, 13(10), 2393. https://doi.org/10.3390/cancers13102393





