Machine Learning Model to Predict Pseudoprogression Versus Progression in Glioblastoma Using MRI: A Multi-Institutional Study (KROG 18-07)
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Population and Definition of PsPD and PD
2.2. Model Structure and New Dataset Collection
2.3. Hyperparameter Optimization and Finalizing the Model
2.4. Selection of Calibration Model Selection and Implementation of User Interface
2.5. Statistical Analysis
3. Results
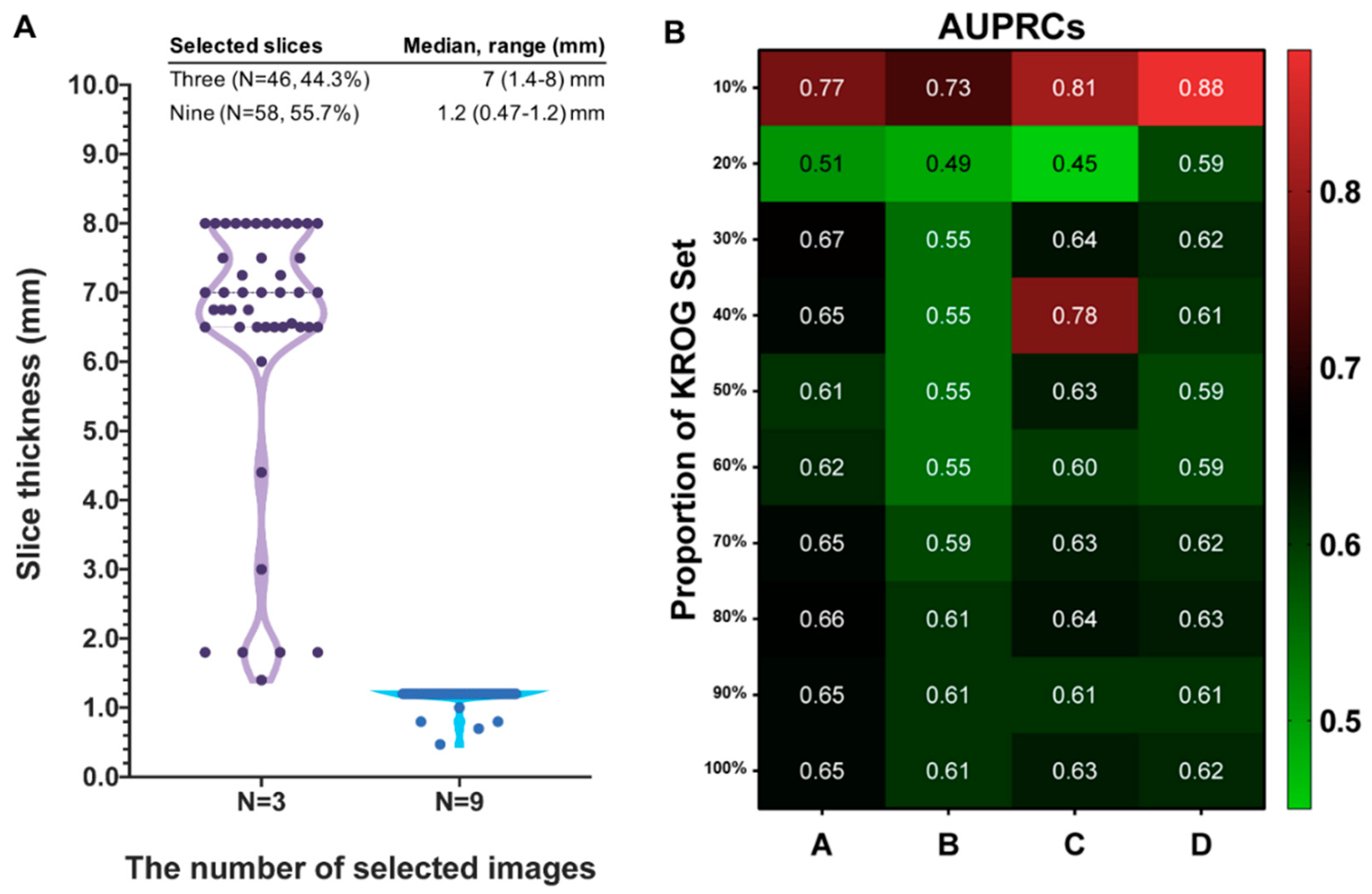
3.1. Characteristics of KROG Dataset
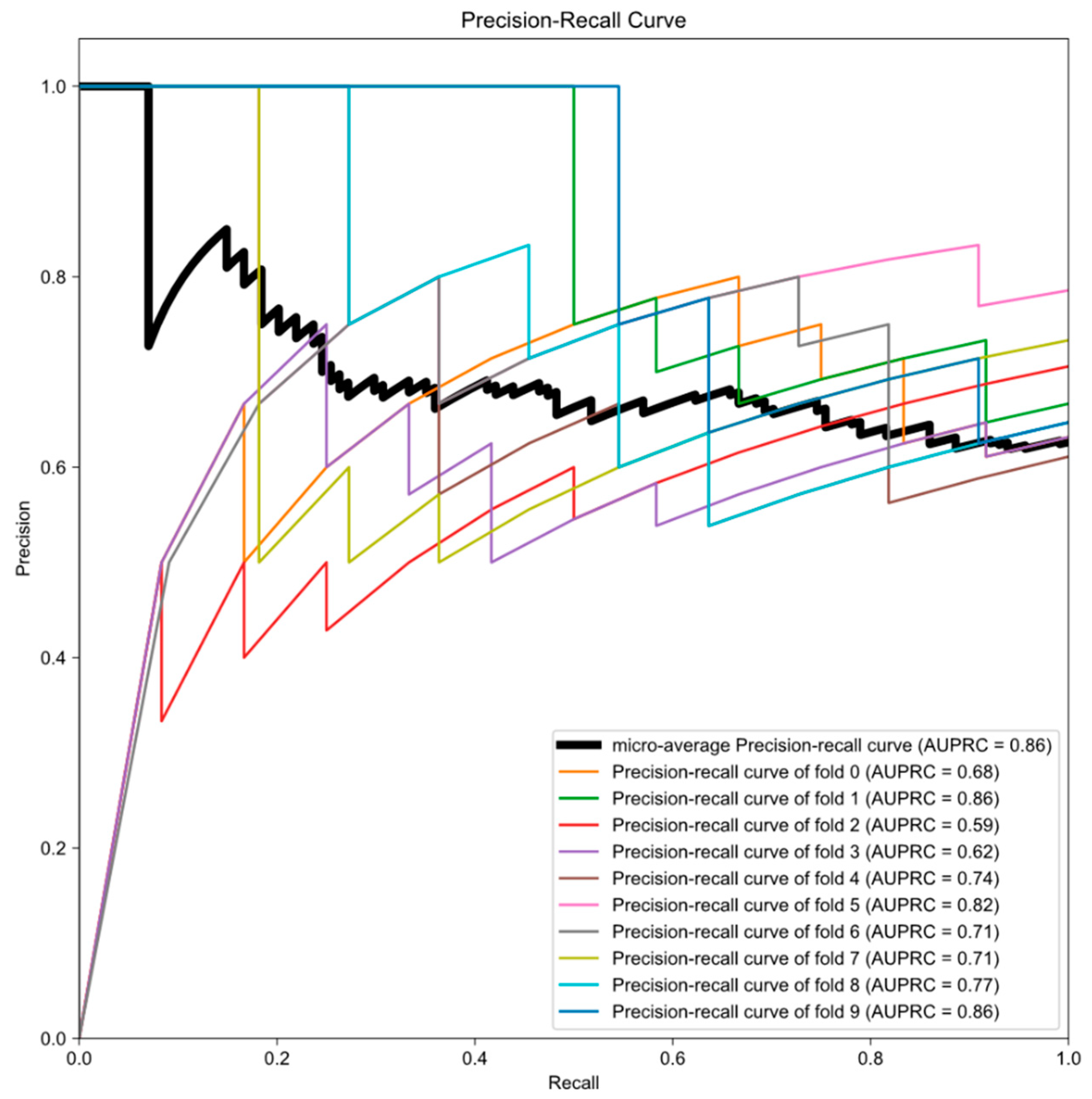
3.2. Testing Results of Previous Model in KROG Dataset
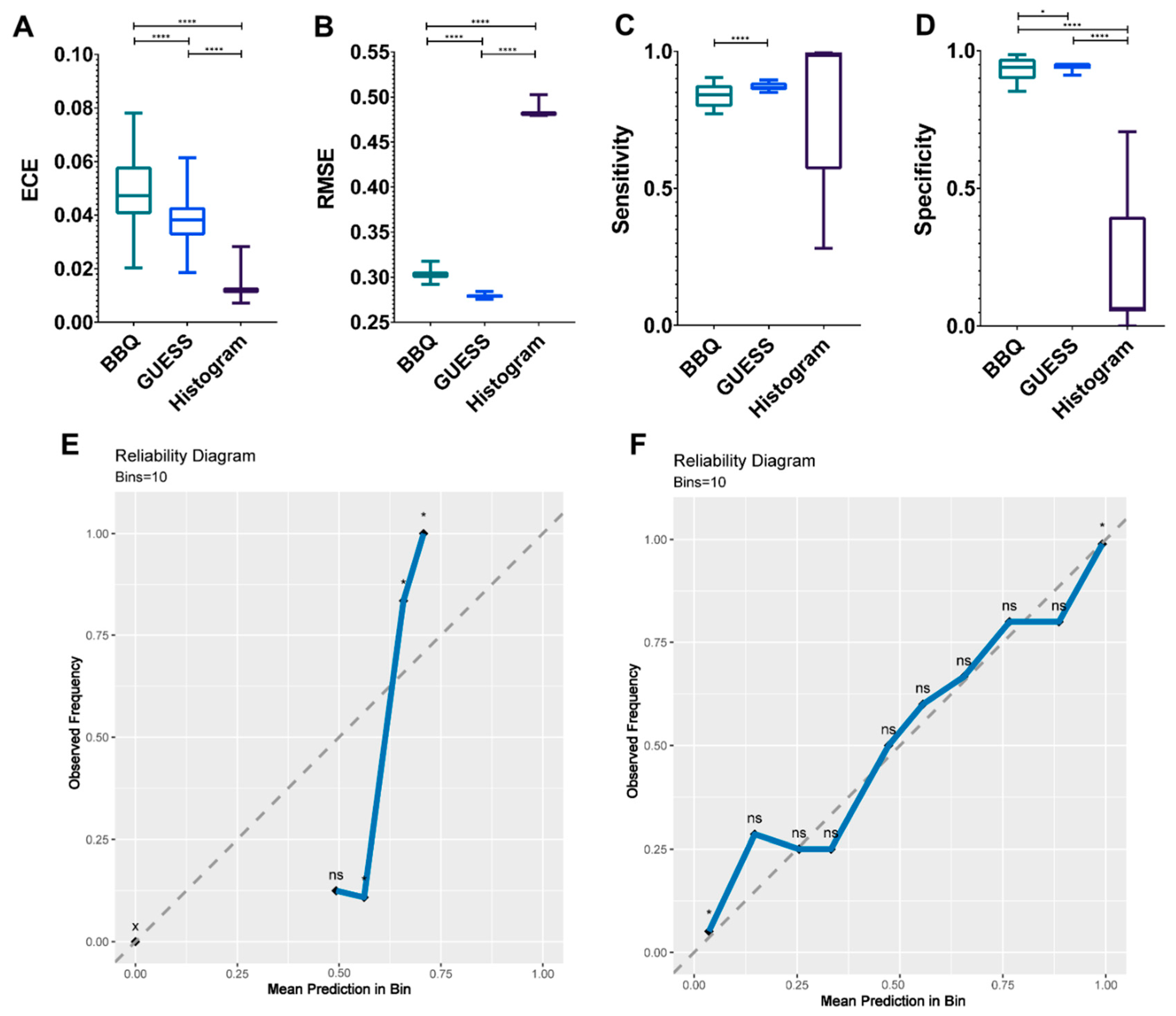
3.3. Establishment of Final Model with Calibration
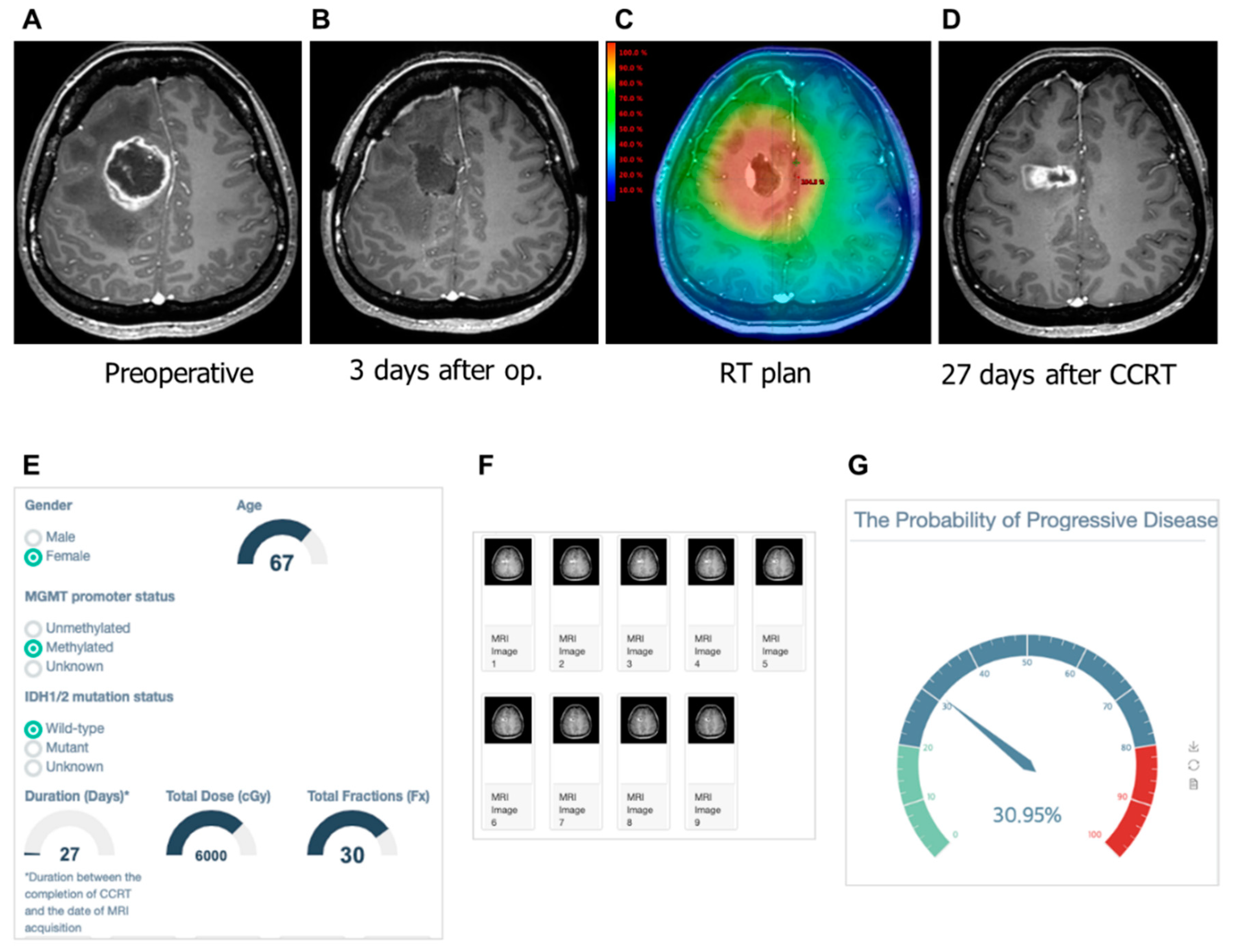
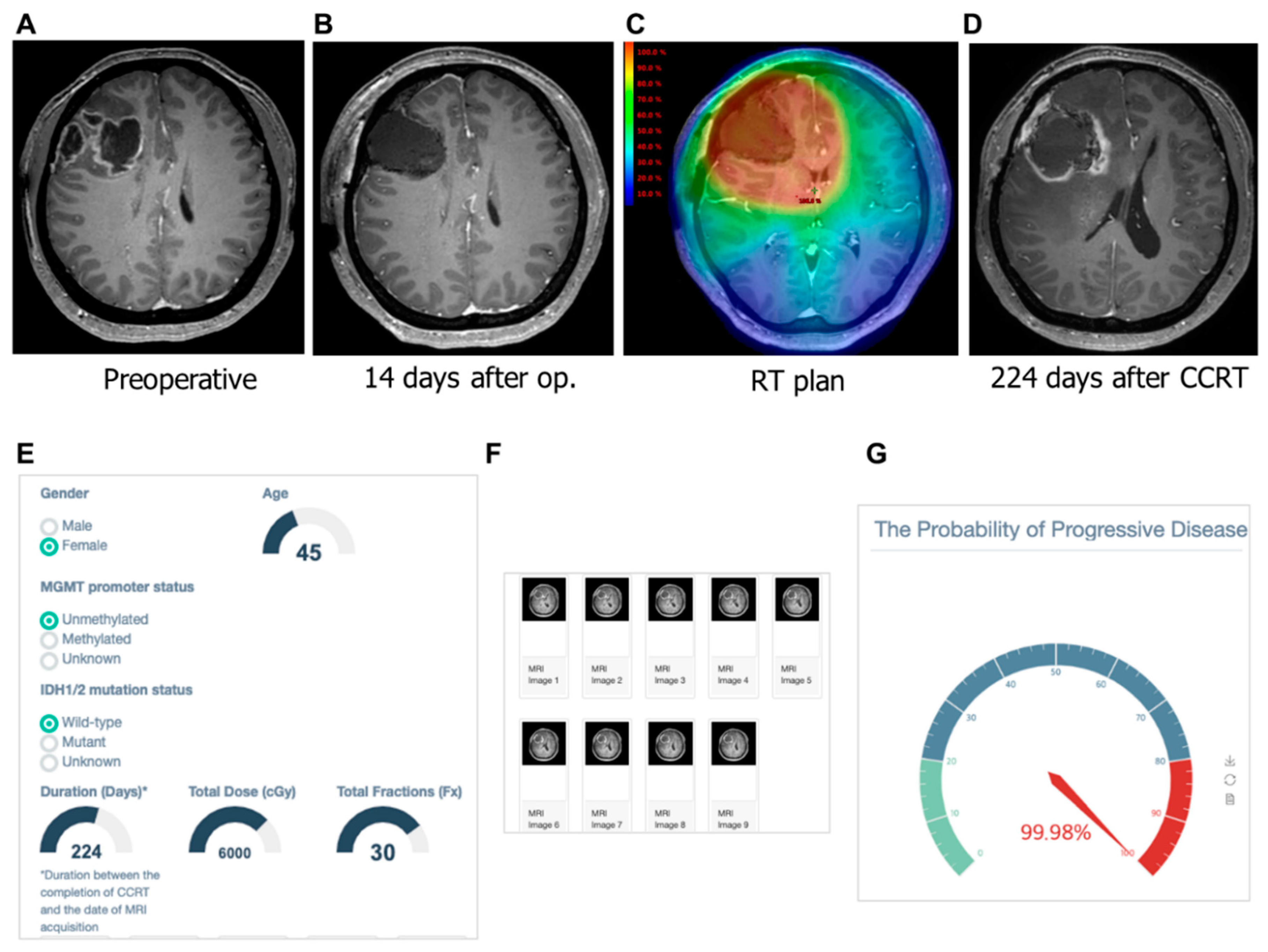
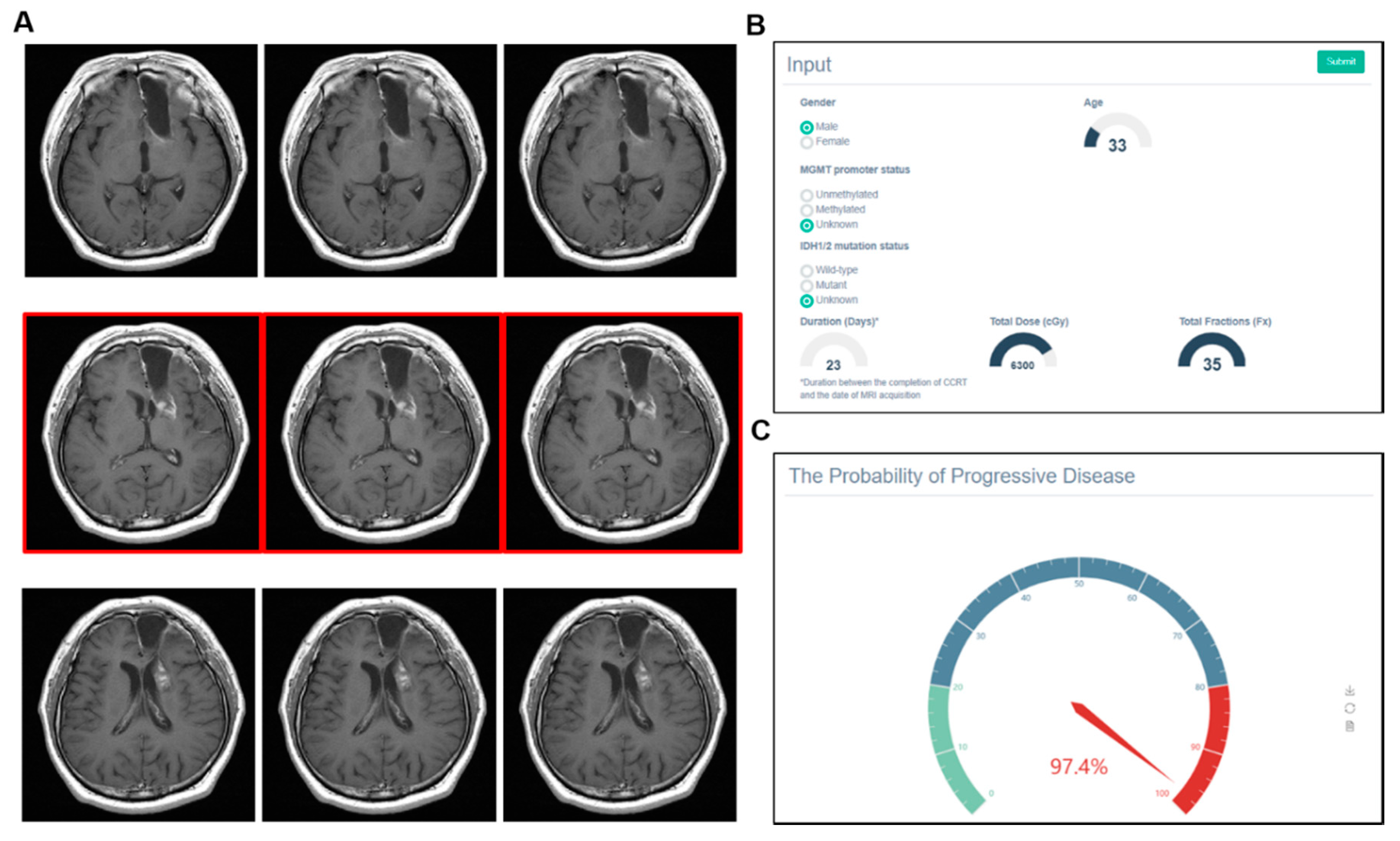
3.4. Examples of Correct and Incorrect Cases
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Taal, W.; Brandsma, D.; De Bruin, H.G.; Bromberg, J.E.; Swaak-Kragten, A.T.; Sillevis Smitt, P.A.E.; Van Es, C.A.; Van Den Bent, M.J. Incidence of early pseudo-progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer 2008. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, A.W.; Westerlaan, H.E.; Holtman, G.A.; Aden, K.M.; van Laar, P.J.; van der Hoorn, A. Incidence of Tumour Progression and Pseudoprogression in High-Grade Gliomas: A Systematic Review and Meta-Analysis. Clin. Neuroradiol. 2018, 28, 401–411. [Google Scholar] [CrossRef]
- Chaskis, C.; Neyns, B.; Michotte, A.; De Ridder, M.; Everaert, H. Pseudoprogression after radiotherapy with concurrent temozolomide for high-grade glioma: Clinical observations and working recommendations. Surg. Neurol. 2009. [Google Scholar] [CrossRef] [PubMed]
- De Wit, M.C.Y.; De Bruin, H.G.; Eijkenboom, W.; Sillevis Smitt, P.A.E.; Van Den Bent, M.J. Immediate post-radiotherapy changes in malignant glioma can mimic tumor progression. Neurology 2004. [Google Scholar] [CrossRef] [PubMed]
- Brandsma, D.; Stalpers, L.; Taal, W.; Sminia, P.; van den Bent, M.J. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol. 2008. [Google Scholar] [CrossRef]
- Rowe, L.S.; Butman, J.A.; Mackey, M.; Shih, J.H.; Cooley-Zgela, T.; Ning, H.; Gilbert, M.R.; Smart, D.D.K.; Camphausen, K.; Krauze, A.V. Differentiating pseudoprogression from true progression: Analysis of radiographic, biologic, and clinical clues in GBM. J. Neurooncol. 2018. [Google Scholar] [CrossRef]
- Brandes, A.A.; Franceschi, E.; Tosoni, A.; Blatt, V.; Pession, A.; Tallini, G.; Bertorelle, R.; Bartolini, S.; Calbucci, F.; Andreoli, A.; et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J. Clin. Oncol. 2008. [Google Scholar] [CrossRef]
- Nasseri, M.; Gahramanov, S.; Netto, J.P.; Fu, R.; Muldoon, L.L.; Varallyay, C.; Hamilton, B.E.; Neuwelt, E.A. Evaluation of pseudoprogression in patients with glioblastoma multiforme using dynamic magnetic resonance imaging with ferumoxytol calls RANO criteria into question. Neuro Oncol. 2014. [Google Scholar] [CrossRef]
- Thomas, A.A.; Arevalo-Perez, J.; Kaley, T.; Lyo, J.; Peck, K.K.; Shi, W.; Zhang, Z.; Young, R.J. Dynamic contrast enhanced T1 MRI perfusion differentiates pseudoprogression from recurrent glioblastoma. J. Neurooncol. 2015. [Google Scholar] [CrossRef]
- Wang, S.; Martinez-Lage, M.; Sakai, Y.; Chawla, S.; Kim, S.G.; Alonso-Basanta, M.; Lustig, R.A.; Brem, S.; Mohan, S.; Wolf, R.L.; et al. Differentiating tumor progression from pseudoprogression in patients with glioblastomas using diffusion tensor imaging and dynamic susceptibility contrast MRI. Am. J. Neuroradiol. 2016. [Google Scholar] [CrossRef]
- Muoio, B.; Giovanella, L.; Treglia, G. Recent Developments of 18F-FET PET in Neuro-oncology. Curr. Med. Chem. 2017, 25, 3061–3073. [Google Scholar] [CrossRef] [PubMed]
- Parent, E.E.; Sharma, A.; Jain, M. Amino Acid PET Imaging of Glioma. Curr. Radiol. Rep. 2019, 7, 1–11. [Google Scholar] [CrossRef]
- Jang, B.S.; Jeon, S.H.; Kim, I.H.; Kim, I.A. Prediction of Pseudoprogression versus Progression using Machine Learning Algorithm in Glioblastoma. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. Supplemental information. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Macdonald, D.R.; Reardon, D.A.; Cloughesy, T.F.; Sorensen, A.G.; Galanis, E.; DeGroot, J.; Wick, W.; Gilbert, M.R.; Lassman, A.B.; et al. Updated response assessment criteria for high-grade gliomas: Response assessment in neuro-oncology working group. J. Clin. Oncol. 2010, 28, 1963–1972. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Bendszus, M.; Boxerman, J.; Barboriak, D.; Erickson, B.J.; Smits, M.; Nelson, S.J.; Gerstner, E.; Alexander, B.; Goldmacher, G.; et al. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials. Neuro. Oncol. 2015. [Google Scholar] [CrossRef]
- Zadrozny, B.; Elkan, C. Obtaining calibrated probability estimates from decision trees and naive Bayesian classifiers. In Proceedings of the Icml, Williamstown, MA, USA, 28 June–1 July 2001; Citeseer: University Park, PA, USA, 2001; Volume 1, pp. 609–616. [Google Scholar]
- Naeini, M.P.; Cooper, G.F. Binary classifier calibration using an ensemble of piecewise linear regression models. Knowl. Inf. Syst. 2018, 54, 151–170. [Google Scholar]
- Schwarz, J.; Heider, D. GUESS: Projecting machine learning scores to well-calibrated probability estimates for clinical decision-making. Bioinformatics 2018. [Google Scholar] [CrossRef]
- Naeini, M.P.; Cooper, G.F.; Hauskrecht, M. Obtaining Well Calibrated Probabilities Using Bayesian Binning. Proc. Conf. Aaai Artif. Intell. 2015, 2015, 2901. [Google Scholar]
- Booth, T.C.; Larkin, T.J.; Yuan, Y.; Kettunen, M.I.; Dawson, S.N.; Scoffings, D.; Canuto, H.C.; Vowler, S.L.; Kirschenlohr, H.; Hobson, M.P.; et al. Analysis of heterogeneity in T2-weighted MR images can differentiate pseudoprogression from progression in glioblastoma. PLoS ONE 2017, e0176528. [Google Scholar] [CrossRef] [PubMed]
- Kebir, S.; Khurshid, Z.; Gaertner, F.C.; Essler, M.; Hattingen, E.; Fimmers, R.; Scheffler, B.; Herrlinger, U.; Bundschuh, R.A.; Glas, M. Unsupervised consensus cluster analysis of [18F]-fluoroethyl-L-tyrosine positron emission tomography identified textural features for the diagnosis of pseudoprogression in high-grade glioma. Oncotarget 2017, 8, 8294–8304. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wong, K.K.; Young, G.S.; Guo, L.; Wong, S.T. Support vector machine multiparametric MRI identification of pseudoprogression from tumor recurrence in patients with resected glioblastoma. J. Magn. Reson. Imaging 2011, 33, 296–305. [Google Scholar] [CrossRef] [PubMed]
- DSouza, A.M.; Chen, L.; Wu, Y.; Abidin, A.Z.; Xu, C.; Wismüller, A. MRI tumor segmentation with densely connected 3D CNN. Int. Soc. Opt. Photonics 2018, 10574, 105741F. [Google Scholar] [CrossRef]
- Qamar, S.; Jin, H.; Zheng, R.; Ahmad, P. 3D Hyper-Dense Connected Convolutional Neural Network for Brain Tumor Segmentation. In Proceedings of the 2018 14th International Conference on Semantics, Knowledge and Grids, SKG 2018, Guangzhou, China, 12–14 September 2018. [Google Scholar]
- Sun, J.; Chen, W.; Peng, S.; Liu, B. DRRNet: Dense Residual Refine Networks for Automatic Brain Tumor Segmentation. J. Med. Syst. 2019. [Google Scholar] [CrossRef]
- Lee, J.K.; Zong, W.; Dai, Z.; Liu, C.; Snyder, J.; Wen, N. Overall Survival Prediction in Glioblastoma Using Clinical Factors Combined with Texture Features Extracted from 3D Convolutional Neural Networks. Int. J. Radiat. Oncol. 2019. [Google Scholar] [CrossRef]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A.; et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from mr imaging. Clin. Cancer Res. 2018. [Google Scholar] [CrossRef]
- Chang, P.; Grinband, J.; Weinberg, B.D.; Bardis, M.; Khy, M.; Cadena, G.; Su, M.Y.; Cha, S.; Filippi, C.G.; Bota, D.; et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. Am. J. Neuroradiol. 2018. [Google Scholar] [CrossRef]
- Acharya, G.; Shah, N.; Krishnamurthi, G.; Khened, M.; Anand, V.K. 3D convolution neural networks for molecular subtype prediction in glioblastoma multiforme. Int. Soc. Opt. Photonics 2019. [Google Scholar] [CrossRef]
- Shaver, M.; Kohanteb, P.; Chiou, C.; Bardis, M.; Chantaduly, C.; Bota, D.; Filippi, C.; Weinberg, B.; Grinband, J.; Chow, D.; et al. Optimizing Neuro-Oncology Imaging: A Review of Deep Learning Approaches for Glioma Imaging. Cancers 2019, 11, 829. [Google Scholar] [CrossRef]
- Booth, T.C.; Williams, M.; Luis, A.; Cardoso, J.; Ashkan, K.; Shuaib, H. Machine learning and glioma imaging biomarkers. Clin. Radiol. 2020, 75, 20–32. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Wen, P.Y.; Cloughesy, T.F. Modified Criteria for Radiographic Response Assessment in Glioblastoma Clinical Trials. Neurotherapeutics 2017, 14, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Trunk, G.V. A Problem of Dimensionality: A Simple Example. IEEE Trans. Pattern Anal. Mach. Intell. 1979. [Google Scholar] [CrossRef] [PubMed]
- Sanghera, P.; Rampling, R.; Haylock, B.; Jefferies, S.; McBain, C.; Rees, J.H.; Soh, C.; Whittle, I.R. The Concepts, Diagnosis and Management of Early Imaging Changes after Therapy for Glioblastomas. Clin. Oncol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Cheng, G.; Zhang, J.; Li, X. IDH mutation and MGMT promoter methylation are associated with the pseudoprogression and improved prognosis of glioblastoma multiforme patients who have undergone concurrent and adjuvant temozolomide-based chemoradiotherapy. Clin. Neurol. Neurosurg. 2016, 151, 31–36. [Google Scholar] [CrossRef]
- Wick, W.; Chinot, O.L.; Bendszus, M.; Mason, W.; Henriksson, R.; Saran, F.; Nishikawa, R.; Revil, C.; Kerloeguen, Y.; Cloughesy, T. Evaluation of pseudoprogression rates and tumor progression patterns in a phase III trial of bevacizumab plus radiotherapy/temozolomide for newly diagnosed glioblastoma. Neuro. Oncol. 2016, 18, 1434–1441. [Google Scholar] [CrossRef]

| KROG Dataset | PsPD (N = 38) | PD (N = 66) | Total (N = 104) | |||
|---|---|---|---|---|---|---|
| N | % | N | % | p | ||
| Age (median, range) | 56.5 (23–75) | 55 (25–76) | 0.79 * | 55 (25–75) | ||
| Gender | 0.02 † | |||||
| Female | 22 | 57.9 | 23 | 34.8 | 45 | |
| Male | 16 | 42.1 | 43 | 65.2 | 59 | |
| MGMT promoter status | 0.40 † | |||||
| Methylated | 13 | 34.2 | 18 | 27.3 | 31 | |
| Unmethylated | 9 | 23.7 | 24 | 36.4 | 33 | |
| Unknown | 16 | 42.1 | 24 | 36.3 | 40 | |
| IDH mutational status | 0.22 ‡ | |||||
| Mutated | 2 | 5.2 | 0 | 0.0 | 2 | |
| Wild-type | 15 | 39.5 | 30 | 45.4 | 45 | |
| Unknown | 21 | 55.3 | 36 | 54.6 | 57 | |
| Dose schedule of RT | 0.65 † | |||||
| Hypofractionated | 2 | 5.3 | 5 | 7.6 | 7 | |
| Conventional | 36 | 94.7 | 61 | 92.4 | 97 | |
| Interval (days), median (range) | 28 (19–700) | 95 (8–744) | 0.21 * | 52 (8–744) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, B.-S.; Park, A.J.; Jeon, S.H.; Kim, I.H.; Lim, D.H.; Park, S.-H.; Lee, J.H.; Chang, J.H.; Cho, K.H.; Kim, J.H.; et al. Machine Learning Model to Predict Pseudoprogression Versus Progression in Glioblastoma Using MRI: A Multi-Institutional Study (KROG 18-07). Cancers 2020, 12, 2706. https://doi.org/10.3390/cancers12092706
Jang B-S, Park AJ, Jeon SH, Kim IH, Lim DH, Park S-H, Lee JH, Chang JH, Cho KH, Kim JH, et al. Machine Learning Model to Predict Pseudoprogression Versus Progression in Glioblastoma Using MRI: A Multi-Institutional Study (KROG 18-07). Cancers. 2020; 12(9):2706. https://doi.org/10.3390/cancers12092706
Chicago/Turabian StyleJang, Bum-Sup, Andrew J. Park, Seung Hyuck Jeon, Il Han Kim, Do Hoon Lim, Shin-Hyung Park, Ju Hye Lee, Ji Hyun Chang, Kwan Ho Cho, Jin Hee Kim, and et al. 2020. "Machine Learning Model to Predict Pseudoprogression Versus Progression in Glioblastoma Using MRI: A Multi-Institutional Study (KROG 18-07)" Cancers 12, no. 9: 2706. https://doi.org/10.3390/cancers12092706
APA StyleJang, B.-S., Park, A. J., Jeon, S. H., Kim, I. H., Lim, D. H., Park, S.-H., Lee, J. H., Chang, J. H., Cho, K. H., Kim, J. H., Sunwoo, L., Choi, S. H., & Kim, I. A. (2020). Machine Learning Model to Predict Pseudoprogression Versus Progression in Glioblastoma Using MRI: A Multi-Institutional Study (KROG 18-07). Cancers, 12(9), 2706. https://doi.org/10.3390/cancers12092706





