Reduced Lamin A/C Does Not Facilitate Cancer Cell Transendothelial Migration but Compromises Lung Metastasis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Mice
2.3. Imaging and Analysis of Tumor Cell Transendothelial Migration
2.4. Transwell Migration Assays
2.5. Generation of shRNA—Expressing Cancer Cell Lines
2.6. RNA Isolation and Real—Time qRT-PCR
2.7. Imaging of Tumor Cell Nuclear Dynamics
2.8. Light Sheet Fluorescent Microscopy of Tumor Cells and Lung Vasculature
2.9. Image Reconstruction and Analysis
2.10. Determination of Tumor Cell Accumulation in Lungs
2.11. Cell Growth on 2D Culture Plates
2.12. Cell Growth in 3D Soft Agar
2.13. Experimental Lung Metastases
2.14. Patient Survival Analysis
2.15. Statistical Analysis
2.16. Additional Supplementary Methods
3. Results
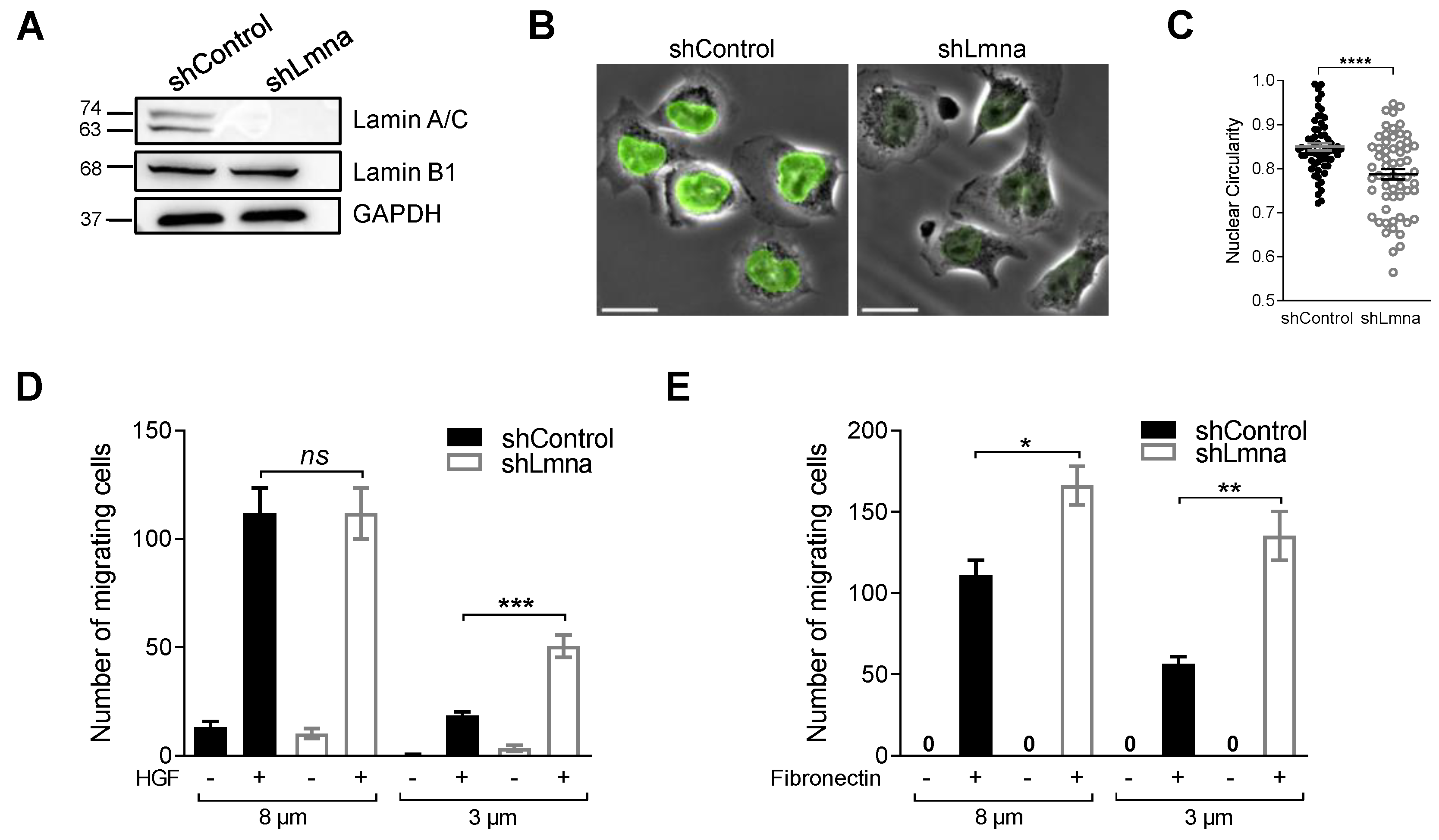
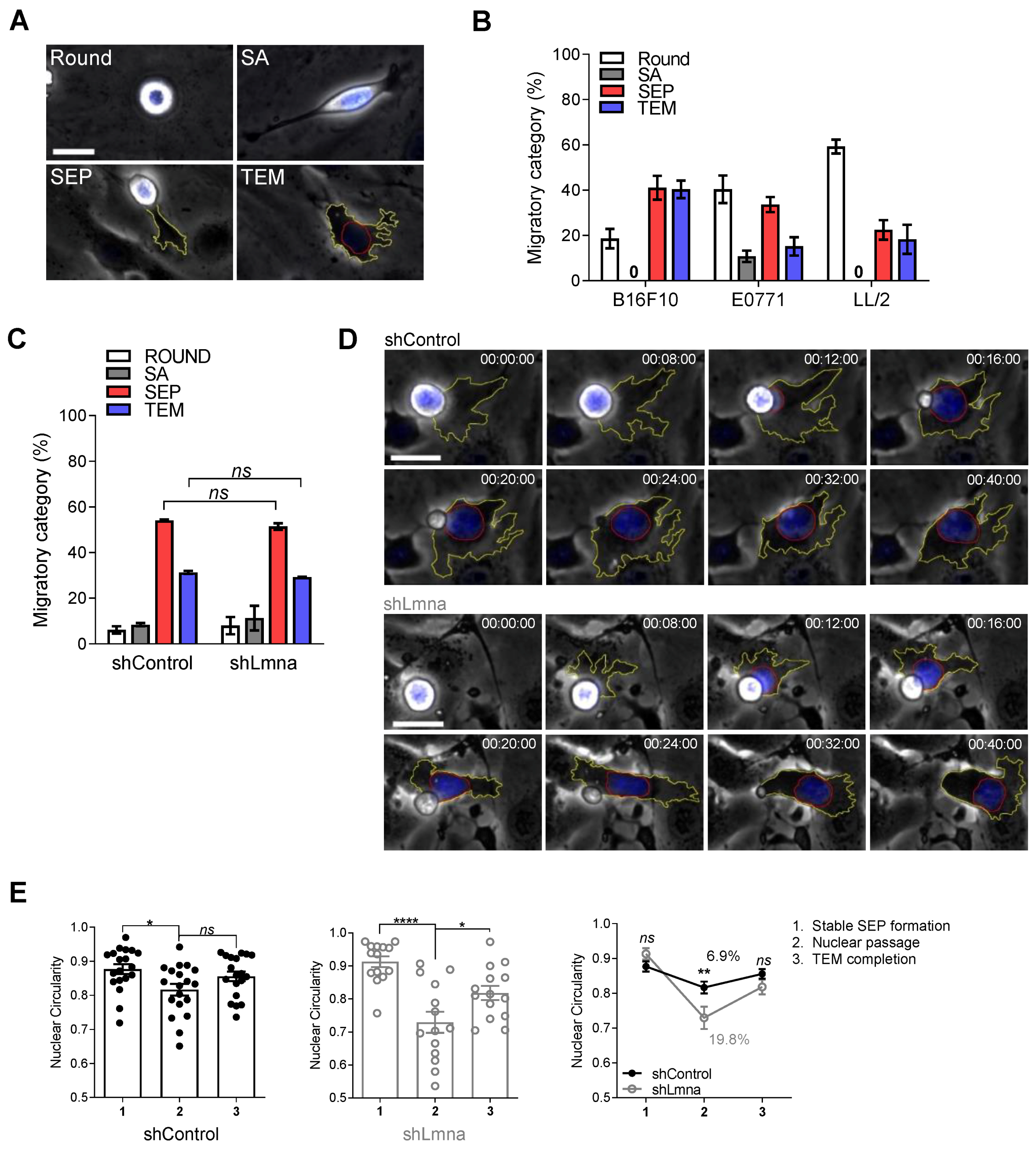
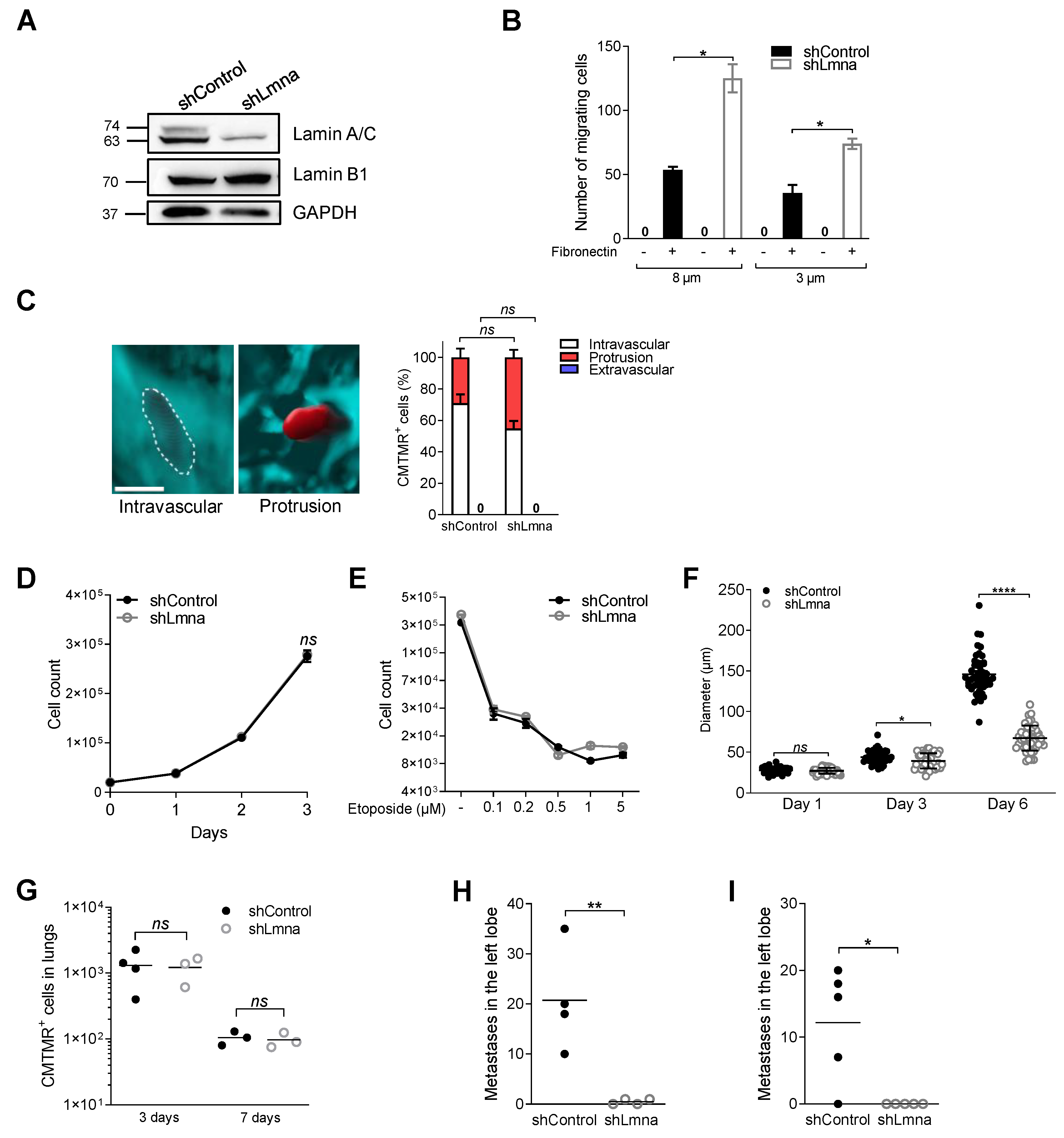
3.1. Lamin A Downregulation in B16F10 Melanoma Increases Nucleus Deformability and Squeezing through Rigid Pores but Does Not Affect Transendothelial Migration
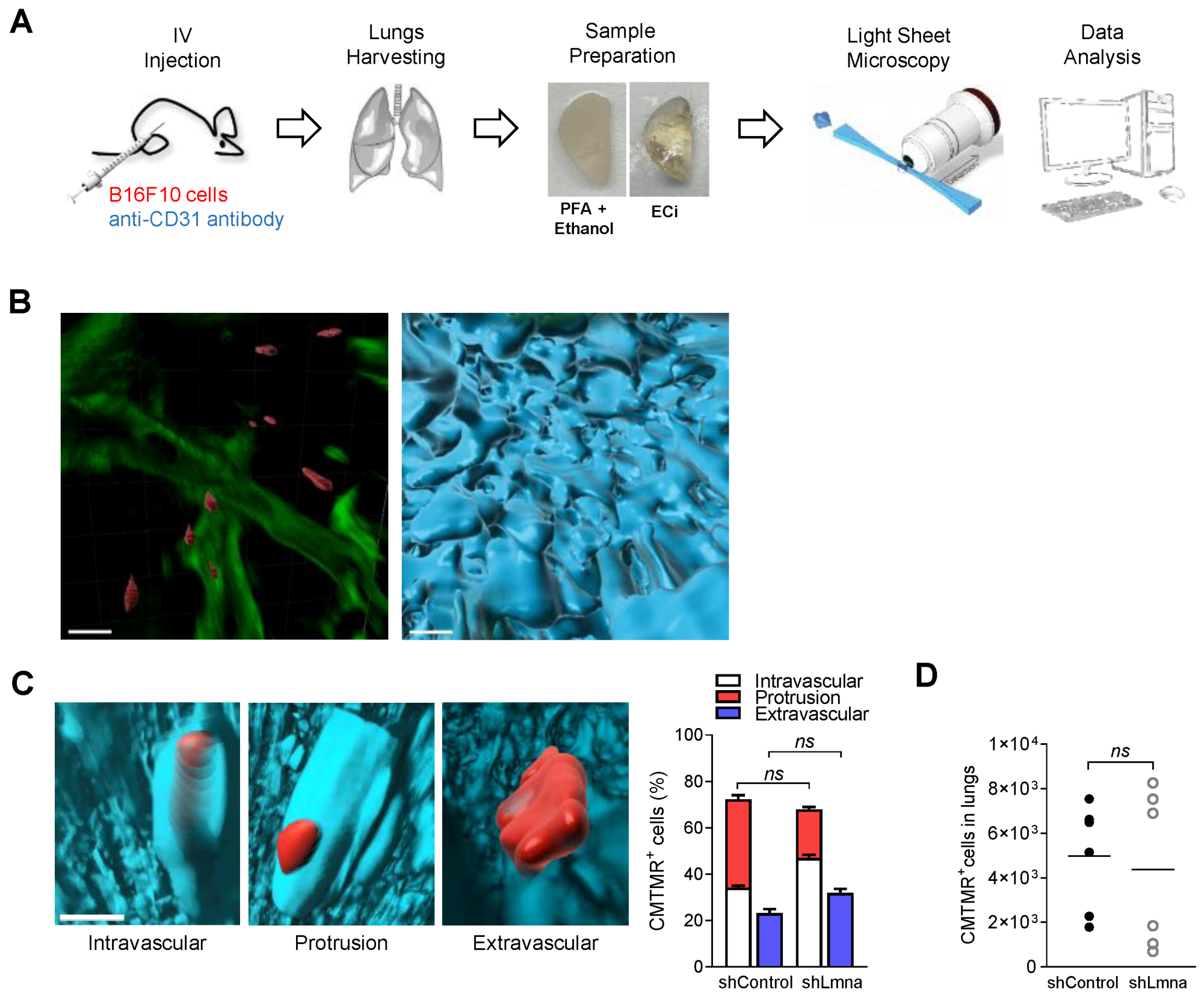
3.2. Lamin A Downregulation Does Not Increase B16F10 Extravasation across Lung Vessels In Vivo and Does Not Accelerate Melanoma Apoptosis in the Lung Parenchyma
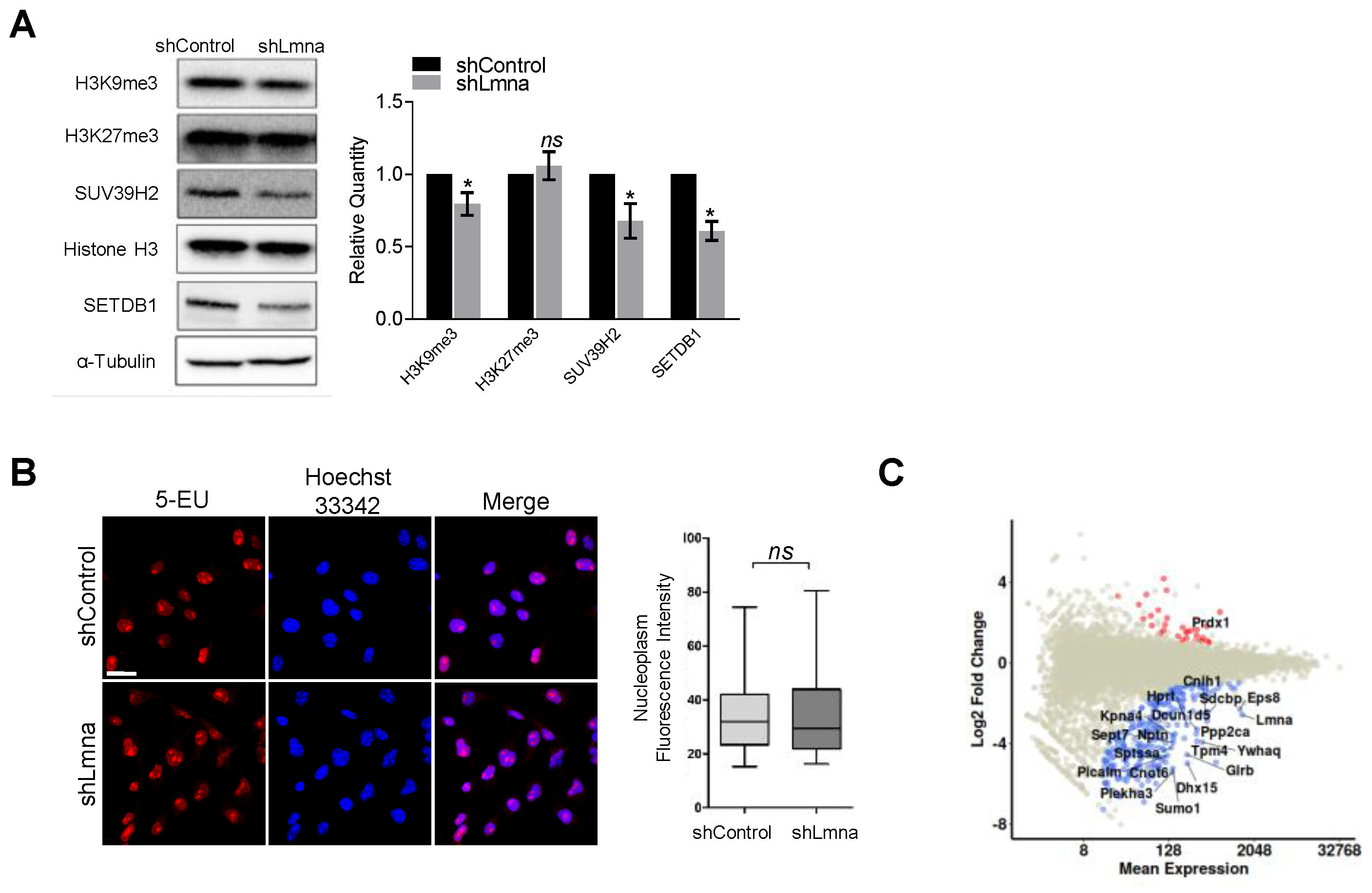
3.3. Lamin A/C Downregulation Slightly Reduces the Content of H3K9Me3 Heterochromatin but Does Not Elevate Transcription
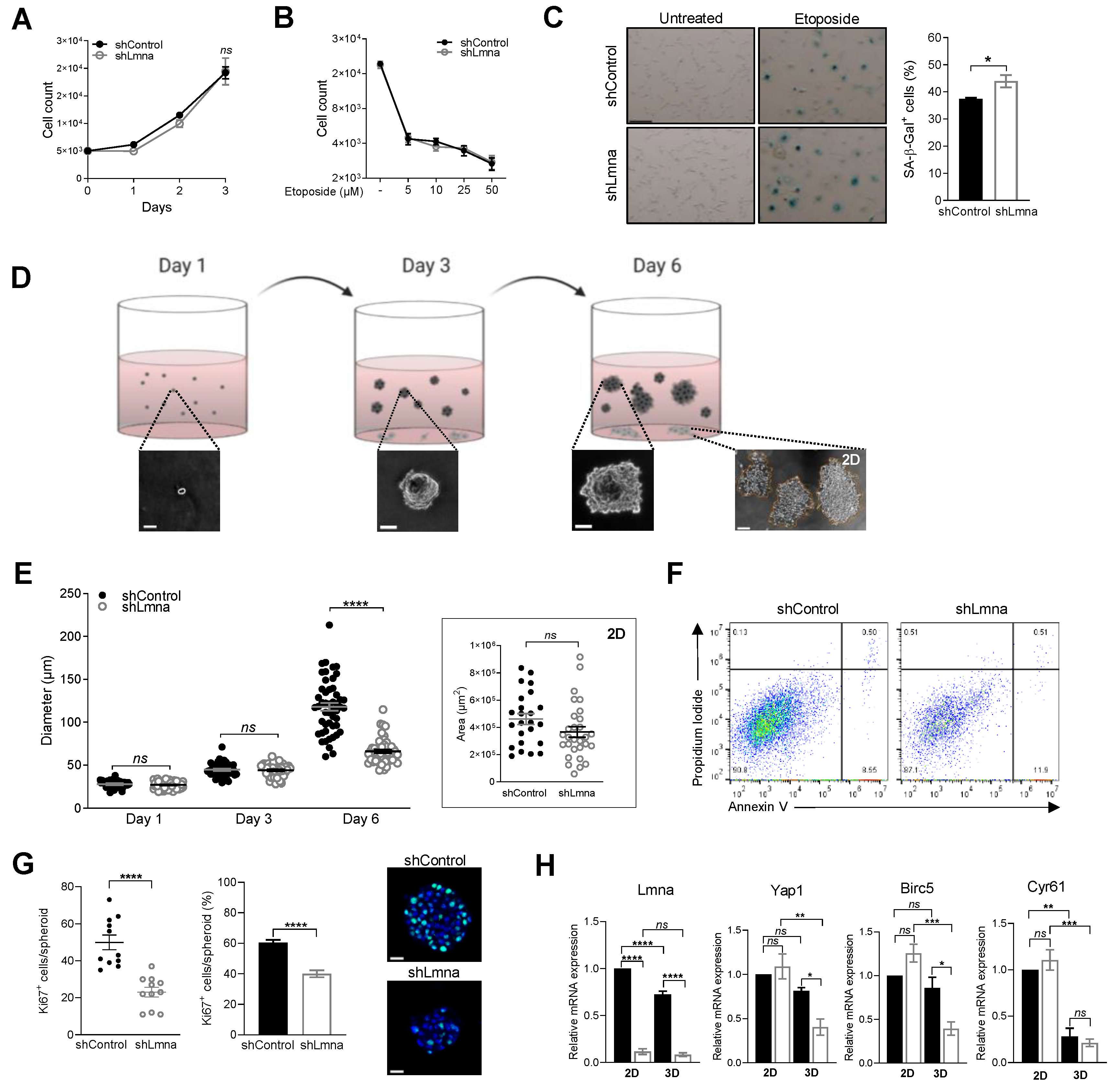
3.4. Lamin A/C Downregulation Does Not Alter Cell Proliferation In Vitro on 2D Surfaces but Impairs 3D Cell Growth in Spheroids
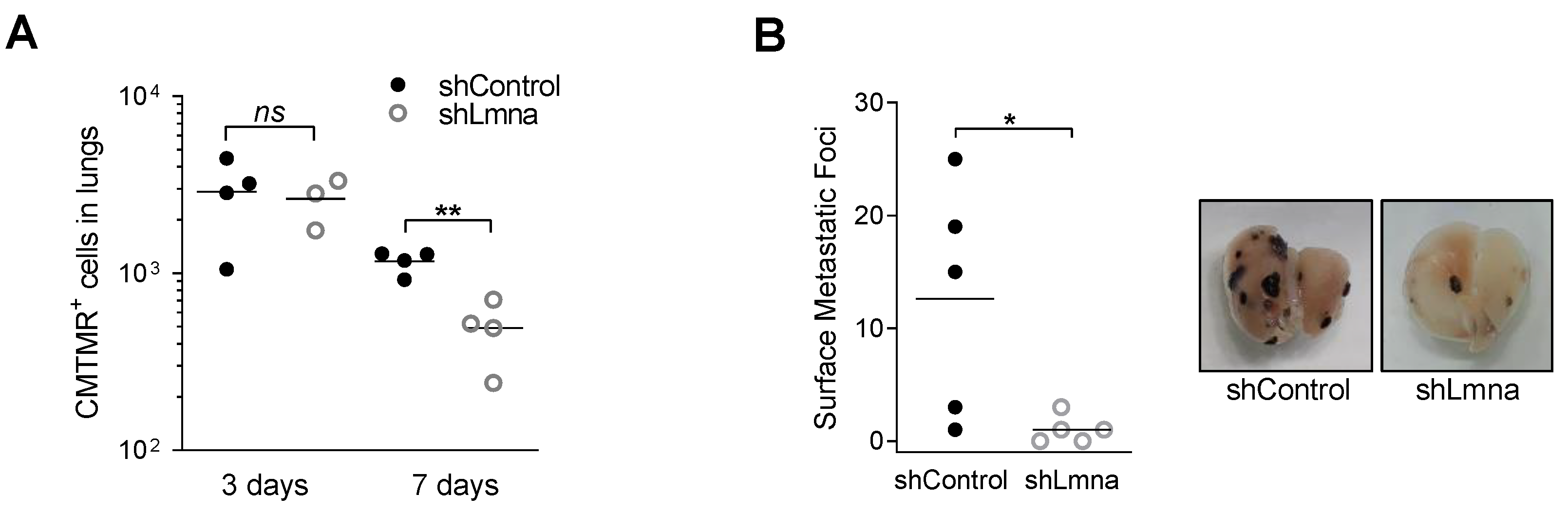
3.5. Lamin A/C Deficiency Results in Reduced B16F10 Lung Metastasis
3.6. Reduced Lamin A/C Levels in E0771 Breast Cancer Cells Recapitulate the In Vitro and In Vivo Migratory Properties and Metastasis Deficiency of Lamin A/C Knockdown B16F10 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Friedl, P.; Wolf, K.; Lammerding, J. Nuclear Mechanics during Cell Migration. Curr. Opin. Cell Biol. 2011, 23, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Burke, B.; Stewart, C.L. The Nuclear Lamins: Flexibility in Function. Nat. Rev. Mol. Cell Biol. 2013, 14, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Lammerding, J.; Fong, L.G.; Ji, J.Y.; Reue, K.; Stewart, C.L.; Young, S.G.; Lee, R.T. Lamins a and C but Not Lamin B1 Regulate Nuclear Mechanics. J. Biol. Chem. 2006, 281, 25768–25780. [Google Scholar] [CrossRef]
- Shin, J.W.; Spinler, K.R.; Swift, J.; Chasis, J.A.; Mohandas, N.; Discher, D.E. Lamins Regulate Cell Trafficking and Lineage Maturation of Adult Human Hematopoietic Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 18892–18897. [Google Scholar] [CrossRef] [PubMed]
- Towbin, B.D.; Meister, P.; Gasser, S.M. The Nuclear Envelope—A Scaffold for Silencing? Curr. Opin. Genet. Dev. 2009, 19, 180–186. [Google Scholar] [CrossRef]
- Bronshtein, I.; Kepten, E.; Kanter, I.; Berezin, S.; Lindner, M.; Redwood, A.B.; Mai, S.; Gonzalo, S.; Foisner, R.; Shav-Tal, Y.; et al. Loss of Lamin A Function Increases Chromatin Dynamics in the Nuclear Interior. Nat. Commun. 2015, 6, 8044. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, M.L.; Jaalouk, D.E.; Shanahan, C.M.; Burke, B.; Roux, K.J.; Lammerding, J. The Interaction between Nesprins and Sun Proteins at the Nuclear Envelope Is Critical for Force Transmission between the Nucleus and Cytoskeleton. J. Biol. Chem. 2011, 286, 26743–26753. [Google Scholar] [CrossRef]
- Chang, W.; Folker, E.S.; Worman, H.J.; Gundersen, G.G. Emerin Organizes Actin Flow for Nuclear Movement and Centrosome Orientation in Migrating Fibroblasts. Mol. Biol. Cell 2013, 24, 3869–3880. [Google Scholar] [CrossRef]
- Etienne-Manneville, S.; Lammerding, J. Connecting the Plasma Membrane to the Nucleus by Intermediate Filaments. Mol. Biol. Cell 2017, 28, 695–696. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.M.; Andersen, T.; Sharek, L.; Uzer, G.; Rothenberg, K.; Hoffman, B.D.; Rubin, J.; Balland, M.; Bear, J.E.; Burridge, K. Enucleated Cells Reveal Differential Roles of the Nucleus in Cell Migration, Polarity, and Mechanotransduction. J. Cell Biol. 2018, 217, 895–914. [Google Scholar] [CrossRef]
- Kirby, T.J.; Lammerding, J. Emerging Views of the Nucleus as a Cellular Mechanosensor. Nat. Cell Biol. 2018, 20, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Wirtz, D. Cytoskeletal Tension Induces the Polarized Architecture of the Nucleus. Biomaterials 2015, 48, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.; Te Lindert, M.; Krause, M.; Alexander, S.; Te Riet, J.; Willis, A.L.; Hoffman, R.M.; Figdor, C.G.; Weiss, S.J.; Friedl, P. Physical Limits of Cell Migration: Control by ECM Space and Nuclear Deformation and Tuning by Proteolysis and Traction Force. J. Cell Biol 2013, 201, 1069–1084. [Google Scholar] [CrossRef] [PubMed]
- Raab, M.; Gentili, M.; de Belly, H.; Thiam, H.R.; Vargas, P.; Jimenez, A.J.; Lautenschlaeger, F.; Voituriez, R.; Lennon-Dumenil, A.M.; Manel, N.; et al. ESCRT III Repairs Nuclear Envelope Ruptures during Cell Migration to Limit DNA Damage and Cell Death. Science 2016, 352, 359–362. [Google Scholar] [CrossRef]
- Denais, C.M.; Gilbert, R.M.; Isermann, P.; McGregor, A.L.; Te Lindert, M.; Weigelin, B.; Davidson, P.M.; Friedl, P.; Wolf, K.; Lammerding, J. Nuclear Envelope Rupture and Repair during Cancer Cell Migration. Science 2016, 352, 353–358. [Google Scholar] [CrossRef]
- Yadav, S.K.; Feigelson, S.W.; Roncato, F.; Antman-Passig, M.; Shefi, O.; Lammerding, J.; Alon, R. Frontline Science: Elevated Nuclear Lamin A Is Permissive for Granulocyte Transendothelial Migration but Not for Motility through Collagen I Barriers. J. Leukoc. Biol. 2018, 104, 239–251. [Google Scholar] [CrossRef]
- Cao, X.; Moeendarbary, E.; Isermann, P.; Davidson, P.M.; Wang, X.; Chen, M.B.; Burkart, A.K.; Lammerding, J.; Kamm, R.D.; Shenoy, V.B. A Chemomechanical Model for Nuclear Morphology and Stresses during Cell Transendothelial Migration. Biophys. J. 2016, 111, 1541–1552. [Google Scholar] [CrossRef]
- Davidson, P.M.; Denais, C.; Bakshi, M.C.; Lammerding, J. Nuclear Deformability Constitutes a Rate-Limiting Step during Cell Migration in 3-D Environments. Cell Mol. Bioeng. 2014, 7, 293–306. [Google Scholar] [CrossRef]
- Heemskerk, N.; Schimmel, L.; Oort, C.; van Rijssel, J.; Yin, T.; Ma, B.; van Unen, J.; Pitter, B.; Huveneers, S.; Goedhart, J.; et al. F-Actin-Rich Contractile Endothelial Pores Prevent Vascular Leakage during Leukocyte Diapedesis through Local RhoA Signalling. Nat. Commun. 2016, 7, 10493. [Google Scholar] [CrossRef]
- Infante, E.; Castagnino, A.; Ferrari, R.; Monteiro, P.; Agüera-González, S.; Paul-Gilloteaux, P.; Domingues, M.J.; Maiuri, P.; Raab, M.; Shanahan, C.M.; et al. LINC Complex-Lis1 Interplay Controls MT1-MMP Matrix Digest-on-Demand Response for Confined Tumor Cell Migration. Nat. Commun. 2018, 9, 2443. [Google Scholar] [CrossRef]
- Broers, J.L.V.; Ramaekers, F.C.S. The Role of the Nuclear Lamina in Cancer and Apoptosis. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2014; Volume 773, pp. 27–48. [Google Scholar] [CrossRef]
- Hutchison, C.J. Do Lamins Influence Disease Progression in Cancer? Adv. Exp. Med. Biol. 2014, 773, 593–604. [Google Scholar] [CrossRef]
- Dubik, N.; Mai, S. Lamin A/C: Function in Normal and Tumor Cells. Cancers 2020, 12, 3688. [Google Scholar] [CrossRef]
- Capo-chichi, C.D.; Cai, K.Q.; Smedberg, J.; Ganjei-Azar, P.; Godwin, A.K.; Xu, X.X. Loss of A-Type Lamin Expression Compromises Nuclear Envelope Integrity in Breast Cancer. Chin. J. Cancer 2011, 30, 415–425. [Google Scholar] [CrossRef]
- Broers, J.L.; Raymond, Y.; Rot, M.K.; Kuijpers, H.; Wagenaar, S.S.; Ramaekers, F.C. Nuclear A-Type Lamins Are Differentially Expressed in Human Lung Cancer Subtypes. Am. J. Pathol. 1993, 143, 211–220. [Google Scholar] [PubMed]
- Venables, R.S.; McLean, S.; Luny, D.; Moteleb, E.; Morley, S.; Quinlan, R.A.; Lane, E.B.; Hutchison, C.J. Expression of Individual Lamins in Basal Cell Carcinomas of the Skin. Br. J. Cancer 2001, 84, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Brady, G.F.; Kwan, R.; Bragazzi Cunha, J.; Elenbaas, J.S.; Omary, M.B. Lamins and Lamin-Associated Proteins in Gastrointestinal Health and Disease. Gastroenterology 2018, 154, 1602–1619.e1. [Google Scholar] [CrossRef] [PubMed]
- Foster, C.R.; Robson, J.L.; Simon, W.J.; Twigg, J.; Cruikshank, D.; Wilson, R.G.; Hutchison, C.J. The Role of Lamin A in Cytoskeleton Organization in Colorectal Cancer Cells: A Proteomic Investigation. Nucleus 2011, 2, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Schäfer, G.; Bu, H.; Zhang, Y.Y.; Zhang, Y.Y.; Klocker, H.; Schafer, G.; Bu, H.; Zhang, Y.Y.; Zhang, Y.Y.; et al. Lamin A/C Protein Is Overexpressed in Tissue-Invading Prostate Cancer and Promotes Prostate Cancer Cell Growth, Migration and Invasion through the PI3K/AKT/PTEN Pathway. Carcinogenesis 2012, 33, 751–759. [Google Scholar] [CrossRef]
- Cho, S.; Vashisth, M.; Abbas, A.; Majkut, S.; Vogel, K.; Xia, Y.; Ivanovska, I.L.; Irianto, J.; Tewari, M.; Zhu, K.; et al. Mechanosensing by the Lamina Protects against Nuclear Rupture, DNA Damage, and Cell-Cycle Arrest. Dev. Cell 2019, 49, 920–935.e5. [Google Scholar] [CrossRef]
- Le Naour, A.; Koffi, Y.; Diab, M.; Le Guennec, D.; Rougé, S.; Aldekwer, S.; Goncalves-Mendes, N.; Talvas, J.; Farges, M.C.; Caldefie-Chezet, F.; et al. EO771, the First Luminal B Mammary Cancer Cell Line from C57BL/6 Mice. Cancer Cell Int. 2020, 20, 328. [Google Scholar] [CrossRef] [PubMed]
- Reymond, N.; Im, J.H.; Garg, R.; Vega, F.M.; Borda d’Agua, B.; Riou, P.; Cox, S.; Valderrama, F.; Muschel, R.J.; Ridley, A.J. Cdc42 Promotes Transendothelial Migration of Cancer Cells through Β1 Integrin. J. Cell Biol. 2012, 199, 653–668. [Google Scholar] [CrossRef]
- Klingberg, A.; Hasenberg, A.; Ludwig-Portugall, I.; Medyukhina, A.; Männ, L.; Brenzel, A.; Engel, D.R.; Figge, M.T.; Kurts, C.; Gunzer, M. Fully Automated Evaluation of Total Glomerular Number and Capillary Tuft Size in Nephritic Kidneys Using Lightsheet Microscopy. J. Am. Soc. Nephrol. 2017, 28, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Horibata, S.; Vo, T.V.; Subramanian, V.; Thompson, P.R.; Coonrod, S.A. Utilization of the Soft Agar Colony Formation Assay to Identify Inhibitors of Tumorigenicity in Breast Cancer Cells. J. Vis. Exp. 2015, 2015, 52727. [Google Scholar] [CrossRef] [PubMed]
- Roman, W.; Martins, J.P.; Carvalho, F.A.; Voituriez, R.; Abella, J.V.G.; Santos, N.C.; Cadot, B.; Way, M.; Gomes, E.R. Myofibril Contraction and Crosslinking Drive Nuclear Movement to the Periphery of Skeletal Muscle. Nat. Cell Biol. 2017, 19, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Jaitin, D.A.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A.; et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science 2014, 343, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Kohen, R.; Barlev, J.; Hornung, G.; Stelzer, G.; Feldmesser, E.; Kogan, K.; Safran, M.; Leshkowitz, D. UTAP: User-Friendly Transcriptome Analysis Pipeline. BMC Bioinform. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting Batch Effects in Microarray Expression Data Using Empirical Bayes Methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Krizhanovsky, V.; Yon, M.; Dickins, R.A.; Hearn, S.; Simon, J.; Miething, C.; Yee, H.; Zender, L.; Lowe, S.W. Senescence of Activated Stellate Cells Limits Liver Fibrosis. Cell 2008, 134, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Spandidos, A.; Wang, X.; Wang, H.; Dragnev, S.; Thurber, T.; Seed, B. A Comprehensive Collection of Experimentally Validated Primers for Polymerase Chain Reaction Quantitation of Murine Transcript Abundance. BMC Genomics 2008, 9, 633. [Google Scholar] [CrossRef]
- Miles, F.L.; Pruitt, F.L.; van Golen, K.L.; Cooper, C.R. Stepping out of the Flow: Capillary Extravasation in Cancer Metastasis. Clin. Exp. Metastasis 2008, 25, 305–324. [Google Scholar] [CrossRef]
- Valastyan, S.; Weinberg, R.A. Tumor Metastasis: Molecular Insights and Evolving Paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef]
- Gorelik, L.; Flavell, R.A. Immune-Mediated Eradication of Tumors through the Blockade of Transforming Growth Factor-β Signaling in T Cells. Nat. Med. 2001, 7, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.J.; Denais, C.; Chan, M.F.; Wang, Z.; Lammerding, J.; King, M.R. Lamin A/C Deficiency Reduces Circulating Tumor Cell Resistance to Fluid Shear Stress. Am. J. Physiol. Physiol. 2015, 309, C736–C746. [Google Scholar] [CrossRef]
- Liu, B.; Wang, Z.; Zhang, L.; Ghosh, S.; Zheng, H.; Zhou, Z. Depleting the Methyltransferase Suv39h1 Improves DNA Repair and Extends Lifespan in a Progeria Mouse Model. Nat. Commun. 2013, 4, 1868. [Google Scholar] [CrossRef]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and Lamin A/C Sequentially Tether Peripheral Heterochromatin and Inversely Regulate Differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef]
- Dai, W.; Jiang, Y.; Chen, K.; Qiu, J.; Sun, J.; Zhang, W.; Zhou, X.; Huang, N.; Li, Y.; Li, W. Effect of Etoposide-Induced Alteration of the Mdm2-Rb Signaling Pathway on Cellular Senescence in A549 Lung Adenocarcinoma Cells. Oncol. Lett. 2017, 14, 3935–3940. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell. 2019, 73, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Pocaterra, A.; Romani, P.; Dupont, S. YAP/TAZ Functions and Their Regulation at a Glance. J. Cell Sci. 2020, 133, jcs230425. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, T.; Imoto, I.; Matsui, T.; Kozaki, K.; Haruki, S.; Sudol, M.; Shimada, Y.; Tsuda, H.; Kawano, T.; Inazawa, J. YAP Is a Candidate Oncogene for Esophageal Squamous Cell Carcinoma. Carcinogenesis 2011, 32, 389–398. [Google Scholar] [CrossRef]
- Zhao, B.; Li, L.; Wang, L.; Wang, C.Y.; Yu, J.; Guan, K.L. Cell Detachment Activates the Hippo Pathway via Cytoskeleton Reorganization to Induce Anoikis. Genes Dev. 2012, 26, 54–68. [Google Scholar] [CrossRef]
- Swift, J.; Ivanovska, I.L.; Buxboim, A.; Harada, T.; Dingal, P.C.; Pinter, J.; Pajerowski, J.D.; Spinler, K.R.; Shin, J.W.; Tewari, M.; et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 2013, 341, 1240104. [Google Scholar] [CrossRef]
- Rowat, A.C.; Jaalouk, D.E.; Zwerger, M.; Ung, W.L.; Eydelnant, I.A.; Olins, D.E.; Olins, A.L.; Herrmann, H.; Weitz, D.A.; Lammerding, J. Nuclear Envelope Composition Determines the Ability of Neutrophil-Type Cells to Passage through Micron-Scale Constrictions. J. Biol. Chem. 2013, 288, 8610–8618. [Google Scholar] [CrossRef]
- Naetar, N.; Ferraioli, S.; Foisner, R. Lamins in the Nuclear Interior—Life Outside the Lamina. J. Cell Sci. 2017, 130, 2087–2096. [Google Scholar] [CrossRef]
- Wazir, U.; Ahmed, M.H.; Bridger, J.M.; Harvey, A.; Jiang, W.G.; Sharma, A.K.; Mokbel, K. The Clinicopathological Significance of Lamin A/C, Lamin B1 and Lamin B Receptor MRNA Expression in Human Breast Cancer. Cell. Mol. Biol. Lett. 2013, 18, 595–611. [Google Scholar] [CrossRef]
- Harada, T.; Swift, J.; Irianto, J.; Shin, J.W.; Spinler, K.R.; Athirasala, A.; Diegmiller, R.; Dingal, P.C.D.P.; Ivanovska, I.L.; Discher, D.E. Nuclear Lamin Stiffness Is a Barrier to 3D Migration, but Softness Can Limit Survival. J. Cell Biol. 2014, 204, 669–682. [Google Scholar] [CrossRef]
- Khuon, S.; Liang, L.; Dettman, R.W.; Sporn, P.H.S.; Wysolmerski, R.B.; Chew, T.-L. Myosin Light Chain Kinase Mediates Transcellular Intravasation of Breast Cancer Cells through the Underlying Endothelial Cells: A Three-Dimensional FRET Study. J. Cell Sci. 2010, 123, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan Sulphate Proteoglycans Fine-Tune Mammalian Physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef]
- Vargas, J.D.; Hatch, E.M.; Anderson, D.J.; Hetzer, M.W. Transient Nuclear Envelope Rupturing during Interphase in Human Cancer Cells. Nucleus 2012, 3, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Pollard, J.W. Microenvironmental Regulation of Metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, S. DNA Damage and Lamins. Adv. Exp. Med. Biol. 2014, 773, 377–399. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roncato, F.; Regev, O.; Feigelson, S.W.; Yadav, S.K.; Kaczmarczyk, L.; Levi, N.; Drago-Garcia, D.; Ovadia, S.; Kizner, M.; Addadi, Y.; et al. Reduced Lamin A/C Does Not Facilitate Cancer Cell Transendothelial Migration but Compromises Lung Metastasis. Cancers 2021, 13, 2383. https://doi.org/10.3390/cancers13102383
Roncato F, Regev O, Feigelson SW, Yadav SK, Kaczmarczyk L, Levi N, Drago-Garcia D, Ovadia S, Kizner M, Addadi Y, et al. Reduced Lamin A/C Does Not Facilitate Cancer Cell Transendothelial Migration but Compromises Lung Metastasis. Cancers. 2021; 13(10):2383. https://doi.org/10.3390/cancers13102383
Chicago/Turabian StyleRoncato, Francesco, Ofer Regev, Sara W. Feigelson, Sandeep Kumar Yadav, Lukasz Kaczmarczyk, Nehora Levi, Diana Drago-Garcia, Samuel Ovadia, Marina Kizner, Yoseph Addadi, and et al. 2021. "Reduced Lamin A/C Does Not Facilitate Cancer Cell Transendothelial Migration but Compromises Lung Metastasis" Cancers 13, no. 10: 2383. https://doi.org/10.3390/cancers13102383
APA StyleRoncato, F., Regev, O., Feigelson, S. W., Yadav, S. K., Kaczmarczyk, L., Levi, N., Drago-Garcia, D., Ovadia, S., Kizner, M., Addadi, Y., Sabino, J. C., Ovadya, Y., de Almeida, S. F., Feldmesser, E., Gerlitz, G., & Alon, R. (2021). Reduced Lamin A/C Does Not Facilitate Cancer Cell Transendothelial Migration but Compromises Lung Metastasis. Cancers, 13(10), 2383. https://doi.org/10.3390/cancers13102383






