The Lung Microbiome: A Central Mediator of Host Inflammation and Metabolism in Lung Cancer Patients?
Abstract
:Simple Summary
Abstract
1. Introduction
2. The Lung Microbiome in Health
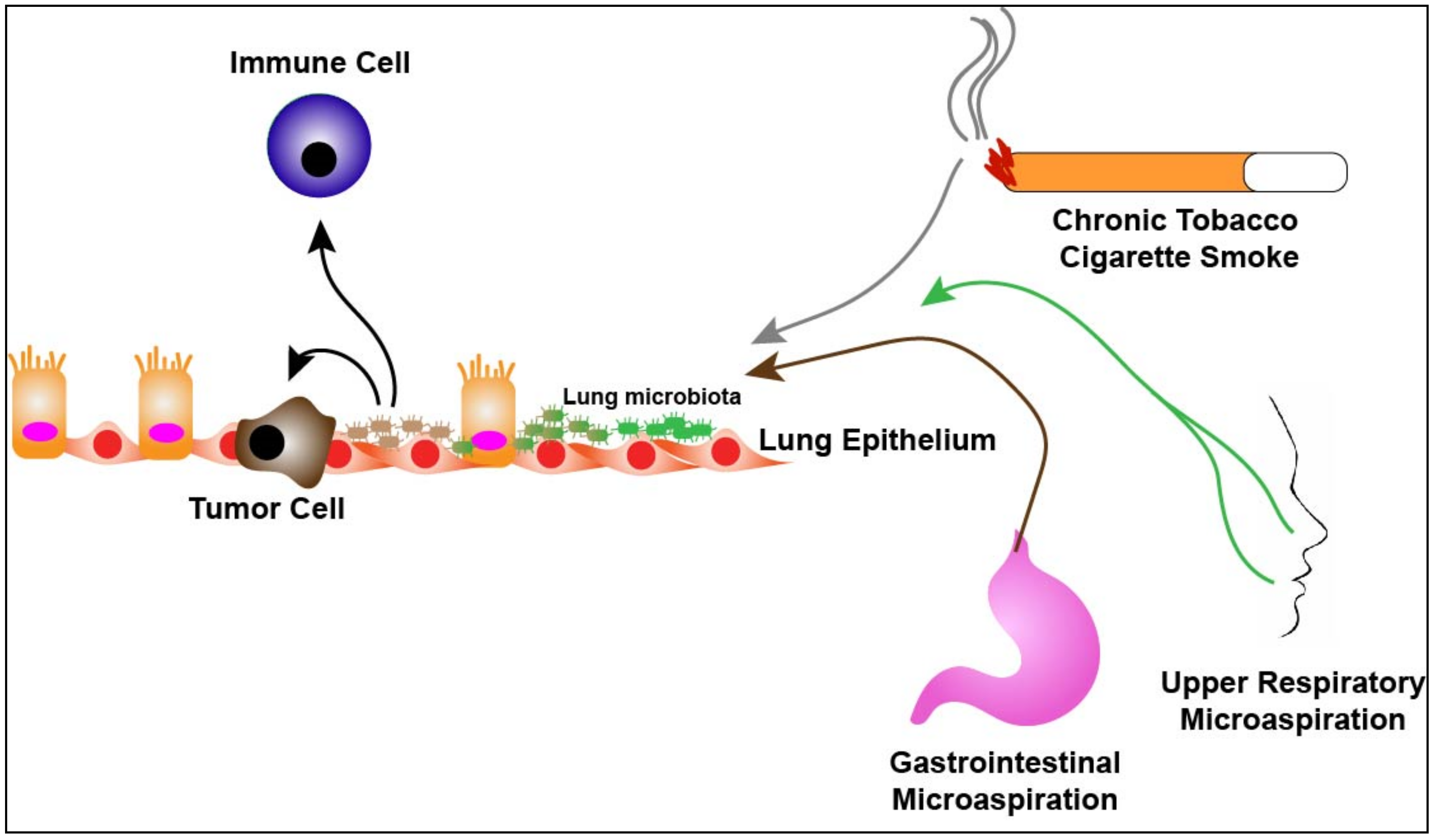
3. The Lung Microbiome and Lung Cancer
4. The Lung Microbiome and the Immune System
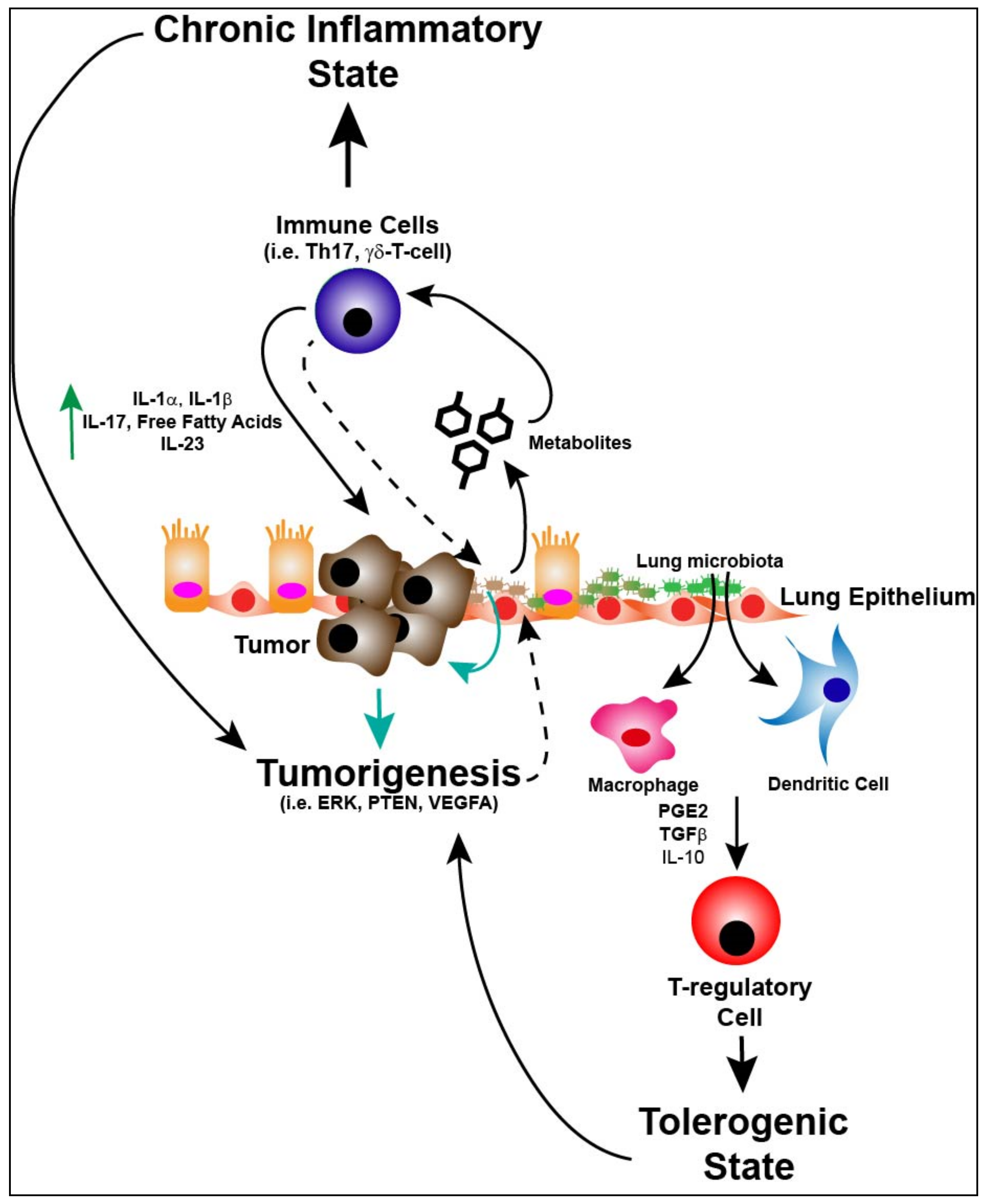
5. Immunity, Lung Microbiome and Tumorigenesis
6. Immune Response, Lung Microbiome and Metabolism
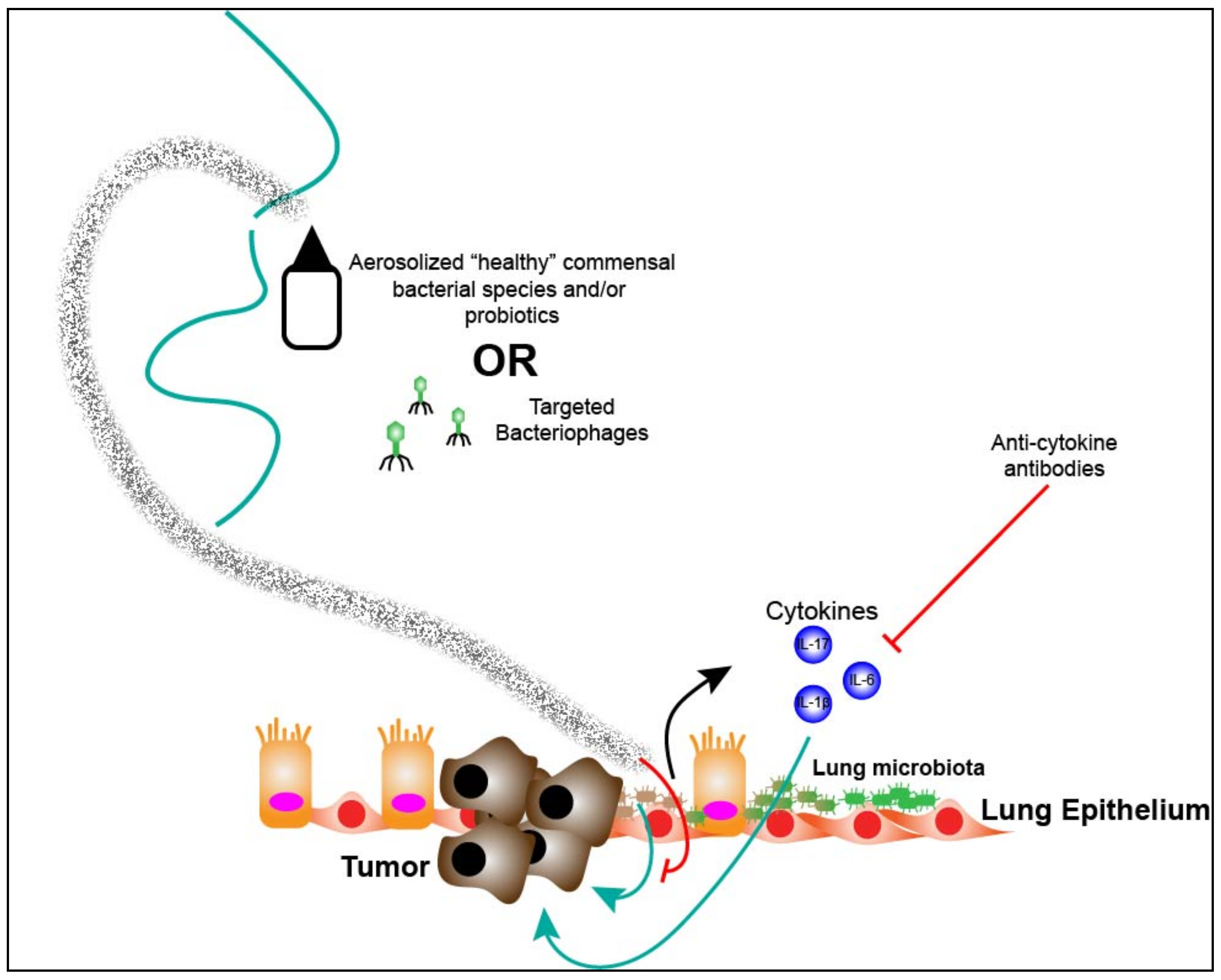
7. Targeting the Lung Microbiota
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Beck, J.M.; Huffnagle, G.B.; Curtis, J.L. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann. Am. Thorac. Soc. 2015, 12, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef] [Green Version]
- Venkataraman, A.; Bassis, C.M.; Beck, J.M.; Young, V.B.; Curtis, J.L.; Huffnagle, G.B.; Schmidt, T.M. Application of a neutral community model to assess structuring of the human lung microbiome. mBio 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Lighthart, B. Mini-review of the concentration variations found inthe alfresco atmospheric bacterial populations. Aerobiologia 2000, 16, 7–16. [Google Scholar] [CrossRef]
- Gleeson, K.; Eggli, D.F.; Maxwell, S.L. Quantitative aspiration during sleep in normal subjects. Chest 1997, 111, 1266–1272. [Google Scholar] [CrossRef] [PubMed]
- Huxley, E.J.; Viroslav, J.; Gray, W.R.; Pierce, A.K. Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am. J. Med. 1978, 64, 564–568. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. Towards an ecology of the lung: New conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir. Med. 2014, 2, 238–246. [Google Scholar] [CrossRef] [Green Version]
- Hatch, T.F. Distribution and deposition of inhaled particles in respiratory tract. Bacteriol. Rev. 1961, 25, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Ingenito, E.P.; Solway, J.; McFadden, E.R., Jr.; Pichurko, B.; Bowman, H.F.; Michaels, D.; Drazen, J.M. Indirect assessment of mucosal surface temperatures in the airways: Theory and tests. J. Appl. Physiol. (1985) 1987, 63, 2075–2083. [Google Scholar] [CrossRef] [PubMed]
- West, J.B. Regional differences in the lung. Chest 1978, 74, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; Walker, N.; Scales, B.S.; Beck, J.M.; Martinez, F.J.; Curtis, J.L.; Lama, V.N.; Huffnagle, G.B. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PLoS ONE 2014, 9, e97214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segal, L.N.; Alekseyenko, A.V.; Clemente, J.C.; Kulkarni, R.; Wu, B.; Gao, Z.; Chen, H.; Berger, K.I.; Goldring, R.M.; Rom, W.N.; et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013, 1, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6, e00037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, C.E.; Murphy, E.; Kozar, L.F.; Phillipson, E.A. Waking and ventilatory responses to laryngeal stimulation in sleeping dogs. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1978, 45, 681–689. [Google Scholar] [CrossRef]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef] [Green Version]
- Gori, S.; Inno, A.; Belluomini, L.; Bocus, P.; Bisoffi, Z.; Russo, A.; Arcaro, G. Gut microbiota and cancer: How gut microbiota modulates activity, efficacy and toxicity of antitumoral therapy. Crit. Rev. Oncol. Hematol. 2019, 143, 139–147. [Google Scholar] [CrossRef]
- Carbone, C.; Piro, G.; Di Noia, V.; D’Argento, E.; Vita, E.; Ferrara, M.G.; Pilotto, S.; Milella, M.; Cammarota, G.; Gasbarrini, A.; et al. Lung and Gut Microbiota as Potential Hidden Driver of Immunotherapy Efficacy in Lung Cancer. Mediators Inflamm. 2019, 2019, 7652014. [Google Scholar] [CrossRef]
- Elkrief, A.; Derosa, L.; Kroemer, G.; Zitvogel, L.; Routy, B. The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: A new independent prognostic factor? Ann. Oncol. 2019, 30, 1572–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voynow, J.A.; Hendricks-Munoz, K.D. The Gut-Lung Axis and Pulmonary Responses to Ozone. Am. J. Respir. Cell Mol. Biol. 2018, 59, 281–282. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Lei, Y.; Tang, L.; Hu, S.; Yang, M.; Wu, L.; Yang, S.; Tang, B. Gut microbiota: The emerging link to lung homeostasis and disease. J. Bacteriol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, F.; Wei, H.; Lian, Z.X.; Sun, R.; Tian, Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 2014, 211, 2397–2410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Tian, Z. How lung infection leads to gut injury. Oncotarget 2015, 6, 42394–42395. [Google Scholar] [CrossRef] [PubMed]
- Pilaniya, V.; Gera, K.; Kunal, S.; Shah, A. Pulmonary tuberculosis masquerading as metastatic lung disease. Eur. Respir. Rev. 2016, 25, 97–98. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.Y.; Li, X.L.; Yu, X.S.; Guan, P.; Yin, Z.H.; He, Q.C.; Zhou, B.S. Facts and fiction of the relationship between preexisting tuberculosis and lung cancer risk: A systematic review. Int. J. Cancer 2009, 125, 2936–2944. [Google Scholar] [CrossRef]
- Pallis, A.G.; Syrigos, K.N. Lung cancer in never smokers: Disease characteristics and risk factors. Crit. Rev. Oncol. Hematol. 2013, 88, 494–503. [Google Scholar] [CrossRef]
- Yu, G.; Gail, M.H.; Consonni, D.; Carugno, M.; Humphrys, M.; Pesatori, A.C.; Caporaso, N.E.; Goedert, J.J.; Ravel, J.; Landi, M.T. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016, 17, 163. [Google Scholar] [CrossRef] [Green Version]
- Morgan, X.C.; Huttenhower, C. Chapter 12: Human microbiome analysis. PLoS Comput. Biol. 2012, 8, e1002808. [Google Scholar] [CrossRef] [Green Version]
- Laroumagne, S.; Lepage, B.; Hermant, C.; Plat, G.; Phelippeau, M.; Bigay-Game, L.; Lozano, S.; Guibert, N.; Segonds, C.; Mallard, V.; et al. Bronchial colonisation in patients with lung cancer: A prospective study. Eur. Respir. J. 2013, 42, 220–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosgood, H.D., 3rd; Sapkota, A.R.; Rothman, N.; Rohan, T.; Hu, W.; Xu, J.; Vermeulen, R.; He, X.; White, J.R.; Wu, G.; et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ. Mol. Mutagen. 2014, 55, 643–651. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Yang, M.; Liu, J.; Gao, R.; Hu, J.; Li, J.; Zhang, L.; Shi, Y.; Guo, H.; Cheng, J.; et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 2015, 5, 3111–3122. [Google Scholar]
- Cameron, S.J.S.; Lewis, K.E.; Huws, S.A.; Hegarty, M.J.; Lewis, P.D.; Pachebat, J.A.; Mur, L.A.J. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS ONE 2017, 12, e0177062. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Sung, J.Y.; Yong, D.; Chun, J.; Kim, S.Y.; Song, J.H.; Chung, K.S.; Kim, E.Y.; Jung, J.Y.; Kang, Y.A.; et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 2016, 102, 89–95. [Google Scholar] [CrossRef]
- Tsay, J.J.; Wu, B.G.; Badri, M.H.; Clemente, J.C.; Shen, N.; Meyn, P.; Li, Y.; Yie, T.A.; Lhakhang, T.; Olsen, E.; et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am. J. Respir. Crit. Care Med. 2018, 198, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.X.; Tao, L.L.; Zhang, J.; Zhu, Y.G.; Zheng, Y.; Liu, D.; Zhou, M.; Ke, H.; Shi, M.M.; Qu, J.M. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int. J. Cancer 2018, 142, 769–778. [Google Scholar] [CrossRef] [Green Version]
- Patnaik, S.K.; Cortes, E.G.; Kannisto, E.D.; Punnanitinont, A.; Dhillon, S.S.; Liu, S.; Yendamuri, S. Lower airway bacterial microbiome may influence recurrence after resection of early-stage non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2020. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.A.; Hayes, R.B.; Goparaju, C.; Reid, C.; Pass, H.I.; Ahn, J. The Microbiome in Lung Cancer Tissue and Recurrence-Free Survival. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 731–740. [Google Scholar] [CrossRef] [Green Version]
- Munck, C.; Helby, J.; Westergaard, C.G.; Porsbjerg, C.; Backer, V.; Hansen, L.H. Smoking Cessation and the Microbiome in Induced Sputum Samples from Cigarette Smoking Asthma Patients. PLoS ONE 2016, 11, e0158622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Einarsson, G.G.; Comer, D.M.; McIlreavey, L.; Parkhill, J.; Ennis, M.; Tunney, M.M.; Elborn, J.S. Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax 2016, 71, 795–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Chen, L.; Cao, L.; Li, K.J.; Huang, Y.; Luan, X.Q.; Li, G. Effects of smoking on the lower respiratory tract microbiome in mice. Respir. Res. 2018, 19, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, F.; Liang, C.L.; Liu, H.; Zeng, Y.Q.; Hou, S.; Huang, S.; Lai, X.; Dai, Z. Impacts of cigarette smoking on immune responsiveness: Up and down or upside down? Oncotarget 2017, 8, 268–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strzelak, A.; Ratajczak, A.; Adamiec, A.; Feleszko, W. Tobacco Smoke Induces and Alters Immune Responses in the Lung Triggering Inflammation, Allergy, Asthma and Other Lung Diseases: A Mechanistic Review. Int. J. Environ. Res. Public Health 2018, 15, 1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, T.B. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect. Immun. 2014, 82, 4596–4606. [Google Scholar] [CrossRef] [Green Version]
- Sassone-Corsi, M.; Raffatellu, M. No vacancy: How beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J. Immunol. 2015, 194, 4081–4087. [Google Scholar] [CrossRef] [Green Version]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013, 339, 708–711. [Google Scholar] [CrossRef] [Green Version]
- Winter, S.E.; Baumler, A.J. Dysbiosis in the inflamed intestine: Chance favors the prepared microbe. Gut Microbes 2014, 5, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Scales, B.S.; Dickson, R.P.; Huffnagle, G.B. A tale of two sites: How inflammation can reshape the microbiomes of the gut and lungs. J. Leukoc. Biol. 2016, 100, 943–950. [Google Scholar] [CrossRef] [Green Version]
- Hussell, T.; Bell, T.J. Alveolar macrophages: Plasticity in a tissue-specific context. Nat. Rev. Immunol. 2014, 14, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Soroosh, P.; Doherty, T.A.; Duan, W.; Mehta, A.K.; Choi, H.; Adams, Y.F.; Mikulski, Z.; Khorram, N.; Rosenthal, P.; Broide, D.H.; et al. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J. Exp. Med. 2013, 210, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Sellge, G.; Kufer, T.A. PRR-signaling pathways: Learning from microbial tactics. Semin. Immunol. 2015, 27, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Baral, P.; Batra, S.; Zemans, R.L.; Downey, G.P.; Jeyaseelan, S. Divergent functions of Toll-like receptors during bacterial lung infections. Am. J. Respir. Crit. Care Med. 2014, 190, 722–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steimle, A.; Autenrieth, I.B.; Frick, J.S. Structure and function: Lipid A modifications in commensals and pathogens. Int. J. Med. Microbiol. 2016, 306, 290–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommariva, M.; Le Noci, V.; Bianchi, F.; Camelliti, S.; Balsari, A.; Tagliabue, E.; Sfondrini, L. The lung microbiota: Role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cell. Mol. Life Sci. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swiatczak, B.; Cohen, I.R. Gut feelings of safety: Tolerance to the microbiota mediated by innate immune receptors. Microbiol. Immunol. 2015, 59, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Petersen, F.C.; Shekhar, S. Commensal Bacteria: An Emerging Player in Defense Against Respiratory Pathogens. Front. Immunol. 2019, 10, 1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artis, D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 2008, 8, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Cava, C.F.; Ishihara, S.; Rumi, M.A.; Kawashima, K.; Ishimura, N.; Kazumori, H.; Udagawa, J.; Kadowaki, Y.; Kinoshita, Y. Strategic compartmentalization of Toll-like receptor 4 in the mouse gut. J. Immunol. 2003, 170, 3977–3985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornef, M.W.; Normark, B.H.; Vandewalle, A.; Normark, S. Intracellular recognition of lipopolysaccharide by toll-like receptor 4 in intestinal epithelial cells. J. Exp. Med. 2003, 198, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kotani, T.; Konno, T.; Setiawan, J.; Kitamura, Y.; Imada, S.; Usui, Y.; Hatano, N.; Shinohara, M.; Saito, Y.; et al. Promotion of Intestinal Epithelial Cell Turnover by Commensal Bacteria: Role of Short-Chain Fatty Acids. PLoS ONE 2016, 11, e0156334. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog. 2013, 5, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pena, O.M.; Pistolic, J.; Raj, D.; Fjell, C.D.; Hancock, R.E. Endotoxin tolerance represents a distinctive state of alternative polarization (M2) in human mononuclear cells. J. Immunol. 2011, 186, 7243–7254. [Google Scholar] [CrossRef] [Green Version]
- Lea, S.R.; Reynolds, S.L.; Kaur, M.; Simpson, K.D.; Hall, S.R.; Hessel, E.M.; Singh, D. The effects of repeated Toll-like receptors 2 and 4 stimulation in COPD alveolar macrophages. Int. J. Chron. Obstruct. Pulm. Dis. 2018, 13, 771–780. [Google Scholar] [CrossRef] [Green Version]
- Geisel, J.; Kahl, F.; Muller, M.; Wagner, H.; Kirschning, C.J.; Autenrieth, I.B.; Frick, J.S. IL-6 and maturation govern TLR2 and TLR4 induced TLR agonist tolerance and cross-tolerance in dendritic cells. J. Immunol. 2007, 179, 5811–5818. [Google Scholar] [CrossRef]
- Couper, K.N.; Blount, D.G.; Riley, E.M. IL-10: The master regulator of immunity to infection. J. Immunol. 2008, 180, 5771–5777. [Google Scholar] [CrossRef]
- Gollwitzer, E.S.; Saglani, S.; Trompette, A.; Yadava, K.; Sherburn, R.; McCoy, K.D.; Nicod, L.P.; Lloyd, C.M.; Marsland, B.J. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 2014, 20, 642–647. [Google Scholar] [CrossRef]
- Herbst, T.; Sichelstiel, A.; Schar, C.; Yadava, K.; Burki, K.; Cahenzli, J.; McCoy, K.; Marsland, B.J.; Harris, N.L. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med. 2011, 184, 198–205. [Google Scholar] [CrossRef]
- Wang, J.; Li, F.; Sun, R.; Gao, X.; Wei, H.; Li, L.J.; Tian, Z. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat. Commun. 2013, 4, 2106. [Google Scholar] [CrossRef] [Green Version]
- Segal, L.N.; Clemente, J.C.; Tsay, J.C.; Koralov, S.B.; Keller, B.C.; Wu, B.G.; Li, Y.; Shen, N.; Ghedin, E.; Morris, A.; et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016, 1, 16031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, J.M.; Steen-Jensen, D.B.; Laursen, J.M.; Sondergaard, J.N.; Musavian, H.S.; Butt, T.M.; Brix, S. Divergent pro-inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS ONE 2012, 7, e31976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernasconi, E.; Pattaroni, C.; Koutsokera, A.; Pison, C.; Kessler, R.; Benden, C.; Soccal, P.M.; Magnan, A.; Aubert, J.D.; Marsland, B.J.; et al. Airway Microbiota Determines Innate Cell Inflammatory or Tissue Remodeling Profiles in Lung Transplantation. Am. J. Respir. Crit. Care Med. 2016, 194, 1252–1263. [Google Scholar] [CrossRef]
- Bao, Z.; Lu, G.; Cui, D.; Yao, Y.; Yang, G.; Zhou, J. IL-17A-producing T cells are associated with the progression of lung adenocarcinoma. Oncol. Rep. 2016, 36, 641–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palucka, A.K.; Coussens, L.M. The Basis of Oncoimmunology. Cell 2016, 164, 1233–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elinav, E.; Nowarski, R.; Thaiss, C.A.; Hu, B.; Jin, C.; Flavell, R.A. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 2013, 13, 759–771. [Google Scholar] [CrossRef]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef] [Green Version]
- Akinosoglou, K.S.; Karkoulias, K.; Marangos, M. Infectious complications in patients with lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 8–18. [Google Scholar]
- Qiao, D.; Wang, Z.; Lu, Y.; Wen, X.; Li, H.; Zhao, H. A retrospective study of risk and prognostic factors in relation to lower respiratory tract infection in elderly lung cancer patients. Am. J. Cancer Res. 2015, 5, 423–432. [Google Scholar]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 2019, 176, 998–1013.e1016. [Google Scholar] [CrossRef] [Green Version]
- Le Noci, V.; Guglielmetti, S.; Arioli, S.; Camisaschi, C.; Bianchi, F.; Sommariva, M.; Storti, C.; Triulzi, T.; Castelli, C.; Balsari, A.; et al. Modulation of Pulmonary Microbiota by Antibiotic or Probiotic Aerosol Therapy: A Strategy to Promote Immunosurveillance against Lung Metastases. Cell Rep. 2018, 24, 3528–3538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirayama, K.; Baranczewski, P.; Akerlund, J.E.; Midtvedt, T.; Moller, L.; Rafter, J. Effects of human intestinal flora on mutagenicity of and DNA adduct formation from food and environmental mutagens. Carcinogenesis 2000, 21, 2105–2111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geuking, M.B.; McCoy, K.D.; Macpherson, A.J. Metabolites from intestinal microbes shape Treg. Cell Res. 2013, 23, 1339–1340. [Google Scholar] [CrossRef] [Green Version]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Memari, B.; Bouttier, M.; Dimitrov, V.; Ouellette, M.; Behr, M.A.; Fritz, J.H.; White, J.H. Engagement of the Aryl Hydrocarbon Receptor in Mycobacterium tuberculosis-Infected Macrophages Has Pleiotropic Effects on Innate Immune Signaling. J. Immunol. 2015, 195, 4479–4491. [Google Scholar] [CrossRef] [Green Version]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [Green Version]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Mendez, R.; Banerjee, S.; Bhattacharya, S.K.; Banerjee, S. Lung inflammation and disease: A perspective on microbial homeostasis and metabolism. IUBMB Life 2019, 71, 152–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, E.L.; Heaver, S.L.; Waters, J.L.; Kim, B.I.; Bretin, A.; Goodman, A.L.; Gewirtz, A.T.; Worgall, T.S.; Ley, R.E. Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. Nat. Commun. 2020, 11, 2471. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.L.; Spiller, O.B.; Beeton, M.L.; Maxwell, N.C.; Remold-O’Donnell, E.; Kotecha, S. Relationship of proteinases and proteinase inhibitors with microbial presence in chronic lung disease of prematurity. Thorax 2010, 65, 246–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cribbs, S.K.; Park, Y.; Guidot, D.M.; Martin, G.S.; Brown, L.A.; Lennox, J.; Jones, D.P. Metabolomics of bronchoalveolar lavage differentiate healthy HIV-1-infected subjects from controls. AIDS Res. Hum. Retrovir. 2014, 30, 579–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Kwon, O.; Ryu, T.Y.; Jung, C.R.; Kim, J.; Min, J.K.; Kim, D.S.; Son, M.Y.; Cho, H.S. Propionate of a microbiota metabolite induces cell apoptosis and cell cycle arrest in lung cancer. Mol. Med. Rep. 2019, 20, 1569–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, J.M.; Musavian, H.S.; Butt, T.M.; Ingvorsen, C.; Thysen, A.H.; Brix, S. Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp., promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology 2015, 144, 333–342. [Google Scholar] [CrossRef] [Green Version]
- Zarogoulidis, P.; Kioumis, I.; Porpodis, K.; Spyratos, D.; Tsakiridis, K.; Huang, H.; Li, Q.; Turner, J.F.; Browning, R.; Hohenforst-Schmidt, W.; et al. Clinical experimentation with aerosol antibiotics: Current and future methods of administration. Drug Des. Dev. Ther. 2013, 7, 1115–1134. [Google Scholar] [CrossRef] [Green Version]
- Le Noci, V.; Sommariva, M.; Tortoreto, M.; Zaffaroni, N.; Campiglio, M.; Tagliabue, E.; Balsari, A.; Sfondrini, L. Reprogramming the lung microenvironment by inhaled immunotherapy fosters immune destruction of tumor. Oncoimmunology 2016, 5, e1234571. [Google Scholar] [CrossRef] [Green Version]
- Storti, C.; Le Noci, V.; Sommariva, M.; Tagliabue, E.; Balsari, A.; Sfondrini, L. Aerosol Delivery in the Treatment of Lung Cancer. Curr. Cancer Drug Targets 2015, 15, 604–612. [Google Scholar] [CrossRef]
- Le Noci, V.; Tortoreto, M.; Gulino, A.; Storti, C.; Bianchi, F.; Zaffaroni, N.; Tripodo, C.; Tagliabue, E.; Balsari, A.; Sfondrini, L. Poly(I:C) and CpG-ODN combined aerosolization to treat lung metastases and counter the immunosuppressive microenvironment. Oncoimmunology 2015, 4, e1040214. [Google Scholar] [CrossRef] [Green Version]
- Marchisio, P.; Santagati, M.; Scillato, M.; Baggi, E.; Fattizzo, M.; Rosazza, C.; Stefani, S.; Esposito, S.; Principi, N. Streptococcus salivarius 24SMB administered by nasal spray for the prevention of acute otitis media in otitis-prone children. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 2377–2383. [Google Scholar] [CrossRef] [PubMed]
- Barfod, K.K.; Vrankx, K.; Mirsepasi-Lauridsen, H.C.; Hansen, J.S.; Hougaard, K.S.; Larsen, S.T.; Ouwenhand, A.C.; Krogfelt, K.A. The Murine Lung Microbiome Changes During Lung Inflammation and Intranasal Vancomycin Treatment. Open Microbiol. J. 2015, 9, 167–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youn, H.N.; Lee, D.H.; Lee, Y.N.; Park, J.K.; Yuk, S.S.; Yang, S.Y.; Lee, H.J.; Woo, S.H.; Kim, H.M.; Lee, J.B.; et al. Intranasal administration of live Lactobacillus species facilitates protection against influenza virus infection in mice. Antivir. Res. 2012, 93, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Geller, D.E. Aerosol antibiotics in cystic fibrosis. Respir. Care 2009, 54, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Morozova, V.V.; Vlassov, V.V.; Tikunova, N.V. Applications of Bacteriophages in the Treatment of Localized Infections in Humans. Front. Microbiol. 2018, 9, 1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Principi, N.; Silvestri, E.; Esposito, S. Advantages and Limitations of Bacteriophages for the Treatment of Bacterial Infections. Front. Pharmacol. 2019, 10, 513. [Google Scholar] [CrossRef] [Green Version]
- Summers, W.C. Cholera and plague in India: The bacteriophage inquiry of 1927-1936. J. Hist. Med. Allied Sci. 1993, 48, 275–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Essoh, C.; Blouin, Y.; Loukou, G.; Cablanmian, A.; Lathro, S.; Kutter, E.; Thien, H.V.; Vergnaud, G.; Pourcel, C. The susceptibility of Pseudomonas aeruginosa strains from cystic fibrosis patients to bacteriophages. PLoS ONE 2013, 8, e60575. [Google Scholar] [CrossRef] [Green Version]
- Hoyle, N.; Zhvaniya, P.; Balarjishvili, N.; Bolkvadze, D.; Nadareishvili, L.; Nizharadze, D.; Wittmann, J.; Rohde, C.; Kutateladze, M. Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: A case report. Res. Microbiol. 2018, 169, 540–542. [Google Scholar] [CrossRef]
- Kutateladze, M.; Adamia, R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 2010, 28, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Murima, P.; McKinney, J.D.; Pethe, K. Targeting bacterial central metabolism for drug development. Chem. Biol. 2014, 21, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Koul, A.; Dendouga, N.; Vergauwen, K.; Molenberghs, B.; Vranckx, L.; Willebrords, R.; Ristic, Z.; Lill, H.; Dorange, I.; Guillemont, J.; et al. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat. Chem. Biol. 2007, 3, 323–324. [Google Scholar] [CrossRef]
- Haagsma, A.C.; Abdillahi-Ibrahim, R.; Wagner, M.J.; Krab, K.; Vergauwen, K.; Guillemont, J.; Andries, K.; Lill, H.; Koul, A.; Bald, D. Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob. Agents Chemother. 2009, 53, 1290–1292. [Google Scholar] [CrossRef] [Green Version]
- O’Dwyer, D.N.; Ashley, S.L.; Gurczynski, S.J.; Xia, M.; Wilke, C.; Falkowski, N.R.; Norman, K.C.; Arnold, K.B.; Huffnagle, G.B.; Salisbury, M.L.; et al. Lung Microbiota Contribute to Pulmonary Inflammation and Disease Progression in Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 199, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, C.E.; Mirabolfathinejad, S.G.; Ruiz, V.A.; Evans, S.E.; Gagea, M.; Evans, C.M.; Dickey, B.F.; Moghaddam, S.J. Interleukin 6, but not T helper 2 cytokines, promotes lung carcinogenesis. Cancer Prev. Res. 2011, 4, 51–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salisbury, M.L.; Han, M.K.; Dickson, R.P.; Molyneaux, P.L. Microbiome in interstitial lung disease: From pathogenesis to treatment target. Curr. Opin. Pulm. Med. 2017, 23, 404–410. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weinberg, F.; Dickson, R.P.; Nagrath, D.; Ramnath, N. The Lung Microbiome: A Central Mediator of Host Inflammation and Metabolism in Lung Cancer Patients? Cancers 2021, 13, 13. https://doi.org/10.3390/cancers13010013
Weinberg F, Dickson RP, Nagrath D, Ramnath N. The Lung Microbiome: A Central Mediator of Host Inflammation and Metabolism in Lung Cancer Patients? Cancers. 2021; 13(1):13. https://doi.org/10.3390/cancers13010013
Chicago/Turabian StyleWeinberg, Frank, Robert P. Dickson, Deepak Nagrath, and Nithya Ramnath. 2021. "The Lung Microbiome: A Central Mediator of Host Inflammation and Metabolism in Lung Cancer Patients?" Cancers 13, no. 1: 13. https://doi.org/10.3390/cancers13010013



