Single-Cell NGS-Based Analysis of Copy Number Alterations Reveals New Insights in Circulating Tumor Cells Persistence in Early-Stage Breast Cancer
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Isolation of CTCs
2.2. Single-Cell CNA Analysis of CTCs with Whole-Genome NGS
2.3. Identification of Aberrations Shared Between CTCs and Matched Tumor Tissue
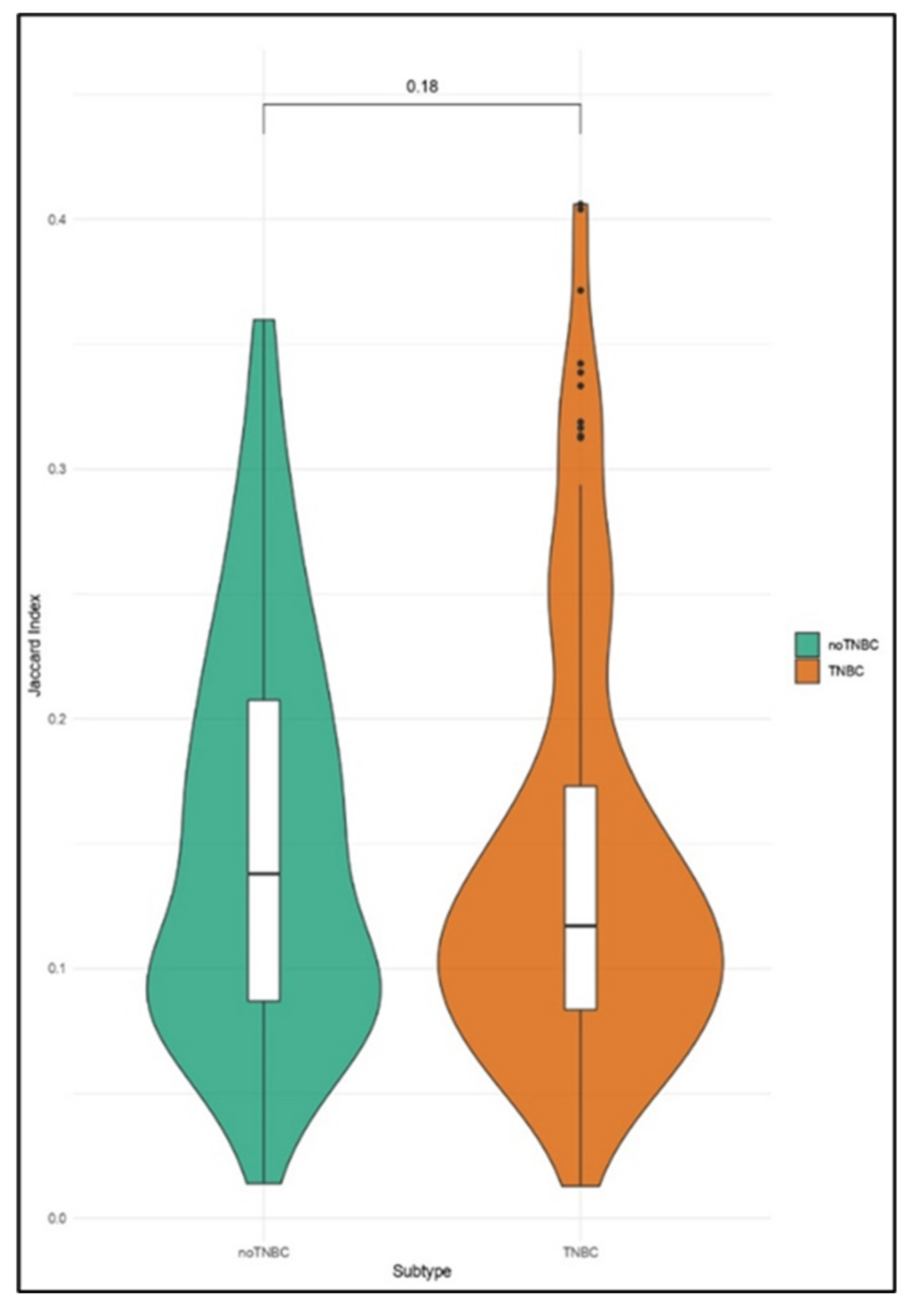
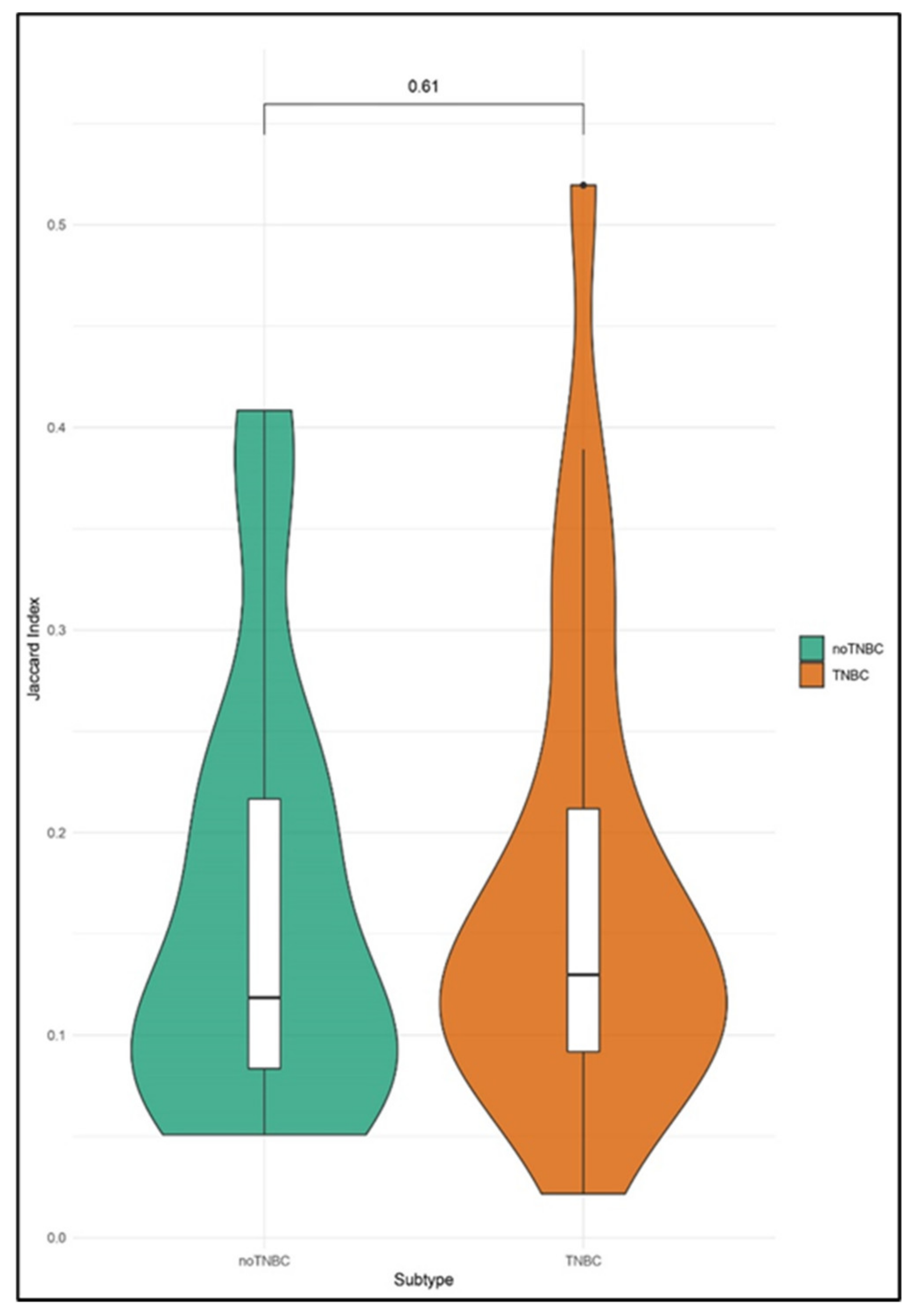
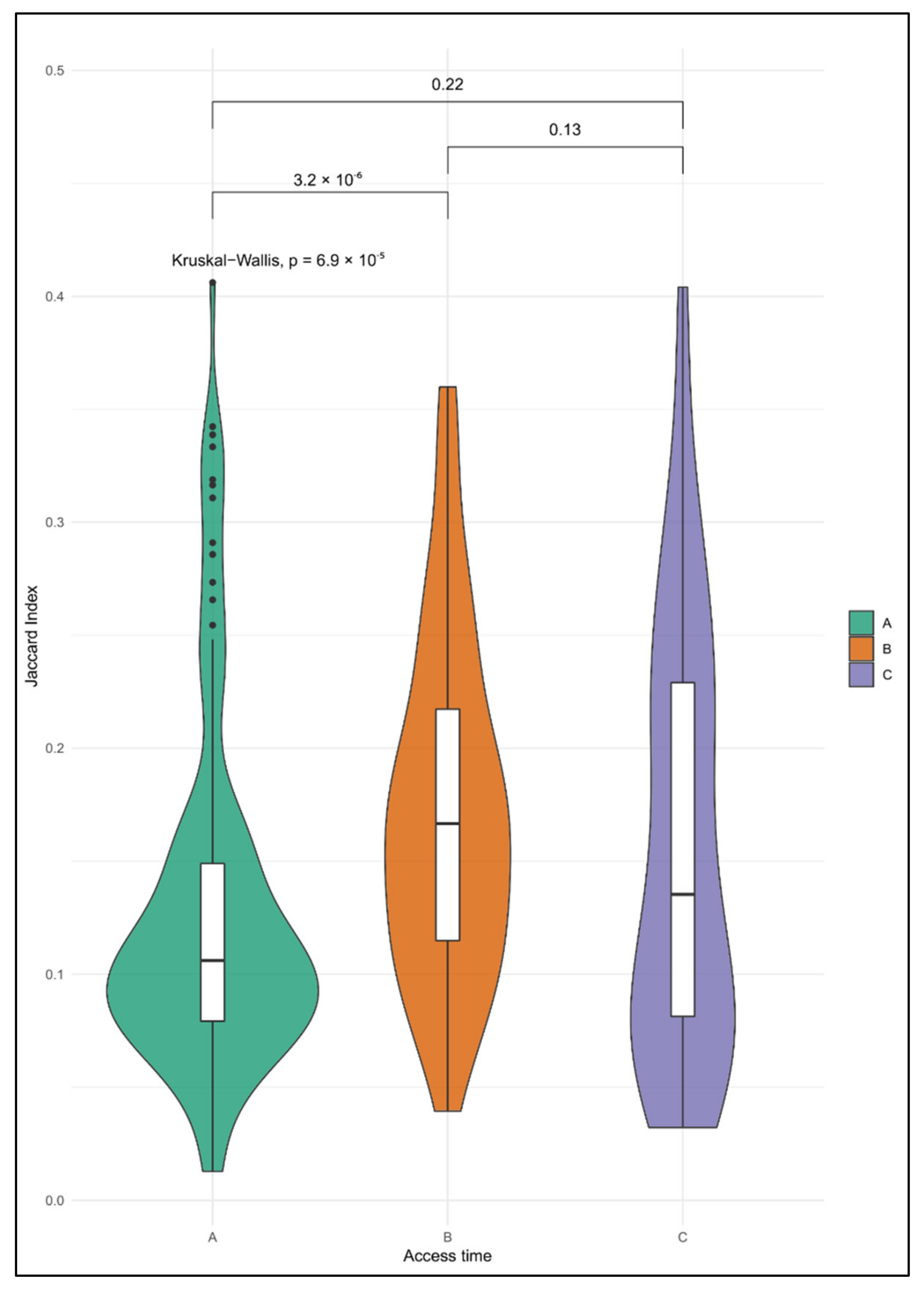
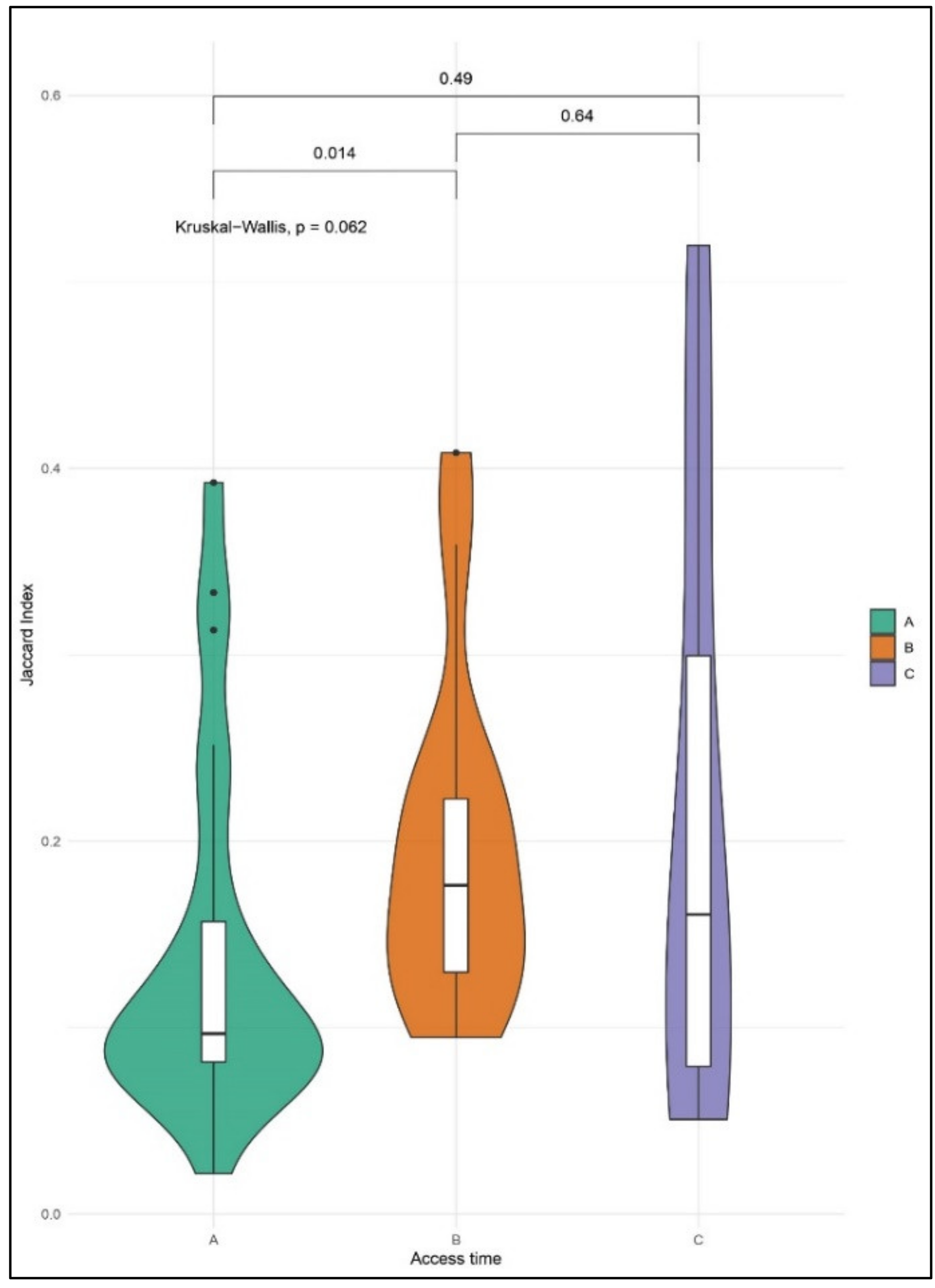
2.4. Enrichment Analyses of CNA in Single CTCs
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. CTC Enrichment and Isolation at The Single-Cell Level From Peripheral Whole Blood
4.3. DNA Extraction From FFPE Specimens
4.4. Library Preparation and Whole-Genome Low-Coverage Sequencing
4.5. Bioinformatic Analyses and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pantel, K.; Speicher, M.R. The biology of circulating tumor cells. Oncogene 2016, 35, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Sarangi, S.; Mosalpuria, K.; Higgins, M.J.; Bardia, A. The Evolving Role of Circulating Tumor Cells in the Personalized Management of Breast Cancer: From Enumeration to Molecular Characterization. Curr. Breast Cancer Rep. 2014, 6, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, S.; Keilholz, U.; Rossi, C.R.; Nitti, D. Circulating tumor cells: The ‘leukemic phase’ of solid cancers. Trends Mol. Med. 2006, 12, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Zamarchi, R. Single-Cell Analysis of Circulating Tumor Cells: How Far Have We Come in the -Omics Era? Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Maltoni, R.; Fici, P.; Amadori, D.; Gallerani, G.; Cocchi, C.; Zoli, M.; Rocca, A.; Cecconetto, L.; Folli, S.; Scarpi, E.; et al. Circulating tumor cells in early breast cancer: A connection with vascular invasion. Cancer Lett. 2015, 367, 43–48. [Google Scholar] [CrossRef]
- Riethdorf, S.; O’Flaherty, L.; Hille, C.; Pantel, K. Clinical applications of the CellSearch platform in cancer patients. Adv. Drug Deliv. Rev. 2018, 125, 102–121. [Google Scholar] [CrossRef] [PubMed]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 2005, 365, 1687–1717. [Google Scholar] [CrossRef]
- Sestak, I.; Cuzick, J. Markers for the identification of late breast cancer recurrence. Breast Cancer Res. 2015, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Khatcheressian, J.L.; Hurley, P.; Bantug, E.; Esserman, L.J.; Grunfeld, E.; Halberg, F.; Hantel, A.; Henry, N.L.; Muss, H.B.; Smith, T.J.; et al. Breast Cancer Follow-up and Management after Primary Treatment: American Society of Clinical Oncology Clinical Practice Guideline Update. J. Clin. Oncol. 2013, 31, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Franken, B.; de Groot, M.R.; Mastboom, W.J.; Vermes, I.; van der Palen, J.; Tibbe, A.G.; Terstappen, L.W. Circulating tumor cells, disease recurrence and survival in newly diagnosed breast cancer. Breast Cancer Res. 2012, 14, R133. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Rack, B.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; Hepp, P.; Zwingers, T.; Friedl, T.W.P.; Lorenz, R.; Tesch, H.; Fasching, P.A.; et al. Circulating Tumor Cells Predict Survival in Early Average-to-High Risk Breast Cancer Patients. JNCI J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Menyailo, M.E.; Tretyakova, M.S.; Denisov, E.V. Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds. Int. J. Mol. Sci. 2020, 21, 1696. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, C.; Cani, A.K.; Larios, J.M.; Hovelson, D.H.; Aung, K.; Darga, E.P.; Cannell, E.M.; Baratta, P.J.; Liu, C.-J.; Chu, D.; et al. Comprehensive Mutation and Copy Number Profiling in Archived Circulating Breast Cancer Tumor Cells Documents Heterogeneous Resistance Mechanisms. Cancer Res. 2018, 78, 1110–1122. [Google Scholar] [CrossRef]
- Shao, X.; Lv, N.; Liao, J.; Long, J.; Xue, R.; Ai, N.; Xu, D.; Fan, X. Copy number variation is highly correlated with differential gene expression: A pan-cancer study. BMC Med. Genet. 2019, 20, 175. [Google Scholar] [CrossRef]
- Riebensahm, C.; Joosse, S.A.; Mohme, M.; Hanssen, A.; Matschke, J.; Goy, Y.; Witzel, I.; Lamszus, K.; Kropidlowski, J.; Petersen, C.; et al. Clonality of circulating tumor cells in breast cancer brain metastasis patients. Breast Cancer Res. 2019, 21, 101. [Google Scholar] [CrossRef]
- Kanwar, N.; Hu, P.; Bedard, P.; Clemons, M.; McCready, D.; Done, S.J. Identification of genomic signatures in circulating tumor cells from breast cancer. Int. J. Cancer 2015, 137, 332–344. [Google Scholar] [CrossRef]
- Hieronymus, H.; Murali, R.; Tin, A.; Yadav, K.; Abida, W.; Moller, H.; Berney, D.; Scher, H.; Carver, B.; Scardino, P.; et al. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. Elife 2018, 7, e37294. [Google Scholar] [CrossRef]
- Hwang, K.T.; Han, W.; Cho, J.; Jong, W.L.; Ko, E.; Eun, K.K.; Jung, S.Y.; Jeong, E.M.; Bae, J.Y.; Kang, J.J.; et al. Genomic copy number alterations as predictive markers of systemic recurrence in breast cancer. Int. J. Cancer 2008, 123, 1807–1815. [Google Scholar] [CrossRef] [PubMed]
- Goh, J.Y.; Feng, M.; Wang, W.; Oguz, G.; Yatim, S.M.J.M.; Lee, P.L.; Bao, Y.; Lim, T.H.; Wang, P.; Tam, W.L.; et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med. 2017, 23, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Milioli, H.H.; Tishchenko, I.; Riveros, C.; Berretta, R.; Moscato, P. Basal-like breast cancer: Molecular profiles, clinical features and survival outcomes. BMC Med. Genom. 2017, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Orsetti, B.; Nugoli, M.; Cervera, N.; Lasorsa, L.; Chuchana, P.; Rougé, C.; Ursule, L.; Nguyen, C.; Bibeau, F.; Rodriguez, C.; et al. Genetic profiling of chromosome 1 in breast cancer: Mapping of regions of gains and losses and identification of candidate genes on 1q. Br. J. Cancer 2006, 95, 1439–1447. [Google Scholar] [CrossRef]
- Thompson, P.A.; Brewster, A.M.; Kim-Anh, D.; Baladandayuthapani, V.; Broom, B.M.; Edgerton, M.E.; Hahn, K.M.; Murray, J.L.; Sahin, A.; Tsavachidis, S.; et al. Selective genomic copy number imbalances and probability of recurrence in early-stage breast cancer. PLoS ONE 2011, 6, e23543. [Google Scholar] [CrossRef]
- Munro, A.F.; Twelves, C.; Thomas, J.S.; Cameron, D.A.; Bartlett, J.M.S. Chromosome instability and benefit from adjuvant anthracyclines in breast cancer. Br. J. Cancer 2012, 107, 71–74. [Google Scholar] [CrossRef]
- Wikman, H.; Vessella, R.; Pantel, K. Cancer micrometastasis and tumour dormancy. APMIS 2008, 116, 754–770. [Google Scholar] [CrossRef]
- Trapp, E.; Janni, W.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; de Gregorio, A.; Alunni-Fabbroni, M.; Tzschaschel, M.; Polasik, A.; Koch, J.G.; et al. Presence of Circulating Tumor Cells in High-Risk Early Breast Cancer During Follow-Up and Prognosis. J. Natl. Cancer Inst. 2019, 111, 380–387. [Google Scholar] [CrossRef]
- Sparano, J.; O’Neill, A.; Alpaugh, K.; Wolff, A.C.; Northfelt, D.W.; Dang, C.T.; Sledge, G.W.; Miller, K.D. Association of Circulating Tumor Cells With Late Recurrence of Estrogen Receptor–Positive Breast Cancer. JAMA Oncol. 2018, 4, 1700. [Google Scholar] [CrossRef]
- Pan, H.; Gray, R.; Braybrooke, J.; Davies, C.; Taylor, C.; McGale, P.; Peto, R.; Pritchard, K.I.; Bergh, J.; Dowsett, M.; et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N. Engl. J. Med. 2017, 377, 1836–1846. [Google Scholar] [CrossRef]
- Burstein, H.J.; Curigliano, G.; Loibl, S.; Dubsky, P.; Gnant, M.; Poortmans, P.; Colleoni, M.; Denkert, C.; Piccart-Gebhart, M.; Regan, M.; et al. Estimating the benefits of therapy for early-stage breast cancer: The St. Gallen International Consensus Guidelines for the primary therapy of early breast cancer 2019. Ann. Oncol. 2019, 30, 1541–1557. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Reinholz, M. Minimal residual disease and circulating tumor cells in breast cancer. Breast Cancer Res. 2011, 13, 222. [Google Scholar] [CrossRef] [PubMed]
- Gay-Bellile, M.; Véronèse, L.; Combes, P.; Eymard-Pierre, E.; Kwiatkowski, F.; Dauplat, M.-M.; Cayre, A.; Privat, M.; Abrial, C.; Bignon, Y.-J.; et al. TERT promoter status and gene copy number gains: Effect on TERT expression and association with prognosis in breast cancer. Oncotarget 2017, 8, 77540–77551. [Google Scholar] [CrossRef] [PubMed]
- Provance, O.K.; Lewis-wambi, J. Deciphering the role of interferon alpha signaling and microenvironment crosstalk in inflammatory breast cancer. Breast Cancer Res. 2019, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Medrano, R.F.V.; Hunger, A.; Mendonça, S.A.; Barbuto, J.A.M.; Strauss, B.E. Immunomodulatory and antitumor effects of type I interferons and their application in cancer therapy. Oncotarget 2017, 8, 71249–71284. [Google Scholar] [CrossRef] [PubMed]
- Leplina, O.Y.; Tyrinova, T.V.; Tikhonova, M.A.; Ostanin, A.A.; Chernykh, E.R. Interferon alpha induces generation of semi-mature dendritic cells with high pro-inflammatory and cytotoxic potential. Cytokine 2015, 71, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ferrarini, A.; Forcato, C.; Buson, G.; Tononi, P.; del Monaco, V.; Terracciano, M.; Bolognesi, C.; Fontana, F.; Medoro, G.; Neves, R.; et al. A streamlined workflow for single-cells genome-wide copy-number profiling by low-pass sequencing of LM-PCR whole-genome amplification products. PLoS ONE 2018, 13, e0193689. [Google Scholar] [CrossRef]
- Boeva, V.; Popova, T.; Bleakley, K.; Chiche, P.; Cappo, J.; Schleiermacher, G.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012, 28, 423–425. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes—Release 72.1, 1 December 2014. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
| Patient Code | BC Subtype | Access Time | Total | ||
|---|---|---|---|---|---|
| A | B | C | |||
| 1 | TNBC | 6 | 0 | 0 | 6 |
| 2 | TNBC | 1 | 2 | 1 | 4 |
| 3 | TNBC | 0 | 1 | 3 | 4 |
| 4 | TNBC | 6 | 3 | 3 | 12 |
| TNBC total | 13 | 6 | 7 | 26 | |
| 6 | HER2+ | 1 | 1 | 1 | 3 |
| 7 | Luminal A | 1 | 1 | 1 | 3 |
| 8 | Luminal A | 2 | 3 | 0 | 5 |
| 9 | Luminal A | 1 | 1 | 1 | 3 |
| 10 | Luminal A | 1 | 0 | 2 | 3 |
| 11 | Luminal A | 3 | 0 | 0 | 3 |
| 12 | Luminal A | 2 | 1 | 0 | 3 |
| Non-TNBC total | 11 | 7 | 5 | 23 | |
| Total | 24 | 13 | 12 | 49 | |
| Chromosome:Start–End (bp) | Region/s | CTC ID Timepoint A | CTC ID Timepoint B | CTC ID Timepoint C | Reference |
|---|---|---|---|---|---|
| Patient P02 | |||||
| 16:81368788–82403137 | 16q23.3 | P02A__2 | P02B_1 | P02C_1, P02C_2 | |
| 21:40355250–43442658 | 21q22.2, 21q22.3 | - | P02B_1 | P02C_1 | [21] |
| Patient 04 | |||||
| 1:33248192–58929280 | 1p35.1, 1p34.3, 1p34.2, 1p34.1, 1p33, 1p32.3, 1p32.2 | P04A_5 | - | P04C_3 | |
| 1:156891657–162239674 | 1q23.1, 1q23.2, 1q23.3 | P04A_2, P04A_5 | P04B_2 | P04C_1, P04C_3 | [19,22,23,24] |
| 1:162239674–167163992 | 1q23.3, 1q24.1 | - | P04B_1 | P04C_1 | [19,22,23,24] |
| 1:231729524–234612196 | 1q42.2 | - | - | P04C_1, P04C_3 | [22,23,24] |
| 2:36278338–54693738 | 2p22.2, 2p22.1, 2p21, 2p16.3, 2p16.2 | P04A_6 | - | P04C_1, P04C_3 | |
| 3:72930104–74214062 | 3p13 | - | - | P04C_1, P04C_3 | |
| 5:0–2025694 | 5p15.33 | P04A_7, P04A_9 | - | P04C_3 | [21,25] |
| 6:19336170–21177710 | 6p22.3 | - | - | P04C_1, P04C_3 | |
| 7:36023910–36646646 | 7p14.2 | 04C458_7 | - | P04C_3 | |
| 7:151606990–156715054 | 7q36.1, 7q36.2, 7q36.3 | - | P04B_2 | P04C_1 | |
| 8:92927068–94286848 | 8q21.2 | P04A_8 | - | P04C_1, P04C_3 | |
| 8:121094607–123567334 | 8q24.12, 8q24.13 | P04A_9 | - | P04C_3 | |
| 9:109019168–109755681 | 9q31.2 | 04A459_9 | - | P04C_1 | |
| 10:124680720–129276108 | 10q26.13 | P04A_6 | - | P04C_3 | |
| 11:17862938–20072786 | 11p15.1 | P04A_2 | - | P04C_1 | |
| 11:112838832–119700100 | 11q23.2, 11q23.3 | P04A_2, P04A_5 | P04C_1 | ||
| 13:24308328–24860790 | 13q12.12 | - | - | P04C_1, P04C_3 | |
| 14:75318986–75871448 | 14q24.3 | P04A_9 | - | P04C_1 | |
| 14:94448360–96128388 | 14q32.13 | - | P04B_2 | P04C_1, P04C_3 | |
| 15:80279738–85999918 | 15q25.1, 15q25.2, 15q25.3 | P04A_2, P04A_5 | - | P04C_3 | |
| 16:84526686–87288996 | 16q24.1 | P04A_5 | P04B_1, P04B_2, P04B_3 | P04C_1 | |
| 17:17494630–19520324 | 17p11.2 | - | P04B_3 | P04C_3 | |
| 17:70159110–70715136 | 17q24.3 | - | P04B_1 | P04C_3 | |
| 18:77160526–78077248 | 18q23 | - | P04B_1 | P04C_1, P04C_3 | [21] |
| 20:42723728–47051680 | 20q13.12, 20q13.13 | P04A_9 | P04B_3 | P04C_1 | |
| 22:34700614–35725876 | 22q12.3 | P04A_5, P04A_7, P04A_9 | P04B_1 | P04C_1 | |
| X:13627396–13995704 | Xp22.2 | P04A_5 | - | P04C_1, P04C_3 | |
| Patient 07 | |||||
| 22:25992468–25992468 | 22q12.1, 22q12.2, 22q12.3 | P07A__1 | P07B_3 | P07C_2 | |
| Patient 09 | |||||
| 15:60636100–64190630 | 15q22.2 | - | P09B__1 | P09C_2 | |
| Patient 10 | |||||
| 16:82480932–85699185 | 16q23.3, 16q24.1 | P10A_1 | P10B__1 | P10C_1 |
| Enriched Terms | Term ID | CTCs | Samples | Genes |
|---|---|---|---|---|
| Natural killer cell activation involved in immune response | GO:0002323 | 10 | P02A__2; P03C_1; P07A__1; P09B__2; P08A__1; P09B__3; P09B__1; P10B_1; P11A_1; P12A_2 | CD244; CORO1A; IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNK; IFNW1; KLRF2; VAMP7 |
| Response to dsRNA | GO:0043331 | 9 | P01A_6; P02A_2; P07A_1; P08A_1; P09B_2; P09B_1; P10B_1; P11A_1; P12A_2 | IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNK; IFNW1; IRAK3; PDE12; PMAIP1; RFTN1; RIOK3; SLC3A2; TICAM1; TLR3 |
| Regulation of peptidyl-serine phosphorylation of STAT protein | GO:0033139 | 9 | P01A_6; P02A_2; P07A_1; P08A_1; P09B_2; P09B_1; P10B_1; P11A_1; P12A_2 | IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNG; IFNK; IFNW1; LIF |
| Positive regulation of peptidyl-serine phosphorylation of STAT protein | GO:0033141 | 9 | P01A_6; P02A_2; P07A_1; P08A_1; P09B_2; P09B_1; P10B_1; P11A_1; P12A_2 | IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNG; IFNK; IFNW1; LIF |
| Lymphocyte activation involved in immune response | GO:0002285 | 9 | P01A_6; P02A_2; P07A_1; P08A_1; P09B_2; P09B_1; P10B_1; P11A_1; P12A_2 | CD1C; CD244; EIF2AK4; F2RL1; IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNK; IFNW1; LCP1 |
| Natural killer cell activation | GO:0030101 | 8 | P02A_2; P07A_1; P08A_1; P09B_2; P09B_3; P09B_1; P11A_1; P12A_2 | BAG6; CASP8; CD2; CD244; ELF4; HNF1A; IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNK; IFNW1; IL12B; IL2; IL21R; KLRK1; NCR1; NCR3; PRDX1; SLAMF7; SNX27; ULBP1; ULBP2; ULBP3 |
| B cell proliferation | GO:0042100 | 8 | P01A_6; P02A_2; P07A_1; P08A_1; P09B_2; P09B_1; P11A_1; P12A_2 | BCL2; CD40; CD40LG; CD79A; CR2; CTPS1; GAPT; HSPD1; IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNK; IFNW1; IL10; LEF1; MEF2C; MS4A1 |
| Cytidine to uridine editing | GO:0016554 | 7 | P01A_5; P04A9; P07A_1; P07C2; P08A_1; P09B_3; P11A_1 | A1CF; AICDA; APOBEC1; APOBEC2; APOBEC3A; APOBEC3B; APOBEC3C; APOBEC3D; APOBEC3F; APOBEC3G; APOBEC3H |
| T cell activation involved in immune response | GO:0002286 | 7 | P01A_6; P07A_1; P08A_1; P09B_2; P09B_1; P11A_1; P12A_2 | CD1C; EIF2AK4; F2RL1; FCER1G; ICAM1; IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNK; IFNW1; ITGAL; LCP1; LILRB1; SLC11A1; TNFSF18 |
| Lymphocyte proliferation | GO:0046651 | 7 | P02A_2; P07A_1; P08A_1; P09B_2; P09B_1; P11A_1; P12A_2 | BCL2; CD40; CD40LG; CD79A; CR2; CTPS1; ELF4; HELLS; HSPD1; IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNK; IFNW1; IL10; MEF2C; MS4A1; MSN; PIK3CG; PPP3CB; TNFRSF4; TNFSF14 |
| Response to exogenous dsRNA | GO:0043330 | 7 | P01A_6; P07A_1; P08A_1; P09B_2; P09B_1; P11A_1; P12A_2 | CAV1; COLEC12; DDX58; DHX9; FLOT1; IFIH1; IFIT1; IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNK; IFNW1; IRAK3; MUL1; PQBP1; RALB; RFTN1; SLC3A2; TICAM1; TLR3; TMEM173 |
| DNA cytosine deamination | GO:0070383 | 6 | P01A_5; P04A9; P07A_1; P07C2 P08A_1; P11A_1 | APOBEC3A; APOBEC3C; APOBEC3D; APOBEC3F; APOBEC3G; APOBEC3H |
| B cell differentiation | GO:0030183 | 6 | P01A_6; P07A_1; P08A_1; P09B_1; P11A_1; P12A_2 | ADAM17; AICDA; BLNK; CD79A; CEBPG; CLCF1; DNAJB9; HDAC5; HDAC9; HHEX; IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNB1; IFNE; IFNK; IFNW1; IL11; ITGA4; JAK3; KIT; KLF6; LYL1; MSH2; NFAM1; NHEJ1; PLCG2; RAG1; RAG2; TCF3; TPD52; VCAM1 |
| Regulation of type I interferon-mediated signaling pathway | GO:0060338 | 6 | P07A_1; P08A_1; P09B_2; P09B_1; P11A_1; P12A_2 | CACTIN; CDC37; FADD; HSP90AB1; IFNA1; IFNA10; IFNA14; IFNA16; IFNA17; IFNA2; IFNA21; IFNA4; IFNA5; IFNA6; IFNA7; IFNA8; IFNAR1; IFNAR2; IFNB1; JAK1; NLRC5; PTPN1; PTPN11; PTPN6; TYK2; WNT5A; ZBP1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, T.; Gallerani, G.; Angeli, D.; Cocchi, C.; Bandini, E.; Fici, P.; Gaudio, M.; Martinelli, G.; Rocca, A.; Maltoni, R.; et al. Single-Cell NGS-Based Analysis of Copy Number Alterations Reveals New Insights in Circulating Tumor Cells Persistence in Early-Stage Breast Cancer. Cancers 2020, 12, 2490. https://doi.org/10.3390/cancers12092490
Rossi T, Gallerani G, Angeli D, Cocchi C, Bandini E, Fici P, Gaudio M, Martinelli G, Rocca A, Maltoni R, et al. Single-Cell NGS-Based Analysis of Copy Number Alterations Reveals New Insights in Circulating Tumor Cells Persistence in Early-Stage Breast Cancer. Cancers. 2020; 12(9):2490. https://doi.org/10.3390/cancers12092490
Chicago/Turabian StyleRossi, Tania, Giulia Gallerani, Davide Angeli, Claudia Cocchi, Erika Bandini, Pietro Fici, Michele Gaudio, Giovanni Martinelli, Andrea Rocca, Roberta Maltoni, and et al. 2020. "Single-Cell NGS-Based Analysis of Copy Number Alterations Reveals New Insights in Circulating Tumor Cells Persistence in Early-Stage Breast Cancer" Cancers 12, no. 9: 2490. https://doi.org/10.3390/cancers12092490
APA StyleRossi, T., Gallerani, G., Angeli, D., Cocchi, C., Bandini, E., Fici, P., Gaudio, M., Martinelli, G., Rocca, A., Maltoni, R., & Fabbri, F. (2020). Single-Cell NGS-Based Analysis of Copy Number Alterations Reveals New Insights in Circulating Tumor Cells Persistence in Early-Stage Breast Cancer. Cancers, 12(9), 2490. https://doi.org/10.3390/cancers12092490








