Current Understanding of Myelomatous Mesenchymal Stromal Cells Extended through Advances in Experimental Methods
Simple Summary
Abstract
1. Introduction
2. Hematopoietic Microenvironment in BM
2.1. Microenvironment for Hematopoietic Stem Cells or Plasma Cells in Normal BM
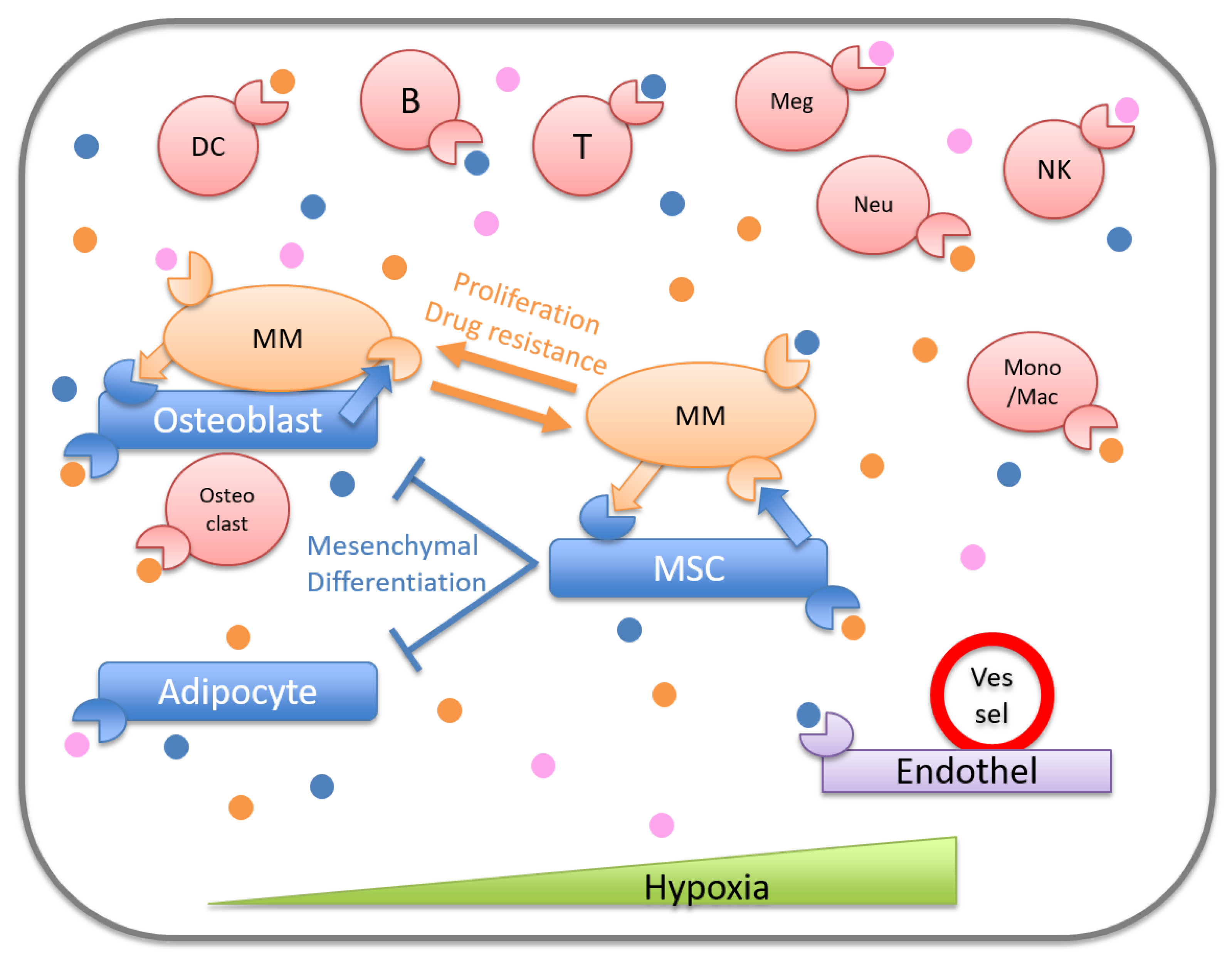
2.2. Microenvironment for Malignant Cells in BM
3. The Role of Mesenchymal Cells in Myeloma Microenvironment
3.1. Studies with Myeloma Cells
3.2. Studies with Human MSCs
3.3. Studies with Mouse Models
3.4. Studies with Humanized Mice
4. Therapeutic Application Targeting the Effects of MM-MSCs
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kyle, R.A.; Rajkumar, S.V. Multiple myeloma. N. Engl. J. Med. 2004, 351, 1860–1873. [Google Scholar] [CrossRef]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.-V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef]
- Pawlyn, C.; Morgan, G.J. Evolutionary biology of high-risk multiple myeloma. Nat. Rev. Cancer 2017, 17, 543–556. [Google Scholar] [CrossRef]
- Manier, S.; Salem, K.Z.; Park, J.; Landau, D.A.; Getz, G.; Ghobrial, I.M. Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Kristinsson, S.Y.; Landgren, O.; Dickman, P.W.; Derolf, A.R.; Björkholm, M. Patterns of survival in multiple myeloma: A population-based study of patients diagnosed in Sweden from 1973 to 2003. J. Clin. Oncol. 2007, 25, 1993–1999. [Google Scholar] [CrossRef] [PubMed]
- Sonneveld, P.; Avet-Loiseau, H.; Lonial, S.; Usmani, S.; Siegel, D.; Anderson, K.C.; Chng, W.J.; Moreau, P.; Attal, M.; Kyle, R.A.; et al. Treatment of multiple myeloma with high-risk cytogenetics: A consensus of the International Myeloma Working Group. Blood 2016, 127, 2955–2962. [Google Scholar] [CrossRef] [PubMed]
- Schofield, R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 1978, 4, 7–25. [Google Scholar]
- Kfoury, Y.; Scadden, D.T. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell 2015, 16, 239–253. [Google Scholar] [CrossRef]
- Pinho, S.; Frenette, P.S. Haematopoietic stem cell activity and interactions with the niche. Nat. Rev. Mol. Cell Biol. 2019, 20, 303–320. [Google Scholar] [CrossRef]
- Crane, G.M.; Jeffery, E.; Morrison, S.J. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 2017, 17, 573–590. [Google Scholar] [CrossRef]
- Kokkaliaris, K.D. Dissecting the spatial bone marrow microenvironment of hematopoietic stem cells. Curr. Opin. Oncol. 2020, 32, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Ferrer, S.; Scadden, D.T.; Sanchez-Aguilera, A. Bone marrow stem cells: Current and emerging concepts. Ann. N. Y. Acad. Sci. 2015, 1335, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Yu, V.W.; Scadden, D.T. Heterogeneity of the bone marrow niche. Curr. Opin. Hematol. 2016, 23, 331–338. [Google Scholar] [CrossRef]
- Wei, Q.; Frenette, P.S. Niches for Hematopoietic Stem Cells and Their Progeny. Immunity 2018, 48, 632–648. [Google Scholar] [CrossRef]
- Yu, V.W.; Lymperi, S.; Oki, T.; Jones, A.; Swiatek, P.; Vasic, R.; Ferraro, F.; Scadden, D.T. Distinctive Mesenchymal-Parenchymal Cell Pairings Govern B Cell Differentiation in the Bone Marrow. Stem Cell Rep. 2016, 7, 220–235. [Google Scholar] [CrossRef]
- Cordeiro Gomes, A.; Hara, T.; Lim, V.Y.; Herndler-Brandstetter, D.; Nevius, E.; Sugiyama, T.; Tani-Ichi, S.; Schlenner, S.; Richie, E.; Rodewald, H.R.; et al. Hematopoietic Stem Cell Niches Produce Lineage-Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity 2016, 45, 1219–1231. [Google Scholar] [CrossRef]
- Tokoyoda, K.; Egawa, T.; Sugiyama, T.; Choi, B.I.; Nagasawa, T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity 2004, 20, 707–718. [Google Scholar] [CrossRef]
- Chang, H.D.; Tokoyoda, K.; Radbruch, A. Immunological memories of the bone marrow. Immunol. Rev. 2018, 283, 86–98. [Google Scholar] [CrossRef]
- Cyster, J.G. Homing of antibody secreting cells. Immunol. Rev. 2003, 194, 48–60. [Google Scholar] [CrossRef]
- Radbruch, A.; Muehlinghaus, G.; Luger, E.O.; Inamine, A.; Smith, K.G.; Dorner, T.; Hiepe, F. Competence and competition: The challenge of becoming a long-lived plasma cell. Nat. Rev. Immunol. 2006, 6, 741–750. [Google Scholar] [CrossRef]
- Belnoue, E.; Tougne, C.; Rochat, A.F.; Lambert, P.H.; Pinschewer, D.D.; Siegrist, C.A. Homing and adhesion patterns determine the cellular composition of the bone marrow plasma cell niche. J. Immunol. 2012, 188, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Chu, V.T.; Berek, C. The establishment of the plasma cell survival niche in the bone marrow. Immunol. Rev. 2013, 251, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.E.; Debes, G.F.; Arce, S.; Cassese, G.; Hamann, A.; Radbruch, A.; Manz, R.A. Chemotactic responsiveness toward ligands for CXCR3 and CXCR4 is regulated on plasma blasts during the time course of a memory immune response. J. Immunol. 2002, 169, 1277–1282. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Hieshima, K.; Izawa, D.; Tatsumi, Y.; Kanamaru, A.; Yoshie, O. Cutting edge: Profile of chemokine receptor expression on human plasma cells accounts for their efficient recruitment to target tissues. J. Immunol. 2003, 170, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- DiLillo, D.J.; Hamaguchi, Y.; Ueda, Y.; Yang, K.; Uchida, J.; Haas, K.M.; Kelsoe, G.; Tedder, T.F. Maintenance of long-lived plasma cells and serological memory despite mature and memory B cell depletion during CD20 immunotherapy in mice. J. Immunol. 2008, 180, 361–371. [Google Scholar] [CrossRef]
- Zehentmeier, S.; Roth, K.; Cseresnyes, Z.; Sercan, O.; Horn, K.; Niesner, R.A.; Chang, H.D.; Radbruch, A.; Hauser, A.E. Static and dynamic components synergize to form a stable survival niche for bone marrow plasma cells. Eur. J. Immunol. 2014, 44, 2306–2317. [Google Scholar] [CrossRef]
- Belnoue, E.; Pihlgren, M.; McGaha, T.L.; Tougne, C.; Rochat, A.F.; Bossen, C.; Schneider, P.; Huard, B.; Lambert, P.H.; Siegrist, C.A. APRIL is critical for plasmablast survival in the bone marrow and poorly expressed by early-life bone marrow stromal cells. Blood 2008, 111, 2755–2764. [Google Scholar] [CrossRef]
- O’Connor, B.P.; Raman, V.S.; Erickson, L.D.; Cook, W.J.; Weaver, L.K.; Ahonen, C.; Lin, L.L.; Mantchev, G.T.; Bram, R.J.; Noelle, R.J. BCMA is essential for the survival of long-lived bone marrow plasma cells. J. Exp. Med. 2004, 199, 91–98. [Google Scholar] [CrossRef]
- Chu, V.T.; Frohlich, A.; Steinhauser, G.; Scheel, T.; Roch, T.; Fillatreau, S.; Lee, J.J.; Lohning, M.; Berek, C. Eosinophils are required for the maintenance of plasma cells in the bone marrow. Nat. Immunol. 2011, 12, 151–159. [Google Scholar] [CrossRef]
- Le, P.M.; Andreeff, M.; Battula, V.L. Osteogenic niche in the regulation of normal hematopoiesis and leukemogenesis. Haematologica 2018, 103, 1945–1955. [Google Scholar] [CrossRef]
- Witkowski, M.T.; Kousteni, S.; Aifantis, I. Mapping and targeting of the leukemic microenvironment. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Ghobrial, I.M.; Detappe, A.; Anderson, K.C.; Steensma, D.P. The bone-marrow niche in MDS and MGUS: Implications for AML and MM. Nat. Rev. Clin. Oncol. 2018, 15, 219–233. [Google Scholar] [CrossRef]
- Krause, D.S.; Fulzele, K.; Catic, A.; Sun, C.C.; Dombkowski, D.; Hurley, M.P.; Lezeau, S.; Attar, E.; Wu, J.Y.; Lin, H.Y.; et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat. Med. 2013, 19, 1513–1517. [Google Scholar] [CrossRef]
- Battula, V.L.; Le, P.M.; Sun, J.C.; Nguyen, K.; Yuan, B.; Zhou, X.; Sonnylal, S.; McQueen, T.; Ruvolo, V.; Michel, K.A.; et al. AML-induced osteogenic differentiation in mesenchymal stromal cells supports leukemia growth. JCI Insight 2017, 2. [Google Scholar] [CrossRef]
- Baryawno, N.; Przybylski, D.; Kowalczyk, M.S.; Kfoury, Y.; Severe, N.; Gustafsson, K.; Kokkaliaris, K.D.; Mercier, F.; Tabaka, M.; Hofree, M.; et al. A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell 2019, 177, 1915–1932. [Google Scholar] [CrossRef]
- Duarte, D.; Hawkins, E.D.; Akinduro, O.; Ang, H.; De Filippo, K.; Kong, I.Y.; Haltalli, M.; Ruivo, N.; Straszkowski, L.; Vervoort, S.J.; et al. Inhibition of Endosteal Vascular Niche Remodeling Rescues Hematopoietic Stem Cell Loss in AML. Cell Stem Cell 2018, 22, 64–77. [Google Scholar] [CrossRef]
- Decker, M.; Martinez-Morentin, L.; Wang, G.; Lee, Y.; Liu, Q.; Leslie, J.; Ding, L. Leptin-receptor-expressing bone marrow stromal cells are myofibroblasts in primary myelofibrosis. Nat. Cell Biol. 2017, 19, 677–688. [Google Scholar] [CrossRef]
- Dong, L.; Yu, W.M.; Zheng, H.; Loh, M.L.; Bunting, S.T.; Pauly, M.; Huang, G.; Zhou, M.; Broxmeyer, H.E.; Scadden, D.T.; et al. Leukaemogenic effects of Ptpn11 activating mutations in the stem cell microenvironment. Nature 2016, 539, 304–308. [Google Scholar] [CrossRef]
- Uccelli, A.; de Rosbo, N.K. The immunomodulatory function of mesenchymal stem cells: Mode of action and pathways. Ann. N. Y. Acad. Sci. 2015, 1351, 114–126. [Google Scholar] [CrossRef]
- Ghannam, S.; Pene, J.; Moquet-Torcy, G.; Jorgensen, C.; Yssel, H. Mesenchymal stem cells inhibit human Th17 cell differentiation and function and induce a T regulatory cell phenotype. J. Immunol. 2010, 185, 302–312. [Google Scholar] [CrossRef]
- Raaijmakers, M.H.; Mukherjee, S.; Guo, S.; Zhang, S.; Kobayashi, T.; Schoonmaker, J.A.; Ebert, B.L.; Al-Shahrour, F.; Hasserjian, R.P.; Scadden, E.O.; et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 2010, 464, 852–857. [Google Scholar] [CrossRef]
- Zambetti, N.A.; Ping, Z.; Chen, S.; Kenswil, K.J.G.; Mylona, M.A.; Sanders, M.A.; Hoogenboezem, R.M.; Bindels, E.M.J.; Adisty, M.N.; Van Strien, P.M.H.; et al. Mesenchymal Inflammation Drives Genotoxic Stress in Hematopoietic Stem Cells and Predicts Disease Evolution in Human Pre-leukemia. Cell Stem Cell 2016, 19, 613–627. [Google Scholar] [CrossRef]
- Kode, A.; Mosialou, I.; Manavalan, S.J.; Rathinam, C.V.; Friedman, R.A.; Teruya-Feldstein, J.; Bhagat, G.; Berman, E.; Kousteni, S. FoxO1-dependent induction of acute myeloid leukemia by osteoblasts in mice. Leukemia 2016, 30, 1–13. [Google Scholar] [CrossRef]
- Kode, A.; Manavalan, J.S.; Mosialou, I.; Bhagat, G.; Rathinam, C.V.; Luo, N.; Khiabanian, H.; Lee, A.; Murty, V.V.; Friedman, R.; et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature 2014, 506, 240–244. [Google Scholar] [CrossRef]
- Reagan, M.R.; Ghobrial, I.M. Multiple myeloma mesenchymal stem cells: Characterization, origin, and tumor-promoting effects. Clin. Cancer Res. 2012, 18, 342–349. [Google Scholar] [CrossRef]
- Xu, S.; De Veirman, K.; De Becker, A.; Vanderkerken, K.; Van Riet, I. Mesenchymal stem cells in multiple myeloma: A therapeutical tool or target? Leukemia 2018, 32, 1500–1514. [Google Scholar] [CrossRef]
- Hu, J.; Van Valckenborgh, E.; Menu, E.; De Bruyne, E.; Vanderkerken, K. Understanding the hypoxic niche of multiple myeloma: Therapeutic implications and contributions of mouse models. Dis. Models Mech. 2012, 5, 763–771. [Google Scholar] [CrossRef]
- Azab, A.K.; Hu, J.; Quang, P.; Azab, F.; Pitsillides, C.; Awwad, R.; Thompson, B.; Maiso, P.; Sun, J.D.; Hart, C.P.; et al. Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition-like features. Blood 2012, 119, 5782–5794. [Google Scholar] [CrossRef]
- Hideshima, T.; Chauhan, D.; Podar, K.; Schlossman, R.L.; Richardson, P.; Anderson, K.C. Novel therapies targeting the myeloma cell and its bone marrow microenvironment. Semin. Oncol. 2001, 28, 607–612. [Google Scholar] [CrossRef]
- Dhodapkar, M.V. MGUS to myeloma: A mysterious gammopathy of underexplored significance. Blood 2016, 128, 2599–2606. [Google Scholar] [CrossRef]
- Standal, T.; Abildgaard, N.; Fagerli, U.M.; Stordal, B.; Hjertner, O.; Borset, M.; Sundan, A. HGF inhibits BMP-induced osteoblastogenesis: Possible implications for the bone disease of multiple myeloma. Blood 2007, 109, 3024–3030. [Google Scholar] [CrossRef]
- Tian, E.; Zhan, F.; Walker, R.; Rasmussen, E.; Ma, Y.; Barlogie, B.; Shaughnessy, J.D., Jr. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N. Engl. J. Med. 2003, 349, 2483–2494. [Google Scholar] [CrossRef]
- Giuliani, N.; Morandi, F.; Tagliaferri, S.; Lazzaretti, M.; Donofrio, G.; Bonomini, S.; Sala, R.; Mangoni, M.; Rizzoli, V. Production of Wnt inhibitors by myeloma cells: Potential effects on canonical Wnt pathway in the bone microenvironment. Cancer Res. 2007, 67, 7665–7674. [Google Scholar] [CrossRef]
- Oshima, T.; Abe, M.; Asano, J.; Hara, T.; Kitazoe, K.; Sekimoto, E.; Tanaka, Y.; Shibata, H.; Hashimoto, T.; Ozaki, S.; et al. Myeloma cells suppress bone formation by secreting a soluble Wnt inhibitor, sFRP-2. Blood 2005, 106, 3160–3165. [Google Scholar] [CrossRef]
- Brunetti, G.; Oranger, A.; Mori, G.; Specchia, G.; Rinaldi, E.; Curci, P.; Zallone, A.; Rizzi, R.; Grano, M.; Colucci, S. Sclerostin is overexpressed by plasma cells from multiple myeloma patients. Ann. N. Y. Acad. Sci. 2011, 1237, 19–23. [Google Scholar] [CrossRef]
- Bisping, G.; Leo, R.; Wenning, D.; Dankbar, B.; Padro, T.; Kropff, M.; Scheffold, C.; Kroger, M.; Mesters, R.M.; Berdel, W.E.; et al. Paracrine interactions of basic fibroblast growth factor and interleukin-6 in multiple myeloma. Blood 2003, 101, 2775–2783. [Google Scholar] [CrossRef]
- Feng, Y.; Wen, J.; Mike, P.; Choi, D.S.; Eshoa, C.; Shi, Z.Z.; Zu, Y.; Chang, C.C. Bone marrow stromal cells from myeloma patients support the growth of myeloma stem cells. Stem Cells Dev. 2010, 19, 1289–1296. [Google Scholar] [CrossRef]
- Xu, S.; Menu, E.; De Becker, A.; Van Camp, B.; Vanderkerken, K.; Van Riet, I. Bone marrow-derived mesenchymal stromal cells are attracted by multiple myeloma cell-produced chemokine CCL25 and favor myeloma cell growth in vitro and in vivo. Stem Cells 2012, 30, 266–279. [Google Scholar] [CrossRef]
- Sanz-Rodríguez, F.; Teixidó, J. VLA-4-dependent myeloma cell adhesion. Leuk. Lymphoma 2001, 41, 239–245. [Google Scholar] [CrossRef]
- Damiano, J.S.; Cress, A.E.; Hazlehurst, L.A.; Shtil, A.A.; Dalton, W.S. Cell adhesion mediated drug resistance (CAM-DR): Role of integrins and resistance to apoptosis in human myeloma cell lines. Blood 1999, 93, 1658–1667. [Google Scholar] [CrossRef]
- Noborio-Hatano, K.; Kikuchi, J.; Takatoku, M.; Shimizu, R.; Wada, T.; Ueda, M.; Nobuyoshi, M.; Oh, I.; Sato, K.; Suzuki, T.; et al. Bortezomib overcomes cell-adhesion-mediated drug resistance through downregulation of VLA-4 expression in multiple myeloma. Oncogene 2009, 28, 231–242. [Google Scholar] [CrossRef]
- Ely, S.A.; Knowles, D.M. Expression of CD56/Neural Cell Adhesion Molecule Correlates with the Presence of Lytic Bone Lesions in Multiple Myeloma and Distinguishes Myeloma from Monoclonal Gammopathy of Undetermined Significance and Lymphomas with Plasmacytoid Differentiation. Am. J. Pathol. 2002, 160, 1293–1299. [Google Scholar] [CrossRef]
- Landowski, T.H.; Olashaw, N.E.; Agrawal, D.; Dalton, W.S. Cell adhesion-mediated drug resistance (CAM-DR) is associated with activation of NF-kappa B (RelB/p50) in myeloma cells. Oncogene 2003, 22, 2417–2421. [Google Scholar] [CrossRef]
- Natoni, A.; Farrell, M.L.; Harris, S.; Falank, C.; Kirkham-McCarthy, L.; Macauley, M.S.; Reagan, M.R.; O’Dwyer, M. Sialyltransferase inhibition leads to inhibition of tumor cell interactions with E-selectin, VCAM1, and MADCAM1, and improves survival in a human multiple myeloma mouse model. Haematologica 2020, 105, 457–467. [Google Scholar] [CrossRef]
- Hosen, N.; Matsunaga, Y.; Hasegawa, K.; Matsuno, H.; Nakamura, Y.; Makita, M.; Watanabe, K.; Yoshida, M.; Satoh, K.; Morimoto, S.; et al. The activated conformation of integrin beta7 is a novel multiple myeloma-specific target for CAR T cell therapy. Nat. Med. 2017, 23, 1436–1443. [Google Scholar] [CrossRef]
- Paiva, B.; Corchete, L.A.; Vidriales, M.B.; Puig, N.; Maiso, P.; Rodriguez, I.; Alignani, D.; Burgos, L.; Sanchez, M.L.; Barcena, P.; et al. Phenotypic and genomic analysis of multiple myeloma minimal residual disease tumor cells: A new model to understand chemoresistance. Blood 2016, 127, 1896–1906. [Google Scholar] [CrossRef]
- Paino, T.; Paiva, B.; Sayagues, J.M.; Mota, I.; Carvalheiro, T.; Corchete, L.A.; Aires-Mejia, I.; Perez, J.J.; Sanchez, M.L.; Barcena, P.; et al. Phenotypic identification of subclones in multiple myeloma with different chemoresistant, cytogenetic and clonogenic potential. Leukemia 2015, 29, 1186–1194. [Google Scholar] [CrossRef]
- Furukawa, M.; Ohkawara, H.; Ogawa, K.; Ikeda, K.; Ueda, K.; Shichishima-Nakamura, A.; Ito, E.; Imai, J.I.; Yanagisawa, Y.; Honma, R.; et al. Autocrine and Paracrine Interactions between Multiple Myeloma Cells and Bone Marrow Stromal Cells by Growth Arrest-specific Gene 6 Cross-talk with Interleukin-6. J. Biol. Chem. 2017, 292, 4280–4292. [Google Scholar] [CrossRef]
- Hurt, E.M.; Wiestner, A.; Rosenwald, A.; Shaffer, A.L.; Campo, E.; Grogan, T.; Bergsagel, P.L.; Kuehl, W.M.; Staudt, L.M. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell 2004, 5, 191–199. [Google Scholar] [CrossRef]
- Moreaux, J.; Hose, D.; Jourdan, M.; Reme, T.; Hundemer, M.; Moos, M.; Robert, N.; Moine, P.; De Vos, J.; Goldschmidt, H.; et al. TACI expression is associated with a mature bone marrow plasma cell signature and C-MAF overexpression in human myeloma cell lines. Haematologica 2007, 92, 803–811. [Google Scholar] [CrossRef]
- Lomas, O.C.; Tahri, S.; Ghobrial, I.M. The microenvironment in myeloma. Curr. Opin. Oncol. 2020, 32, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Terpos, E.; Ntanasis-Stathopoulos, I.; Dimopoulos, M.A. Myeloma bone disease: From biology findings to treatment approaches. Blood 2019, 133, 1534–1539. [Google Scholar] [CrossRef] [PubMed]
- Corre, J.; Mahtouk, K.; Attal, M.; Gadelorge, M.; Huynh, A.; Fleury-Cappellesso, S.; Danho, C.; Laharrague, P.; Klein, B.; Reme, T.; et al. Bone marrow mesenchymal stem cells are abnormal in multiple myeloma. Leukemia 2007, 21, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Corre, J.; Labat, E.; Espagnolle, N.; Hebraud, B.; Avet-Loiseau, H.; Roussel, M.; Huynh, A.; Gadelorge, M.; Cordelier, P.; Klein, B.; et al. Bioactivity and prognostic significance of growth differentiation factor GDF15 secreted by bone marrow mesenchymal stem cells in multiple myeloma. Cancer Res. 2012, 72, 1395–1406. [Google Scholar] [CrossRef]
- Fernando, R.C.; Mazzotti, D.R.; Azevedo, H.; Sandes, A.F.; Rizzatti, E.G.; de Oliveira, M.B.; Alves, V.L.F.; Eugenio, A.I.P.; de Carvalho, F.; Dalboni, M.A.; et al. Transcriptome Analysis of Mesenchymal Stem Cells from Multiple Myeloma Patients Reveals Downregulation of Genes Involved in Cell Cycle Progression, Immune Response, and Bone Metabolism. Sci. Rep. 2019, 9, 1056. [Google Scholar] [CrossRef]
- Alameda, D.; Saez, B.; Lara-Astiaso, D.; Sarvide, S.; Lasa, M.; Alignani, D.; Rodriguez, I.; Garate, S.; Vilas, A.; Paiva, B.; et al. Characterization of freshly isolated bone marrow mesenchymal stromal cells from healthy donors and patients with multiple myeloma: Transcriptional modulation of the microenvironment. Haematologica 2020, 105, e470–e473. [Google Scholar] [CrossRef]
- Garcia-Gomez, A.; De Las Rivas, J.; Ocio, E.M.; Díaz-Rodríguez, E.; Montero, J.C.; Martín, M.; Blanco, J.F.; Sanchez-Guijo, F.M.; Pandiella, A.; San Miguel, J.F.; et al. Transcriptomic profile induced in bone marrow mesenchymal stromal cells after interaction with multiple myeloma cells: Implications in myeloma progression and myeloma bone disease. Oncotarget 2014, 5, 8284–8305. [Google Scholar] [CrossRef]
- Arnulf, B.; Lecourt, S.; Soulier, J.; Ternaux, B.; Lacassagne, M.N.; Crinquette, A.; Dessoly, J.; Sciaini, A.K.; Benbunan, M.; Chomienne, C.; et al. Phenotypic and functional characterization of bone marrow mesenchymal stem cells derived from patients with multiple myeloma. Leukemia 2007, 21, 158–163. [Google Scholar] [CrossRef]
- Andre, T.; Meuleman, N.; Stamatopoulos, B.; De Bruyn, C.; Pieters, K.; Bron, D.; Lagneaux, L. Evidences of early senescence in multiple myeloma bone marrow mesenchymal stromal cells. PLoS ONE 2013, 8, e59756. [Google Scholar] [CrossRef]
- Garderet, L.; Mazurier, C.; Chapel, A.; Ernou, I.; Boutin, L.; Holy, X.; Gorin, N.C.; Lopez, M.; Doucet, C.; Lataillade, J.J. Mesenchymal stem cell abnormalities in patients with multiple myeloma. Leuk. Lymphoma 2007, 48, 2032–2041. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, Y.; Fei, C.; Zhao, S.; Zheng, Q.; Su, J.; Wu, D.; Li, X.; Chang, C. Dicer1 downregulation by multiple myeloma cells promotes the senescence and tumor-supporting capacity and decreases the differentiation potential of mesenchymal stem cells. Cell Death Dis. 2018, 9, 512. [Google Scholar] [CrossRef] [PubMed]
- Kassen, D.; Moore, S.; Percy, L.; Herledan, G.; Bounds, D.; Rodriguez-Justo, M.; Croucher, P.; Yong, K. The bone marrow stromal compartment in multiple myeloma patients retains capability for osteogenic differentiation in vitro: Defining the stromal defect in myeloma. Br. J. Haematol. 2014, 167, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Zdzisinska, B.; Bojarska-Junak, A.; Dmoszynska, A.; Kandefer-Szerszen, M. Abnormal cytokine production by bone marrow stromal cells of multiple myeloma patients in response to RPMI8226 myeloma cells. Arch. Immunol. Ther. Exp. (Warsz) 2008, 56, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Y.; Li, A.; Liu, X.; Xing, X.; Guo, Y.; Xu, Y.; Hao, Y.; Zheng, C. Telomere length is positively associated with the expression of IL6 and MIP1alpha in bone marrow mesenchymal stem cells of multiple myeloma. Mol. Med. Rep. 2017, 16, 2497–2504. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, X.; Pennisi, A.; Yaccoby, S. Role of decorin in the antimyeloma effects of osteoblasts. Blood 2008, 112, 159–168. [Google Scholar] [CrossRef]
- Todoerti, K.; Lisignoli, G.; Storti, P.; Agnelli, L.; Novara, F.; Manferdini, C.; Codeluppi, K.; Colla, S.; Crugnola, M.; Abeltino, M.; et al. Distinct transcriptional profiles characterize bone microenvironment mesenchymal cells rather than osteoblasts in relationship with multiple myeloma bone disease. Exp. Hematol. 2010, 38, 141–153. [Google Scholar] [CrossRef]
- Wallace, S.R.; Oken, M.M.; Lunetta, K.L.; Panoskaltsis-Mortari, A.; Masellis, A.M. Abnormalities of bone marrow mesenchymal cells in multiple myeloma patients. Cancer 2001, 91, 1219–1230. [Google Scholar] [CrossRef]
- McNee, G.; Eales, K.L.; Wei, W.; Williams, D.S.; Barkhuizen, A.; Bartlett, D.B.; Essex, S.; Anandram, S.; Filer, A.; Moss, P.A.; et al. Citrullination of histone H3 drives IL-6 production by bone marrow mesenchymal stem cells in MGUS and multiple myeloma. Leukemia 2017, 31, 373–381. [Google Scholar] [CrossRef]
- Berenstein, R.; Nogai, A.; Waechter, M.; Blau, O.; Kuehnel, A.; Schmidt-Hieber, M.; Kunitz, A.; Pezzutto, A.; Dörken, B.; Blau, I.W. Multiple myeloma cells modify VEGF/IL-6 levels and osteogenic potential of bone marrow stromal cells via Notch/miR-223. Mol. Carcinog. 2016, 55, 1927–1939. [Google Scholar] [CrossRef]
- Kristensen, I.B.; Pedersen, L.; Ro, T.B.; Christensen, J.H.; Lyng, M.B.; Rasmussen, L.M.; Ditzel, H.J.; Borset, M.; Abildgaard, N. Decorin is down-regulated in multiple myeloma and MGUS bone marrow plasma and inhibits HGF-induced myeloma plasma cell viability and migration. Eur. J. Haematol. 2013, 91, 196–200. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Maiso, P.; Azab, A.K.; Tai, Y.T.; Reagan, M.; Azab, F.; Flores, L.M.; Campigotto, F.; Weller, E.; et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J. Clin. Investig. 2013, 123, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- Umezu, T.; Imanishi, S.; Yoshizawa, S.; Kawana, C.; Ohyashiki, J.H.; Ohyashiki, K. Induction of multiple myeloma bone marrow stromal cell apoptosis by inhibiting extracellular vesicle miR-10a secretion. Blood Adv. 2019, 3, 3228–3240. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Hiura, K.; Ozaki, S.; Kido, S.; Matsumoto, T. Vicious cycle between myeloma cell binding to bone marrow stromal cells via VLA-4-VCAM-1 adhesion and macrophage inflammatory protein-1alpha and MIP-1beta production. J. Bone Miner. Metab. 2009, 27, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Kirshner, J.; Thulien, K.J.; Martin, L.D.; Debes Marun, C.; Reiman, T.; Belch, A.R.; Pilarski, L.M. A unique three-dimensional model for evaluating the impact of therapy on multiple myeloma. Blood 2008, 112, 2935–2945. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, J.; Koyama, D.; Wada, T.; Izumi, T.; Hofgaard, P.O.; Bogen, B.; Furukawa, Y. Phosphorylation-mediated EZH2 inactivation promotes drug resistance in multiple myeloma. J. Clin. Investig. 2015, 125, 4375–4390. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, Y.; Kikuchi, J. Epigenetic mechanisms of cell adhesion-mediated drug resistance in multiple myeloma. Int. J. Hematol. 2016, 104, 281–292. [Google Scholar] [CrossRef]
- Ohguchi, H.; Harada, T.; Sagawa, M.; Kikuchi, S.; Tai, Y.T.; Richardson, P.G.; Hideshima, T.; Anderson, K.C. KDM6B modulates MAPK pathway mediating multiple myeloma cell growth and survival. Leukemia 2017, 31, 2661–2669. [Google Scholar] [CrossRef]
- Matsumoto, T.; Abe, M. TGF-β-related mechanisms of bone destruction in multiple myeloma. Bone 2011, 48, 129–134. [Google Scholar] [CrossRef]
- Giuliani, N.; Rizzoli, V.; Roodman, G.D. Multiple myeloma bone disease: Pathophysiology of osteoblast inhibition. Blood 2006, 108, 3992–3996. [Google Scholar] [CrossRef]
- Mehdi, S.J.; Johnson, S.K.; Epstein, J.; Zangari, M.; Qu, P.; Hoering, A.; van Rhee, F.; Schinke, C.; Thanendrarajan, S.; Barlogie, B.; et al. Mesenchymal stem cells gene signature in high-risk myeloma bone marrow linked to suppression of distinct IGFBP2-expressing small adipocytes. Br. J. Haematol. 2019, 184, 578–593. [Google Scholar] [CrossRef]
- Silvestris, F.; Cafforio, P.; Calvani, N.; Dammacco, F. Impaired osteoblastogenesis in myeloma bone disease: Role of upregulated apoptosis by cytokines and malignant plasma cells. Br. J. Haematol. 2004, 126, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, N.; Colla, S.; Morandi, F.; Lazzaretti, M.; Sala, R.; Bonomini, S.; Grano, M.; Colucci, S.; Svaldi, M.; Rizzoli, V. Myeloma cells block RUNX2/CBFA1 activity in human bone marrow osteoblast progenitors and inhibit osteoblast formation and differentiation. Blood 2005, 106, 2472–2483. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Abe, M.; Hiasa, M.; Oda, A.; Amou, H.; Kido, S.; Harada, T.; Tanaka, O.; Miki, H.; Nakamura, S.; et al. Tgf-Beta inhibition restores terminal osteoblast differentiation to suppress myeloma growth. PLoS ONE 2010, 5, e9870. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Lovendorf, M.B.; Park, J.; Salem, K.Z.; Reagan, M.R.; Manier, S.; Zavidij, O.; Rahmat, M.; Huynh, D.; Takagi, S.; et al. Inhibition of microRNA-138 enhances bone formation in multiple myeloma bone marrow niche. Leukemia 2018, 32, 1739–1750. [Google Scholar] [CrossRef]
- Xu, S.; Cecilia Santini, G.; De Veirman, K.; Vande Broek, I.; Leleu, X.; De Becker, A.; Van Camp, B.; Vanderkerken, K.; Van Riet, I. Upregulation of miR-135b is involved in the impaired osteogenic differentiation of mesenchymal stem cells derived from multiple myeloma patients. PLoS ONE 2013, 8, e79752. [Google Scholar] [CrossRef]
- Garayoa, M.; Garcia, J.L.; Santamaria, C.; Garcia-Gomez, A.; Blanco, J.F.; Pandiella, A.; Hernández, J.M.; Sanchez-Guijo, F.M.; del Cañizo, M.C.; Gutiérrez, N.C.; et al. Mesenchymal stem cells from multiple myeloma patients display distinct genomic profile as compared with those from normal donors. Leukemia 2009, 23, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Berenstein, R.; Blau, O.; Nogai, A.; Waechter, M.; Slonova, E.; Schmidt-Hieber, M.; Kunitz, A.; Pezzutto, A.; Doerken, B.; Blau, I.W. Multiple myeloma cells alter the senescence phenotype of bone marrow mesenchymal stromal cells under participation of the DLK1-DIO3 genomic region. BMC Cancer 2015, 15, 68. [Google Scholar] [CrossRef]
- Adamik, J.; Roodman, G.D.; Galson, D.L. Epigenetic-Based Mechanisms of Osteoblast Suppression in Multiple Myeloma Bone Disease. JBMR Plus 2019, 3, e10183. [Google Scholar] [CrossRef]
- Adamik, J.; Jin, S.; Sun, Q.; Zhang, P.; Weiss, K.R.; Anderson, J.L.; Silbermann, R.; Roodman, G.D.; Galson, D.L. EZH2 or HDAC1 Inhibition Reverses Multiple Myeloma-Induced Epigenetic Suppression of Osteoblast Differentiation. Mol. Cancer Res. 2017, 15, 405–417. [Google Scholar] [CrossRef]
- D’Souza, S.; del Prete, D.; Jin, S.; Sun, Q.; Huston, A.J.; Kostov, F.E.; Sammut, B.; Hong, C.S.; Anderson, J.L.; Patrene, K.D.; et al. Gfi1 expressed in bone marrow stromal cells is a novel osteoblast suppressor in patients with multiple myeloma bone disease. Blood 2011, 118, 6871–6880. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Churchman, S.M.; Ponchel, F.; Boxall, S.A.; Cuthbert, R.; Kouroupis, D.; Roshdy, T.; Giannoudis, P.V.; Emery, P.; McGonagle, D.; Jones, E.A. Transcriptional profile of native CD271+ multipotential stromal cells: Evidence for multiple fates, with prominent osteogenic and Wnt pathway signaling activity. Arthritis Rheum. 2012, 64, 2632–2643. [Google Scholar] [CrossRef] [PubMed]
- Harichandan, A.; Sivasubramaniyan, K.; Bühring, H.J. Prospective isolation and characterization of human bone marrow-derived MSCs. Adv. Biochem. Eng. Biotechnol. 2013, 129, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Mabuchi, Y.; Morikawa, S.; Harada, S.; Niibe, K.; Suzuki, S.; Renault-Mihara, F.; Houlihan, D.D.; Akazawa, C.; Okano, H.; Matsuzaki, Y. LNGFR(+)THY-1(+)VCAM-1(hi+) cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Rep. 2013, 1, 152–165. [Google Scholar] [CrossRef]
- Tormin, A.; Li, O.; Brune, J.C.; Walsh, S.; Schutz, B.; Ehinger, M.; Ditzel, N.; Kassem, M.; Scheding, S. CD146 expression on primary nonhematopoietic bone marrow stem cells is correlated with in situ localization. Blood 2011, 117, 5067–5077. [Google Scholar] [CrossRef]
- Rossi, M.; Botta, C.; Arbitrio, M.; Grembiale, R.D.; Tagliaferri, P.; Tassone, P. Mouse models of multiple myeloma: Technologic platforms and perspectives. Oncotarget 2018, 9, 20119–20133. [Google Scholar] [CrossRef]
- Radl, J.; De Glopper, E.D.; Schuit, H.R.; Zurcher, C. Idiopathic paraproteinemia. II. Transplantation of the paraprotein-producing clone from old to young C57BL/KaLwRij mice. J. Immunol. 1979, 122, 609–613. [Google Scholar]
- Radl, J.; Croese, J.W.; Zurcher, C.; Van den Enden-Vieveen, M.H.; de Leeuw, A.M. Animal model of human disease. Multiple myeloma. Am. J. Pathol. 1988, 132, 593–597. [Google Scholar]
- Fowler, J.A.; Lwin, S.T.; Drake, M.T.; Edwards, J.R.; Kyle, R.A.; Mundy, G.R.; Edwards, C.M. Host-derived adiponectin is tumor-suppressive and a novel therapeutic target for multiple myeloma and the associated bone disease. Blood 2011, 118, 5872–5882. [Google Scholar] [CrossRef]
- Fowler, J.A.; Mundy, G.R.; Lwin, S.T.; Edwards, C.M. Bone marrow stromal cells create a permissive microenvironment for myeloma development: A new stromal role for Wnt inhibitor Dkk1. Cancer Res. 2012, 72, 2183–2189. [Google Scholar] [CrossRef]
- Gooding, S.; Olechnowicz, S.W.Z.; Morris, E.V.; Armitage, A.E.; Arezes, J.; Frost, J.; Repapi, E.; Edwards, J.R.; Ashley, N.; Waugh, C.; et al. Transcriptomic profiling of the myeloma bone-lining niche reveals BMP signalling inhibition to improve bone disease. Nat. Commun. 2019, 10, 4533. [Google Scholar] [CrossRef] [PubMed]
- Lawson, M.A.; McDonald, M.M.; Kovacic, N.; Hua Khoo, W.; Terry, R.L.; Down, J.; Kaplan, W.; Paton-Hough, J.; Fellows, C.; Pettitt, J.A.; et al. Osteoclasts control reactivation of dormant myeloma cells by remodelling the endosteal niche. Nat. Commun. 2015, 6, 8983. [Google Scholar] [CrossRef]
- Khoo, W.H.; Ledergor, G.; Weiner, A.; Roden, D.L.; Terry, R.L.; McDonald, M.M.; Chai, R.C.; De Veirman, K.; Owen, K.L.; Opperman, K.S.; et al. A niche-dependent myeloid transcriptome signature defines dormant myeloma cells. Blood 2019, 134, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Morito, N.; Yoh, K.; Maeda, A.; Nakano, T.; Fujita, A.; Kusakabe, M.; Hamada, M.; Kudo, T.; Yamagata, K.; Takahashi, S. A novel transgenic mouse model of the human multiple myeloma chromosomal translocation t(14;16)(q32;q23). Cancer Res. 2011, 71, 339–348. [Google Scholar] [CrossRef]
- Carrasco, D.R.; Sukhdeo, K.; Protopopova, M.; Sinha, R.; Enos, M.; Carrasco, D.E.; Zheng, M.; Mani, M.; Henderson, J.; Pinkus, G.S.; et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 2007, 11, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Chesi, M.; Robbiani, D.F.; Sebag, M.; Chng, W.J.; Affer, M.; Tiedemann, R.; Valdez, R.; Palmer, S.E.; Haas, S.S.; Stewart, A.K.; et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell 2008, 13, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Calcinotto, A.; Brevi, A.; Chesi, M.; Ferrarese, R.; Garcia Perez, L.; Grioni, M.; Kumar, S.; Garbitt, V.M.; Sharik, M.E.; Henderson, K.J.; et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. 2018, 9, 4832. [Google Scholar] [CrossRef]
- Nakamura, K.; Kassem, S.; Cleynen, A.; Chretien, M.L.; Guillerey, C.; Putz, E.M.; Bald, T.; Forster, I.; Vuckovic, S.; Hill, G.R.; et al. Dysregulated IL-18 Is a Key Driver of Immunosuppression and a Possible Therapeutic Target in the Multiple Myeloma Microenvironment. Cancer Cell 2018, 33, 634–648. [Google Scholar] [CrossRef]
- Bordini, J.; Bertilaccio, M.T.; Ponzoni, M.; Fermo, I.; Chesi, M.; Bergsagel, P.L.; Camaschella, C.; Campanella, A. Erythroblast apoptosis and microenvironmental iron restriction trigger anemia in the VK*MYC model of multiple myeloma. Haematologica 2015, 100, 834–841. [Google Scholar] [CrossRef]
- Alsina, M.; Boyce, B.; Devlin, R.D.; Anderson, J.L.; Craig, F.; Mundy, G.R.; Roodman, G.D. Development of an in vivo model of human multiple myeloma bone disease. Blood 1996, 87, 1495–1501. [Google Scholar] [CrossRef]
- Rabin, N.; Kyriakou, C.; Coulton, L.; Gallagher, O.M.; Buckle, C.; Benjamin, R.; Singh, N.; Glassford, J.; Otsuki, T.; Nathwani, A.C.; et al. A new xenograft model of myeloma bone disease demonstrating the efficacy of human mesenchymal stem cells expressing osteoprotegerin by lentiviral gene transfer. Leukemia 2007, 21, 2181–2191. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iriuchishima, H.; Takubo, K.; Miyakawa, Y.; Nakamura-Ishizu, A.; Miyauchi, Y.; Fujita, N.; Miyamoto, K.; Miyamoto, T.; Ikeda, E.; Kizaki, M.; et al. Neovascular niche for human myeloma cells in immunodeficient mouse bone. PLoS ONE 2012, 7, e30557. [Google Scholar] [CrossRef] [PubMed]
- Chaidos, A.; Barnes, C.P.; Cowan, G.; May, P.C.; Melo, V.; Hatjiharissi, E.; Papaioannou, M.; Harrington, H.; Doolittle, H.; Terpos, E.; et al. Clinical drug resistance linked to interconvertible phenotypic and functional states of tumor-propagating cells in multiple myeloma. Blood 2013, 121, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Schueler, J.; Wider, D.; Klingner, K.; Siegers, G.M.; May, A.M.; Wasch, R.; Fiebig, H.H.; Engelhardt, M. Intratibial injection of human multiple myeloma cells in NOD/SCID IL-2Rgamma(null) mice mimics human myeloma and serves as a valuable tool for the development of anticancer strategies. PLoS ONE 2013, 8, e79939. [Google Scholar] [CrossRef]
- Hosen, N.; Matsuoka, Y.; Kishida, S.; Nakata, J.; Mizutani, Y.; Hasegawa, K.; Mugitani, A.; Ichihara, H.; Aoyama, Y.; Nishida, S.; et al. CD138-negative clonogenic cells are plasma cells but not B cells in some multiple myeloma patients. Leukemia 2012, 26, 2135–2141. [Google Scholar] [CrossRef] [PubMed]
- Tassone, P.; Neri, P.; Carrasco, D.R.; Burger, R.; Goldmacher, V.S.; Fram, R.; Munshi, V.; Shammas, M.A.; Catley, L.; Jacob, G.S.; et al. A clinically relevant SCID-hu in vivo model of human multiple myeloma. Blood 2005, 106, 713–716. [Google Scholar] [CrossRef]
- Tsunenari, T.; Koishihara, Y.; Nakamura, A.; Moriya, M.; Ohkawa, H.; Goto, H.; Shimazaki, C.; Nakagawa, M.; Ohsugi, Y.; Kishimoto, T.; et al. New xenograft model of multiple myeloma and efficacy of a humanized antibody against human interleukin-6 receptor. Blood 1997, 90, 2437–2444. [Google Scholar] [CrossRef]
- Sartoris, S.; Mazzocco, M.; Tinelli, M.; Martini, M.; Mosna, F.; Lisi, V.; Indraccolo, S.; Moserle, L.; Cestari, T.; Riviera, A.P.; et al. Efficacy assessment of interferon-alpha-engineered mesenchymal stromal cells in a mouse plasmacytoma model. Stem Cells Dev. 2011, 20, 709–719. [Google Scholar] [CrossRef]
- Das, R.; Strowig, T.; Verma, R.; Koduru, S.; Hafemann, A.; Hopf, S.; Kocoglu, M.H.; Borsotti, C.; Zhang, L.; Branagan, A.; et al. Microenvironment-dependent growth of preneoplastic and malignant plasma cells in humanized mice. Nat. Med. 2016, 22, 1351–1357. [Google Scholar] [CrossRef]
- Lang, J.; Zhang, B.; Kelly, M.; Peterson, J.N.; Barbee, J.; Freed, B.M.; Di Santo, J.P.; Matsuda, J.L.; Torres, R.M.; Pelanda, R. Replacing mouse BAFF with human BAFF does not improve B-cell maturation in hematopoietic humanized mice. Blood Adv. 2017, 1, 2729–2741. [Google Scholar] [CrossRef]
- Yaccoby, S.; Barlogie, B.; Epstein, J. Primary myeloma cells growing in SCID-hu mice: A model for studying the biology and treatment of myeloma and its manifestations. Blood 1998, 92, 2908–2913. [Google Scholar] [CrossRef] [PubMed]
- Urashima, M.; Chen, B.P.; Chen, S.; Pinkus, G.S.; Bronson, R.T.; Dedera, D.A.; Hoshi, Y.; Teoh, G.; Ogata, A.; Treon, S.P.; et al. The development of a model for the homing of multiple myeloma cells to human bone marrow. Blood 1997, 90, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, A.; Ling, W.; Li, X.; Khan, S.; Shaughnessy, J.D., Jr.; Barlogie, B.; Yaccoby, S. The ephrinB2/EphB4 axis is dysregulated in osteoprogenitors from myeloma patients and its activation affects myeloma bone disease and tumor growth. Blood 2009, 114, 1803–1812. [Google Scholar] [CrossRef]
- Yata, K.; Yaccoby, S. The SCID-rab model: A novel in vivo system for primary human myeloma demonstrating growth of CD138-expressing malignant cells. Leukemia 2004, 18, 1891–1897. [Google Scholar] [CrossRef] [PubMed]
- Groen, R.W.; Noort, W.A.; Raymakers, R.A.; Prins, H.J.; Aalders, L.; Hofhuis, F.M.; Moerer, P.; van Velzen, J.F.; Bloem, A.C.; van Kessel, B.; et al. Reconstructing the human hematopoietic niche in immunodeficient mice: Opportunities for studying primary multiple myeloma. Blood 2012, 120, e9–e16. [Google Scholar] [CrossRef] [PubMed]
- Calimeri, T.; Battista, E.; Conforti, F.; Neri, P.; Di Martino, M.T.; Rossi, M.; Foresta, U.; Piro, E.; Ferrara, F.; Amorosi, A.; et al. A unique three-dimensional SCID-polymeric scaffold (SCID-synth-hu) model for in vivo expansion of human primary multiple myeloma cells. Leukemia 2011, 25, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Reinisch, A.; Hernandez, D.C.; Schallmoser, K.; Majeti, R. Generation and use of a humanized bone-marrow-ossicle niche for hematopoietic xenotransplantation into mice. Nat. Protoc. 2017, 12, 2169–2188. [Google Scholar] [CrossRef] [PubMed]
- San-Miguel, J.; Blade, J.; Shpilberg, O.; Grosicki, S.; Maloisel, F.; Min, C.K.; Polo Zarzuela, M.; Robak, T.; Prasad, S.V.; Tee Goh, Y.; et al. Phase 2 randomized study of bortezomib-melphalan-prednisone with or without siltuximab (anti-IL-6) in multiple myeloma. Blood 2014, 123, 4136–4142. [Google Scholar] [CrossRef]
- Brighton, T.A.; Khot, A.; Harrison, S.J.; Ghez, D.; Weiss, B.M.; Kirsch, A.; Magen, H.; Gironella, M.; Oriol, A.; Streetly, M.; et al. Randomized, Double-Blind, Placebo-Controlled, Multicenter Study of Siltuximab in High-Risk Smoldering Multiple Myeloma. Clin. Cancer Res. 2019, 25, 3772–3775. [Google Scholar] [CrossRef]
- Orlowski, R.Z.; Gercheva, L.; Williams, C.; Sutherland, H.; Robak, T.; Masszi, T.; Goranova-Marinova, V.; Dimopoulos, M.A.; Cavenagh, J.D.; Spicka, I.; et al. A phase 2, randomized, double-blind, placebo-controlled study of siltuximab (anti-IL-6 mAb) and bortezomib versus bortezomib alone in patients with relapsed or refractory multiple myeloma. Am. J. Hematol. 2015, 90, 42–49. [Google Scholar] [CrossRef]
- Lam, C.; Ferguson, I.D.; Mariano, M.C.; Lin, Y.T.; Murnane, M.; Liu, H.; Smith, G.A.; Wong, S.W.; Taunton, J.; Liu, J.O.; et al. Repurposing tofacitinib as an anti-myeloma therapeutic to reverse growth-promoting effects of the bone marrow microenvironment. Haematologica 2018, 103, 1218–1228. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.B.; Fook-Alves, V.L.; Eugenio, A.I.P.; Fernando, R.C.; Sanson, L.F.G.; de Carvalho, M.F.; Braga, W.M.T.; Davies, F.E.; Colleoni, G.W.B. Anti-myeloma effects of ruxolitinib combined with bortezomib and lenalidomide: A rationale for JAK/STAT pathway inhibition in myeloma patients. Cancer Lett. 2017, 403, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.P.; Beck, J.T.; Stewart, A.K.; Shah, J.; Kelly, K.R.; Isaacs, R.; Bilic, S.; Sen, S.; Munshi, N.C. A Phase IB multicentre dose-determination study of BHQ880 in combination with anti-myeloma therapy and zoledronic acid in patients with relapsed or refractory multiple myeloma and prior skeletal-related events. Br. J. Haematol. 2014, 167, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Abdulkadyrov, K.M.; Salogub, G.N.; Khuazheva, N.K.; Sherman, M.L.; Laadem, A.; Barger, R.; Knight, R.; Srinivasan, S.; Terpos, E. Sotatercept in patients with osteolytic lesions of multiple myeloma. Br. J. Haematol. 2014, 165, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Ehnert, S.; Culmes, M.; Bachmann, A.; Seeliger, C.; Schyschka, L.; Wang, Z.; Rahmanian-Schwarz, A.; Stockle, U.; De Sousa, P.A.; et al. 5-azacytidine improves the osteogenic differentiation potential of aged human adipose-derived mesenchymal stem cells by DNA demethylation. PLoS ONE 2014, 9, e90846. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, T.M.; Westendorf, J.J. Histone deacetylase inhibitors promote osteoblast maturation. J. Bone Miner. Res. 2005, 20, 2254–2263. [Google Scholar] [CrossRef]
- Cho, H.H.; Park, H.T.; Kim, Y.J.; Bae, Y.C.; Suh, K.T.; Jung, J.S. Induction of osteogenic differentiation of human mesenchymal stem cells by histone deacetylase inhibitors. J. Cell Biochem. 2005, 96, 533–542. [Google Scholar] [CrossRef]
- McGee-Lawrence, M.E.; McCleary-Wheeler, A.L.; Secreto, F.J.; Razidlo, D.F.; Zhang, M.; Stensgard, B.A.; Li, X.; Stein, G.S.; Lian, J.B.; Westendorf, J.J. Suberoylanilide hydroxamic acid (SAHA; vorinostat) causes bone loss by inhibiting immature osteoblasts. Bone 2011, 48, 1117–1126. [Google Scholar] [CrossRef][Green Version]
- Pratap, J.; Akech, J.; Wixted, J.J.; Szabo, G.; Hussain, S.; McGee-Lawrence, M.E.; Li, X.; Bedard, K.; Dhillon, R.J.; van Wijnen, A.J.; et al. The histone deacetylase inhibitor, vorinostat, reduces tumor growth at the metastatic bone site and associated osteolysis, but promotes normal bone loss. Mol. Cancer Ther. 2010, 9, 3210–3220. [Google Scholar] [CrossRef]
- Xu, S.; De Veirman, K.; Evans, H.; Santini, G.C.; Vande Broek, I.; Leleu, X.; De Becker, A.; Van Camp, B.; Croucher, P.; Vanderkerken, K.; et al. Effect of the HDAC inhibitor vorinostat on the osteogenic differentiation of mesenchymal stem cells in vitro and bone formation in vivo. Acta Pharmacol. Sin. 2013, 34, 699–709. [Google Scholar] [CrossRef]
- Giuliani, N.; Morandi, F.; Tagliaferri, S.; Lazzaretti, M.; Bonomini, S.; Crugnola, M.; Mancini, C.; Martella, E.; Ferrari, L.; Tabilio, A.; et al. The proteasome inhibitor bortezomib affects osteoblast differentiation in vitro and in vivo in multiple myeloma patients. Blood 2007, 110, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Podar, K.; Zimmerhackl, A.; Fulciniti, M.; Tonon, G.; Hainz, U.; Tai, Y.T.; Vallet, S.; Halama, N.; Jager, D.; Olson, D.L.; et al. The selective adhesion molecule inhibitor Natalizumab decreases multiple myeloma cell growth in the bone marrow microenvironment: Therapeutic implications. Br. J. Haematol. 2011, 155, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Azab, A.K.; Runnels, J.M.; Pitsillides, C.; Moreau, A.S.; Azab, F.; Leleu, X.; Jia, X.; Wright, R.; Ospina, B.; Carlson, A.L.; et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood 2009, 113, 4341–4351. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, H.; Weisel, K.; Petrucci, M.T.; Leleu, X.; Cafro, A.M.; Garderet, L.; Leitgeb, C.; Foa, R.; Greil, R.; Yakoub-Agha, I.; et al. Olaptesed pegol, an anti-CXCL12/SDF-1 Spiegelmer, alone and with bortezomib-dexamethasone in relapsed/refractory multiple myeloma: A Phase IIa Study. Leukemia 2017, 31, 997–1000. [Google Scholar] [CrossRef] [PubMed]
- Ghobrial, I.M.; Liu, C.J.; Redd, R.A.; Perez, R.P.; Baz, R.; Zavidij, O.; Sklavenitis-Pistofidis, R.; Richardson, P.G.; Anderson, K.C.; Laubach, J.; et al. A Phase Ib/II Trial of the First-in-Class Anti-CXCR4 Antibody Ulocuplumab in Combination with Lenalidomide or Bortezomib Plus Dexamethasone in Relapsed Multiple Myeloma. Clin. Cancer Res. 2020, 26, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Fulciniti, M.; Tassone, P.; Hideshima, T.; Vallet, S.; Nanjappa, P.; Ettenberg, S.A.; Shen, Z.; Patel, N.; Tai, Y.T.; Chauhan, D.; et al. Anti-DKK1 mAb (BHQ880) as a potential therapeutic agent for multiple myeloma. Blood 2009, 114, 371–379. [Google Scholar] [CrossRef]
- Long, M.; Beckwith, K.; Do, P.; Mundy, B.L.; Gordon, A.; Lehman, A.M.; Maddocks, K.J.; Cheney, C.; Jones, J.A.; Flynn, J.M.; et al. Ibrutinib treatment improves T cell number and function in CLL patients. J. Clin. Investig. 2017, 127, 3052–3064. [Google Scholar] [CrossRef]
- Gauthier, J.; Hirayama, A.V.; Purushe, J.; Hay, K.A.; Lymp, J.; Li, D.H.; Yeung, C.C.S.; Sheih, A.; Pender, B.S.; Hawkins, R.M.; et al. Feasibility and efficacy of CD19-targeted CAR T cells with concurrent ibrutinib for CLL after ibrutinib failure. Blood 2020, 135, 1650–1660. [Google Scholar] [CrossRef]
- Regmi, S.; Pathak, S.; Kim, J.O.; Yong, C.S.; Jeong, J.H. Mesenchymal stem cell therapy for the treatment of inflammatory diseases: Challenges, opportunities, and future perspectives. Eur. J. Cell Biol. 2019, 98, 151041. [Google Scholar] [CrossRef]
- Walker, B.A.; Wardell, C.P.; Johnson, D.C.; Kaiser, M.F.; Begum, D.B.; Dahir, N.B.; Ross, F.M.; Davies, F.E.; Gonzalez, D.; Morgan, G.J. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood 2013, 121, 3413–3419. [Google Scholar] [CrossRef]
- Boucher, K.; Parquet, N.; Widen, R.; Shain, K.; Baz, R.; Alsina, M.; Koomen, J.; Anasetti, C.; Dalton, W.; Perez, L.E. Stemness of B-cell progenitors in multiple myeloma bone marrow. Clin. Cancer Res. 2012, 18, 6155–6168. [Google Scholar] [CrossRef] [PubMed]
- Al-Sabah, J.; Baccin, C.; Haas, S. Single-cell and spatial transcriptomics approaches of the bone marrow microenvironment. Curr. Opin. Oncol. 2020, 32, 146–153. [Google Scholar] [CrossRef] [PubMed]
| Cell Type | Effects | Effects | Effects | Regulated Factors | References |
|---|---|---|---|---|---|
| MM cells | Effects on MSCs | Soluble factor secretion | Inhibition of osteoblastogenesis | Wnt inhibitors (DKK1, sFRPs, sclerostin), TNFα | [49,51,52,53,54,55] |
| IL-6 production in MSCs | bFGF1, TNFα | [49,56] | |||
| Localization with MSCs | Proliferation and drug resistance in MM cells | CCL25, CXCR4 Integrin α4, α5, β1, β7, ALCAM, VCAM-1 | [32,48,57,58,59,60,61,62,63,64,65,66,67,68] | ||
| VEGF production in MSCs | Integrin β7 | [69] | |||
| MSCs | Effects on MM cells | Soluble factor secretion | MM cell proliferation | IL-6, GDF15, BAFF, micro vesicles (miRNA-15a,-10a) | [73,74,83,84,85,86,87,88,89,90,91] |
| Localization with MM cells | Proliferation and drug resistance in MM cells | CXCL12, CCL3, ICAM-1, VCAM-1, MADCAM-1 | [63,83,84,85,86,87,88,89,90,91,92] | ||
| MSCs | Effects on MM microenvironment | Inhibition of osteoblastogenesis | DKK1, TNFα, IL-6, TGFβ | [101,102,103,104] | |
| immunomodulation | IL-6, IL-10, IL-1β, IL-3, TNFα, | [75,76,77,78,79,83,84,85] | |||
| angiogenesis | VEGF, HGF | [69,89] |
| Target | Agent | Agent | Agent | Efficacies |
|---|---|---|---|---|
| MM proliferation | IL-6 chimeric antibody [148,149,150] | Siltuximab | Clinical trials | Not clear |
| JAK signal inhibitor [151,152] | Tofacitinib, ruxolitinib | Preclinical studies | Yes | |
| Microvesicle release inhibitor [92] | FTY720 | Preclinical studies | Yes | |
| Osteoblastogenesis suppression | DKK1 inhibitor [153] | BHQ880 | Clinical trials | No |
| TGFβ antagonist [154] | Sotatercept | Clinical trials | No | |
| DNMT inhibitor [155] | 5-Azacytidine | Preclinical studies | Not clear | |
| HDAC inhibitor [156,157,158,159,160] | Vorinostat | Preclinical studies | Not clear | |
| Proteasome inhibitor [161] | Bortezomib | Clinical studies | Yes | |
| Adhesion to MM microenvironment | Integrin α4 inhibition [162] | Natalizumab | Preclinical studies | Yes |
| CXCL12-CXCR4 inhibition [162,163,164,165] | Plerixafor, olaptesed pegol, ulocuplumab | Clinical trials | Ongoing |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ichii, M.; Hosen, N. Current Understanding of Myelomatous Mesenchymal Stromal Cells Extended through Advances in Experimental Methods. Cancers 2021, 13, 25. https://doi.org/10.3390/cancers13010025
Ichii M, Hosen N. Current Understanding of Myelomatous Mesenchymal Stromal Cells Extended through Advances in Experimental Methods. Cancers. 2021; 13(1):25. https://doi.org/10.3390/cancers13010025
Chicago/Turabian StyleIchii, Michiko, and Naoki Hosen. 2021. "Current Understanding of Myelomatous Mesenchymal Stromal Cells Extended through Advances in Experimental Methods" Cancers 13, no. 1: 25. https://doi.org/10.3390/cancers13010025
APA StyleIchii, M., & Hosen, N. (2021). Current Understanding of Myelomatous Mesenchymal Stromal Cells Extended through Advances in Experimental Methods. Cancers, 13(1), 25. https://doi.org/10.3390/cancers13010025





