The CXCR4-Dependent LASP1-Ago2 Interaction in Triple-Negative Breast Cancer
Simple Summary
Abstract
1. Introduction
2. Results
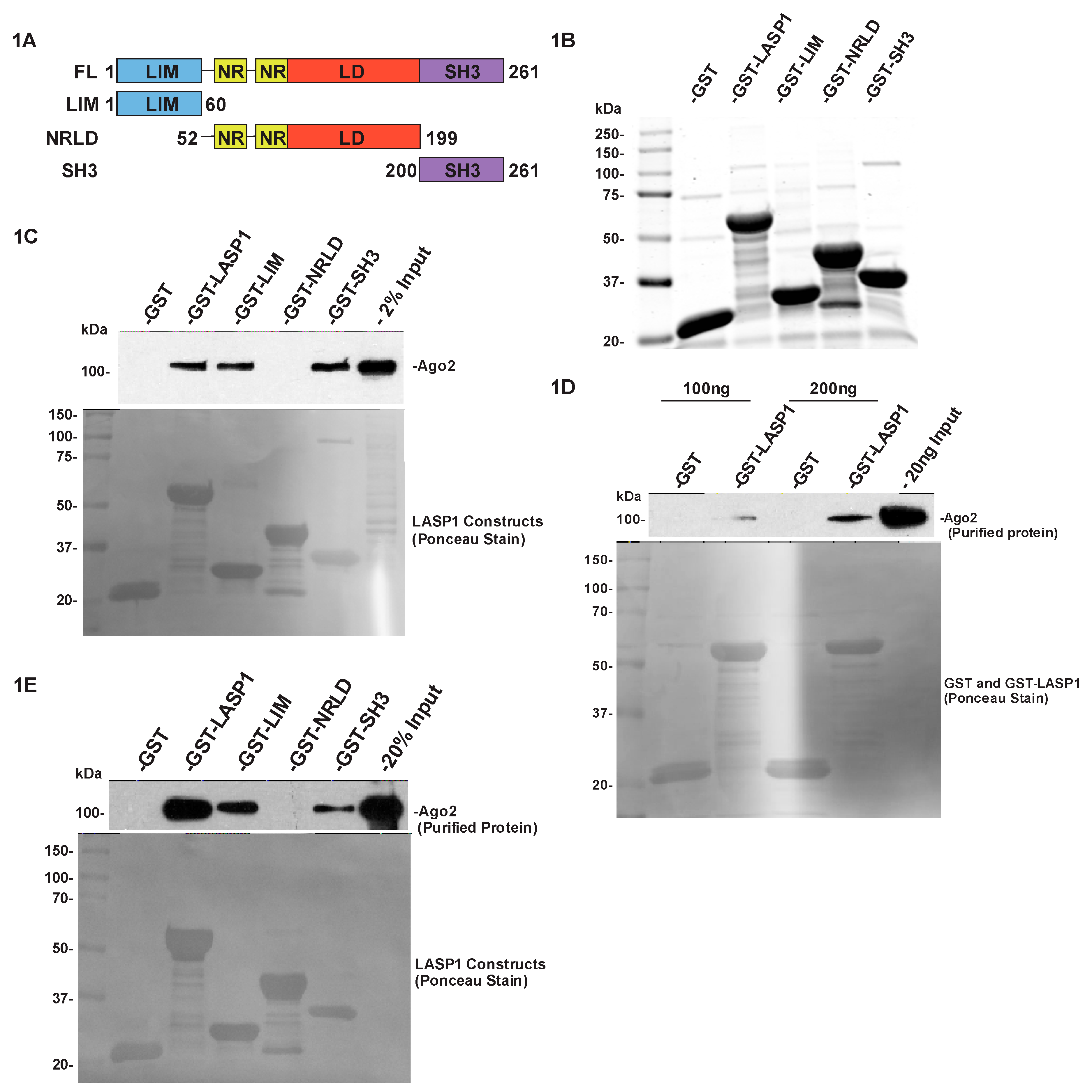
2.1. LASP1 Associates with Ago2 through Its LIM and SH3 Domains
2.2. LASP1 Directly Binds to Ago2 through Its LIM and SH3 Domains
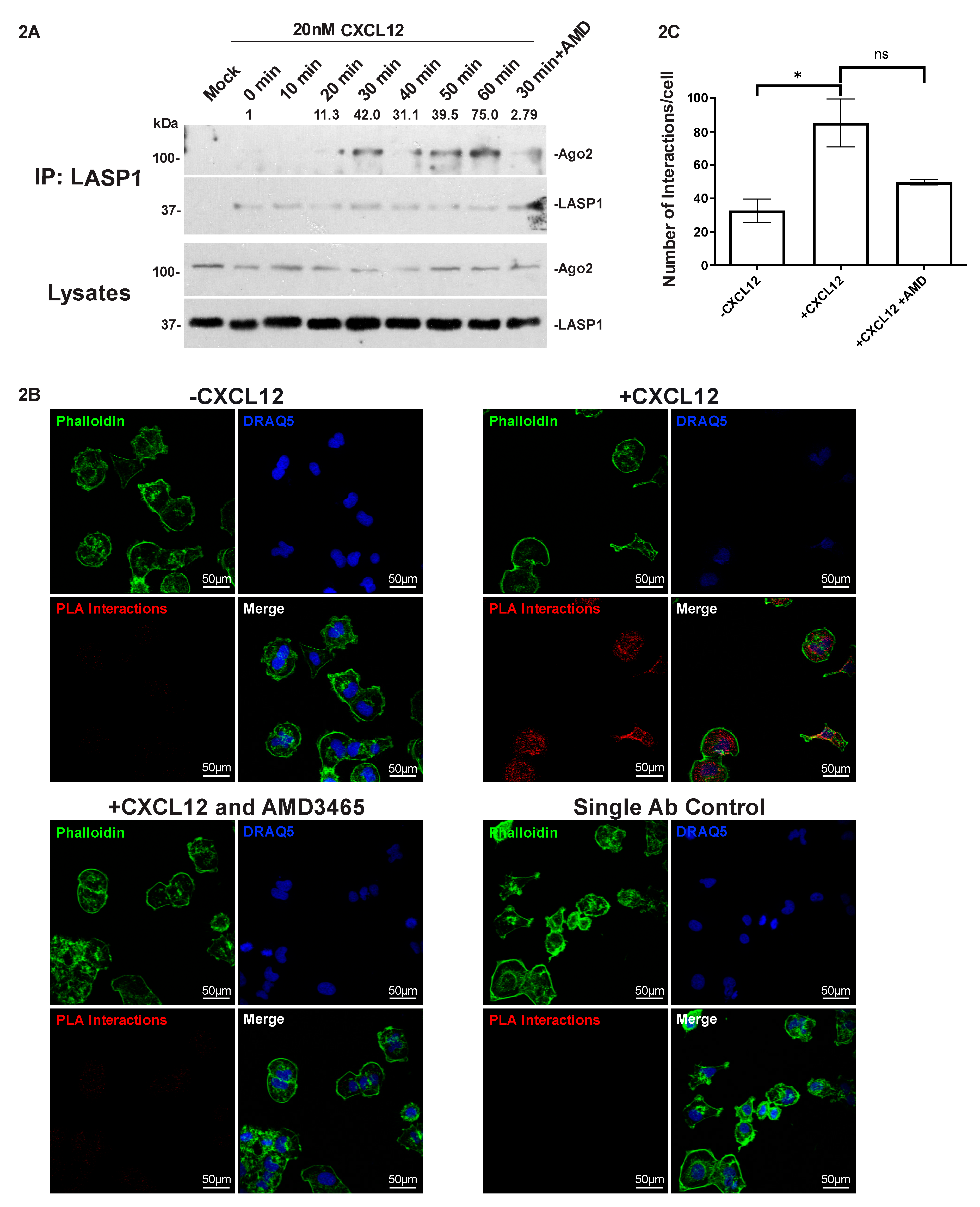
2.3. LASP1 Endogenously Interacts with Ago2 in a CXCL12-Dependent Manner
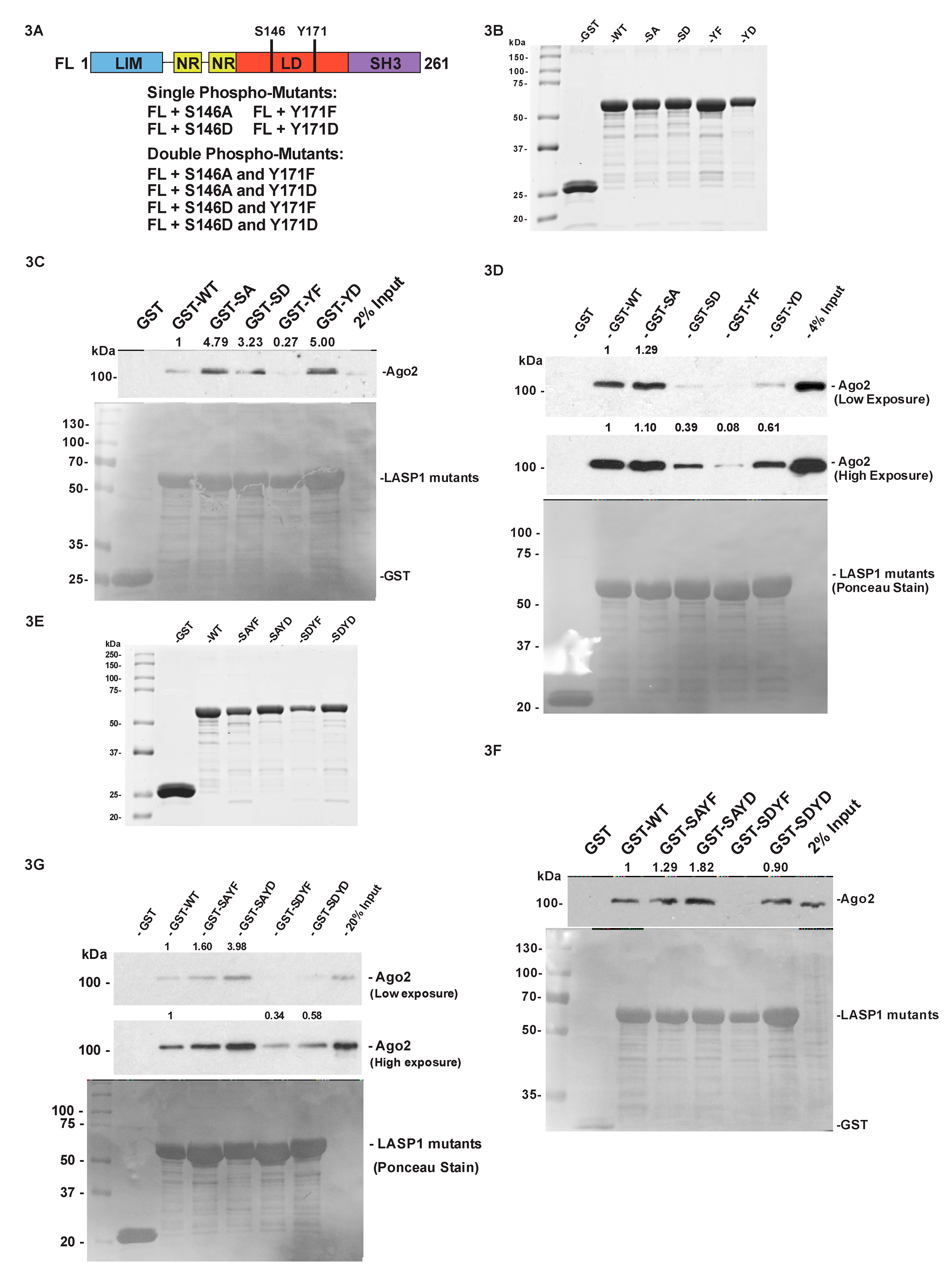
2.4. The Phosphorylation Status of LASP1 Dictates the LASP1-Ago2 Interaction
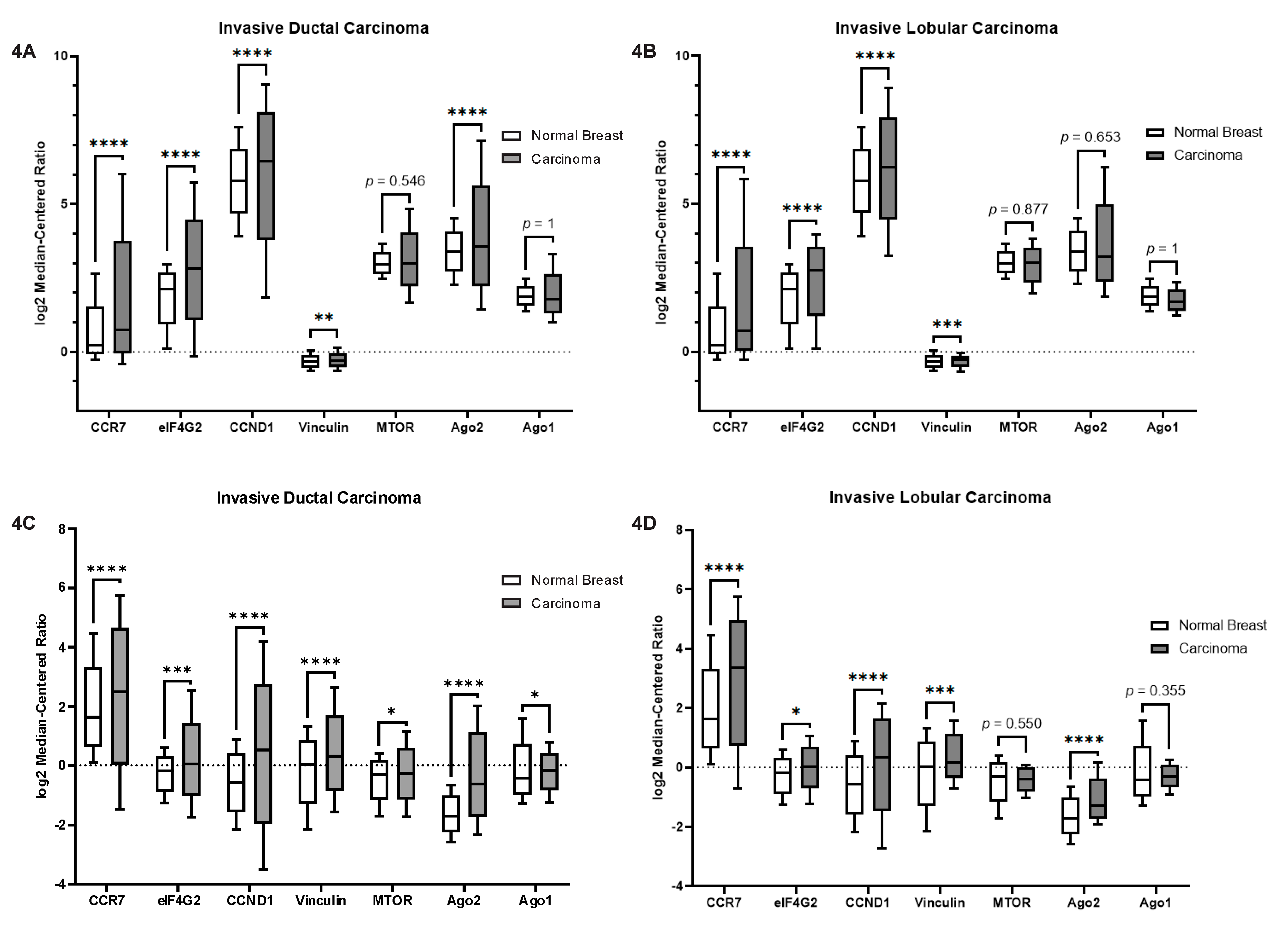
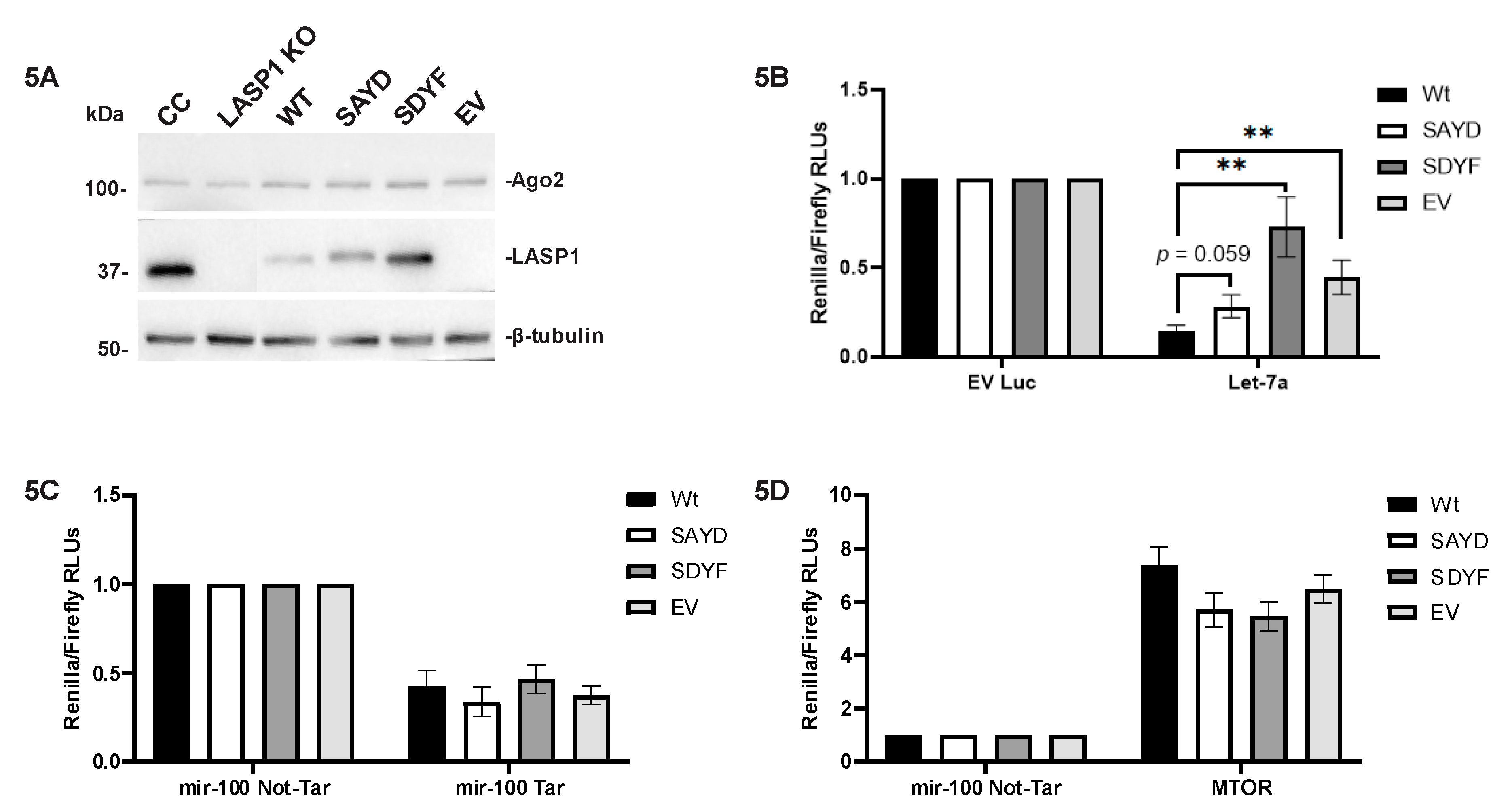
2.5. Let-7a and MiR-100 Targets Are Altered in TNBC
2.6. Let-7a Based Target Repression is Altered in a LASP1-Dependent Manner
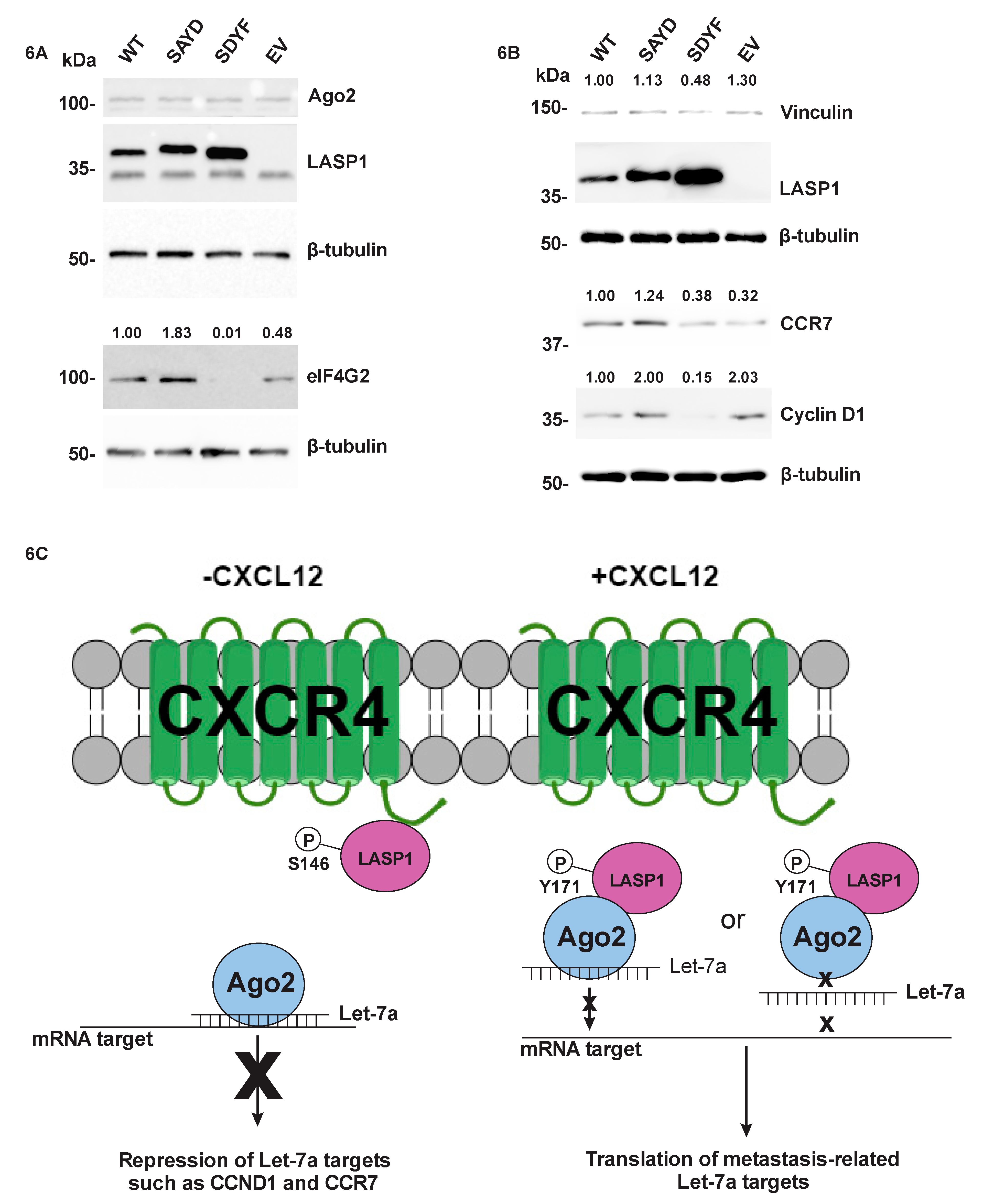
2.7. The LASP1-Ago2 Interaction Impacts Expression of Let-7a Targets
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Engineering of GST-LASP1 Domain Constructs
4.3. GST-Pulldown/Direct Binding Assays
4.4. Co-Immunoprecipitation Assay
4.5. Proximity Ligation Assay
4.6. Generation of LASP1 Phospho-Mutant Constructs and Phospho-Mutant Cell Lines
4.7. Bioinformatic Analysis
4.8. Luciferase Reporter Assays for miRNA Targets
4.9. Western Blotting
4.10. Statistical Analysis, Graph Preparation and Figure Design
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.U.; Vanderplas, A.; Hughes, M.E.; Theriault, R.L.; Edge, S.B.; Wong, Y.N.; Blayney, D.W.; Niland, J.C.; Winer, E.P.; Weeks, J.C. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the National Comprehensive Cancer Network. Cancer 2012, 118, 5463–5472. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into molecular classifications of triple-negative breast cancer: Improving patient selection for treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef]
- Lin, N.U.; Claus, E.; Sohl, J.; Razzak, A.R.; Arnaout, A.; Winer, E.P. Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: High incidence of central nervous system metastases. Cancer 2008, 113, 2638–2645. [Google Scholar] [CrossRef]
- Kim, C.; Gao, R.; Sei, E.; Brandt, R.; Hartman, J.; Hatschek, T.; Crosetto, N.; Foukakis, T.; Navin, N.E. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 2018, 173, 879–893. [Google Scholar] [CrossRef]
- Tomasetto, C.; Régnier, C.; Moog-Lutz, C.; Mattei, M.G.; Chenard, M.P.; Lidereau, R.; Basset, P.; Rio, M.C. Identification of Four Novel Human Genes Amplified and Overexpressed in Breast Carcinoma and Localized to the q11–q21.3 Region of Chromosome 17. Genomics 1995, 28, 367–376. [Google Scholar] [CrossRef]
- Grunewald, T.G.P.; Kammerer, U.; Schulze, E.; Schindler, D.; Honig, A.; Zimmer, M.; Butt, E. Silencing of LASP-1 influences zyxin localization, inhibits proliferation and reduces migration in breast cancer cells. Exp. Cell Res. 2006, 312, 974–982. [Google Scholar] [CrossRef]
- Duvall-Noelle, N.; Karwandyar, A.; Richmond, A.; Raman, D. LASP-1: A nuclear hub for the UHRF1-DNMT1-G9a-Snail1 complex. Oncogene 2016, 35, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Butt, E.; Raman, D. New Frontiers for the Cytoskeletal Protein LASP1. Front Oncol. 2018, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Park, Z.Y.; Lin, D.; Brahmbhatt, A.A.; Rio, M.C.; Yates, J.R., III; Klemke, R.L. Regulation of cell migration and survival by focal adhesion targeting of Lasp-1. J. Cell Biol. 2004, 165, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Mihlan, S.; Reiß, C.; Thalheimer, P.; Herterich, S.; Gaetzner, S.; Kremerskothen, J.; Pavenstädt, H.J.; Lewandrowski, U.; Sickmann, A.; Butt, E. Nuclear import of LASP-1 is regulated by phosphorylation and dynamic protein–protein interactions. Oncogene 2013, 32, 2107–2113. [Google Scholar] [CrossRef]
- Li, B.; Zhuang, L.; Trueb, B. Zyxin Interacts with the SH3 Domains of the Cytoskeletal Proteins LIM-nebulette and Lasp-1. J. Biol. Chem. 2004, 279, 20401–20410. [Google Scholar] [CrossRef]
- Raman, D.; Sai, J.; Neel, N.F.; Chew, C.S.; Richmond, A. LIM and SH3 protein-1 modulates CXCR2-mediated cell migration. PLoS ONE 2010, 5, e10050. [Google Scholar] [CrossRef]
- Salvi, A.; Bongarzone, I.; Ferrari, L.; Abeni, E.; Arici, B.; De Bortoli, M.; Scuri, S.; Bonini, D.; Grossi, I.; Benetti, A.; et al. Molecular characterization of LASP-1 expression reveals vimentin as its new partner in human hepatocellular carcinoma cells. Int. J. Oncol. 2015, 46, 1901–1912. [Google Scholar] [CrossRef]
- Tomasetto, C.; Moog-Lutz, C.; Régnier, C.H.; Schreiber, V.; Basset, P.; Rio, M.-C. Lasp-1 (MLN 50) defines a new LIM protein subfamily characterized by the association of LIM and SH3 domains. Febs Lett. 1995, 373, 245–249. [Google Scholar] [CrossRef]
- Schreiber, V.; Moog-Lutz, C.; Régnier, C.H.; Chenard, M.P.; Boeuf, H.; Vonesch, J.L.; Tomasetto, C.; Rio, M.C. Lasp-1, a novel type of actin-binding protein accumulating in cell membrane extensions. Mol. Med. 1998, 4, 675–687. [Google Scholar] [CrossRef]
- Schreiber, V.; Masson, R.; Linares, J.L.; Mattei, M.G.; Tomasetto, C.; Rio, M.C. Chromosomal assignment and expression pattern of the murine Lasp-1 gene. Gene 1998, 207, 171–175. [Google Scholar] [CrossRef]
- Bach, I. The LIM domain: Regulation by association. Mech. Dev. 2000, 91, 5–17. [Google Scholar] [CrossRef]
- Pawson, T.; Schlessingert, J. SH2 and SH3 domains. Curr. Biol. 1993, 3, 434–442. [Google Scholar] [CrossRef]
- Butt, E.; Gambaryan, S.; Göttfert, N.; Galler, A.; Marcus, K.; Meyer, H.E. Actin Binding of Human LIM and SH3 Protein Is Regulated by cGMP- and cAMP-dependent Protein Kinase Phosphorylation on Serine 146. J. Biol. Chem. 2003, 278, 15601–15607. [Google Scholar] [CrossRef]
- Traenka, J.; Hauck, C.R.; Lewandrowski, U.; Sickmann, A.; Gambaryan, S.; Thalheimer, P.; Butt, E. Integrin-dependent translocation of LASP-1 to the cytoskeleton of activated platelets correlates with LASP-1 phosphorylation at tyrosine 171 by Src-kinase. Thromb. Haemost. 2009, 102, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Frietsch, J.; Grunewald, T.; Jasper, S.; Kammerer, U.; Herterich, S.; Kapp, M.; Honig, A.; Butt, E. Nuclear localisation of LASP-1 correlates with poor long-term survival in female breast cancer. Br. J. Cancer 2010, 102, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Butt, E.; Stempfle, K.; Lister, L.; Wolf, F.; Kraft, M.; Herrmann, B.A.; Viciano, P.C.; Weber, C.; Hochhaus, A.; Ernst, T.; et al. Phosphorylation-Dependent Differences in CXCR4-LASP1-AKT1 Interaction between Breast Cancer and Chronic Myeloid Leukemia. Cells 2020, 9, 444. [Google Scholar] [CrossRef]
- Merino, J.J.; Bellver-Landete, V.; Oset-Gasque, M.J.; Cubelos, B. CXCR4/CXCR7 molecular involvement in neuronal and neural progenitor migration: Focus in CNS repair. J. Cell. Physiol. 2015, 230, 27–42. [Google Scholar] [CrossRef]
- Shimizu, S.; Brown, M.; Sengupta, R.; Penfold, M.E.; Meucci, O. CXCR7 protein expression in human adult brain and differentiated neurons. PLoS ONE 2011, 6, e20680. [Google Scholar] [CrossRef]
- Nie, Y.; Han, Y.C.; Zou, Y.R. CXCR4 is required for the quiescence of primitive hematopoietic cells. J. Exp. Med. 2008, 205, 777–783. [Google Scholar] [CrossRef]
- Bleul, C.C.; Farzan, M.; Choe, H.; Parolin, C.; Clark-Lewis, I.; Sodroski, J.; Springer, T.A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 1996, 382, 829–833. [Google Scholar] [CrossRef]
- Raman, D.; Sobolik-Delmaire, T.; Richmond, A. Chemokines in health and disease. Exp. Cell Res. 2011, 317, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Teicher, B.A.; Fricker, S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 2010, 16, 2927–2931. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cheng, G.; Hao, M.; Zheng, J.; Zhou, X.; Zhang, J.; Taichman, R.S.; Pienta, K.J.; Wang, J. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010, 29, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Allinen, M.; Beroukhim, R.; Cai, L.; Brennan, C.; Lahti-Domenici, J.; Huang, H.; Porter, D.; Hu, M.; Chin, L.; Richardson, A.; et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 2004, 6, 17–32. [Google Scholar] [CrossRef]
- Nakanishi, K. Anatomy of RISC: How do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip. Rev. RNA 2016, 7, 637–660. [Google Scholar] [CrossRef]
- Rivas, F.V.; Tolia, N.H.; Song, J.J.; Aragon, J.P.; Liu, J.; Hannon, G.J.; Joshua-Tor, L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005, 12, 340–349. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1: DCP2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Behm-Ansmant, I.; Gatfield, D.; Izaurralde, E. A crucial role for GW182 and the DCP1: DCP2 decapping complex in miRNA-mediated gene silencing. RNA 2005, 11, 1640–1647. [Google Scholar] [CrossRef]
- Orban, T.I.; Izaurralde, E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 2005, 11, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Till, S.; Lejeune, E.; Thermann, R.; Bortfeld, M.; Hothorn, M.; Enderle, D.; Heinrich, C.; Hentze, M.W.; Ladurner, A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007, 14, 897–903. [Google Scholar] [CrossRef]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell 2011, 44, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Zekri, L.; Huntzinger, E.; Heimstädt, S.; Izaurralde, E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 2009, 29, 6220–6231. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Hatse, S.; Princen, K.; De Clercq, E.; Rosenkilde, M.M.; Schwartz, T.W.; Hernandez-Abad, P.E.; Skerlj, R.T.; Bridger, G.J.; Schols, D. AMD3465, a monomacrocyclic CXCR4 antagonist and potent HIV entry inhibitor. Biochem. Pharmacol. 2005, 70, 752–761. [Google Scholar] [CrossRef]
- Rosenkilde, M.M.; Gerlach, L.O.; Hatse, S.; Skerlj, R.T.; Schols, D.; Bridger, G.J.; Schwartz, T.W. Molecular mechanism of action of monocyclam versus bicyclam non-peptide antagonists in the CXCR4 chemokine receptor. J. Biol. Chem. 2007, 282, 27354–27365. [Google Scholar] [CrossRef]
- Bodart, V.; Anastassov, V.; Darkes, M.C.; Idzan, S.R.; Labrecque, J.; Lau, G.; Mosi, R.M.; Neff, K.S.; Nelson, K.L.; Ruzek, M.C. Pharmacology of AMD3465: A small molecule antagonist of the chemokine receptor CXCR4. Biochem. Pharmacol. 2009, 78, 993–1000. [Google Scholar] [CrossRef]
- Ling, X.; Spaeth, E.; Chen, Y.; Shi, Y.; Zhang, W.; Schober, W.; Hail, N., Jr.; Konopleva, M.; Andreeff, M. The CXCR4 antagonist AMD3465 regulates oncogenic signaling and invasiveness in vitro and prevents breast cancer growth and metastasis in vivo. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Feuerstein, R.; Wang, X.; Song, D.; Cooke, N.E.; Liebhaber, S.A. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc. Natl. Acad. Sci. USA 1994, 91, 10655–10659. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Soosairajah, J.; Harari, D.; Citri, A.; Price, J.; Ng, H.L.; Morton, C.J.; Parker, M.W.; Yarden, Y.; Bernard, O. Hsp90 increases LIM kinase activity by promoting its homo-dimerization. Faseb J. 2006, 20, 1218–1220. [Google Scholar] [CrossRef] [PubMed]
- Matthews, J.M.; Sunde, M. Dimers, Oligomers, Everywhere. In Protein Dimerization and Oligomerization in Biology; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–18. [Google Scholar]
- Keicher, C.; Gambaryan, S.; Schulze, E.; Marcus, K.; Meyer, H.E.; Butt, E. Phosphorylation of mouse LASP-1 on threonine 156 by cAMP-and cGMP-dependent protein kinase. Biochem. Biophys. Res. Commun. 2004, 324, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Orth, M.F.; Cazes, A.; Butt, E.; Grunewald, T.G. An update on the LIM and SH3 domain protein 1 (LASP1): A versatile structural, signaling, and biomarker protein. Oncotarget 2015, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Howard, C.M.; Bearss, N.; Subramaniyan, B.; Tilley, A.; Sridharan, S.; Villa, N.; Fraser, C.S.; Raman, D. The CXCR4-LASP1-eIF4F Axis Promotes Translation of Oncogenic Proteins in Triple-Negative Breast Cancer Cells. Front Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Subramaniyan, B.; Sridharan, S.; C, M.H.; A, M.C.T.; Basuroy, T.; de la Serna, I.; Butt, E.; Raman, D. Role of the CXCR4-LASP1 Axis in the Stabilization of Snail1 in Triple-Negative Breast Cancer. Cancers (Basel) 2020, 12, 2372. [Google Scholar] [CrossRef]
- Bridge, K.S.; Shah, K.M.; Li, Y.; Foxler, D.E.; Wong, S.C.; Miller, D.C.; Davidson, K.M.; Foster, J.G.; Rose, R.; Hodgkinson, M.R. Argonaute utilization for miRNA silencing is determined by phosphorylation-dependent recruitment of LIM-domain-containing proteins. Cell Rep. 2017, 20, 173–187. [Google Scholar] [CrossRef]
- Kim, S.J.; Shin, J.Y.; Lee, K.D.; Bae, Y.K.; Sung, K.W.; Nam, S.J.; Chun, K.H. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of CC chemokine receptor type 7. Breast Cancer Res. 2012, 14, R14. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Feng, J.; Cui, X.; Huang, W.; Li, Y.; Su, F.; Liu, Q.; Zhu, J.; Lv, X. Lin28 induces epithelial-to-mesenchymal transition and stemness via downregulation of let-7a in breast cancer cells. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, C.; Li, T.; Ding, Y.; Tu, T.; Zhou, F.; Qi, W.; Chen, H.; Sun, X. Let-7a inhibits growth and migration of breast cancer cells by targeting HMGA1. Int. J. Oncol. 2015, 46, 2526–2534. [Google Scholar] [CrossRef]
- Chen, D.; Sun, Y.; Yuan, Y.; Han, Z.; Zhang, P.; Zhang, J.; You, M.J.; Teruya-Feldstein, J.; Wang, M.; Gupta, S. miR-100 induces epithelial-mesenchymal transition but suppresses tumorigenesis, migration and invasion. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Gebeshuber, C.; Martinez, J. miR-100 suppresses IGF2 and inhibits breast tumorigenesis by interfering with proliferation and survival signaling. Oncogene 2013, 32, 3306–3310. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; He, M.; Guan, S.; Ma, M.; Wu, H.; Yu, Z.; Jiang, L.; Wang, Y.; Zong, X.; Jin, F. MicroRNA-100 suppresses the migration and invasion of breast cancer cells by targeting FZD-8 and inhibiting Wnt/β-catenin signaling pathway. Tumor Biol. 2016, 37, 5001–5011. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. RNA 2008, 14, 1012–1017. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M.; Network, C.G.A.R. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Cabioglu, N.; Yazici, M.S.; Arun, B.; Broglio, K.R.; Hortobagyi, G.N.; Price, J.E.; Sahin, A. CCR7 and CXCR4 as novel biomarkers predicting axillary lymph node metastasis in T1 breast cancer. Clin. Cancer Res. 2005, 11, 5686–5693. [Google Scholar] [CrossRef]
- Kochetkova, M.; Kumar, S.; McColl, S. Chemokine receptors CXCR4 and CCR7 promote metastasis by preventing anoikis in cancer cells. Cell Death Differ. 2009, 16, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Emmrich, S.; Engeland, F.; El-Khatib, M.; Henke, K.; Obulkasim, A.; Schöning, J.; Katsman-Kuipers, J.; Zwaan, C.M.; Pich, A.; Stary, J. miR-139-5p controls translation in myeloid leukemia through EIF4G2. Oncogene 2016, 35, 1822–1831. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Li, Y.S.; Xiao, W.F.; Deng, Z.H.; He, H.B.; Liu, Q.; Luo, W. MicroRNA-379 inhibits the proliferation, migration and invasion of human osteosarcoma cells by targetting EIF4G2. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Li, J.P.; Liao, X.H.; Xiang, Y.; Yao, A.; Song, R.H.; Zhang, Z.J.; Huang, F.; Dai, Z.T.; Zhang, T.C. Hyperoside and let-7a-5p synergistically inhibits lung cancer cell proliferation via inducing G1/S phase arrest. Gene 2018, 679, 232–240. [Google Scholar] [CrossRef]
- Bostner, J.; Waltersson, M.A.; Fornander, T.; Skoog, L.; Nordenskjöld, B.; Stål, O. Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer. Oncogene 2007, 26, 6997–7005. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Feber, A.; Xi, L.; Pennathur, A.; Wu, M.; Luketich, J.D.; Godfrey, T.E. Association between CCND1 G/A870 polymorphism, allele-specific amplification, cyclin D1 expression, and survival in esophageal and lung carcinoma. Clin. Cancer Res. 2008, 14, 7804–7812. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, Z.; Hao, Q.; Li, W.; Xu, Y.; Zhang, J.; Zhang, W.; Wang, S.; Liu, S.; Li, M. Loss of ERα induces amoeboid-like migration of breast cancer cells by downregulating vinculin. Nat. Commun. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Carey, S.P.; Kraning-Rush, C.M.; Goldblatt, Z.E.; Bordeleau, F.; Lampi, M.C.; Lin, D.Y.; García, A.J.; Reinhart-King, C.A. Vinculin regulates directionality and cell polarity in two-and three-dimensional matrix and three-dimensional microtrack migration. Mol. Biol. Cell 2016, 27, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhao, R.; He, Y.; Fu, X.; Fu, L.; Zhu, Z.; Fu, L.; Dong, J.T. Micro RNA 100 sensitizes luminal A breast cancer cells to paclitaxel treatment in part by targeting mTOR. Oncotarget 2016, 7, 5702. [Google Scholar] [CrossRef]
- Sun, J.; Chen, Z.; Tan, X.; Zhou, F.; Tan, F.; Gao, Y.; Sun, N.; Xu, X.; Shao, K.; He, J. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Med Oncol. 2013, 30, 411. [Google Scholar] [CrossRef]
- Becker, W.R.; Ober-Reynolds, B.; Jouravleva, K.; Jolly, S.M.; Zamore, P.D.; Greenleaf, W.J. High-throughput analysis reveals rules for target RNA binding and cleavage by AGO2. Mol. Cell 2019, 75, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.J.; Hao, H.J.; Ding, Y.H.; Wen, H.; Li, X.F.; Wang, Q.R.; Zhang, B.B. Suppression of EIF 4G2 by miR-379 potentiates the cisplatin chemosensitivity in nonsmall cell lung cancer cells. FEBS Lett. 2017, 591, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Jannie, K.M.; Ellerbroek, S.M.; Zhou, D.W.; Chen, S.; Crompton, D.J.; García, A.J.; DeMali, K.A. Vinculin-dependent actin bundling regulates cell migration and traction forces. Biochem. J. 2015, 465, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, K.T.; Li, L.; Chu, Y.; Janowski, B.A.; Corey, D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014, 6, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Janowski, B.A.; Huffman, K.E.; Schwartz, J.C.; Ram, R.; Nordsell, R.; Shames, D.S.; Minna, J.D.; Corey, D.R. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006, 13, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Younger, S.T.; Corey, D.R. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011, 39, 5682–5691. [Google Scholar] [CrossRef]
- Moshkovich, N.; Nisha, P.; Boyle, P.J.; Thompson, B.A.; Dale, R.K.; Lei, E.P. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011, 25, 1686–1701. [Google Scholar] [CrossRef]
- Ameyar-Zazoua, M.; Rachez, C.; Souidi, M.; Robin, P.; Fritsch, L.; Young, R.; Morozova, N.; Fenouil, R.; Descostes, N.; Andrau, J.C. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012, 19, 998. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.W.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef]
- Diederichs, S.; Haber, D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007, 131, 1097–1108. [Google Scholar] [CrossRef]
- Raman, D.; Neel, N.F.; Sai, J.; Mernaugh, R.L.; Ham, A.J.L.; Richmond, A.J. Characterization of chemokine receptor CXCR2 interacting proteins using a proteomics approach to define the CXCR2 “chemosynapse”. Methods Enzymol. 2009, 460, 315–330. [Google Scholar] [PubMed]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Prism, G. Prism 8 for windows. GraphPad Software Inc. 2019. Available online: https://www.graphpad.com/scientific-software/prism/ (accessed on 29 August 2020).
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tilley, A.M.C.; Howard, C.M.; Sridharan, S.; Subramaniyan, B.; Bearss, N.R.; Alkhalili, S.; Raman, D. The CXCR4-Dependent LASP1-Ago2 Interaction in Triple-Negative Breast Cancer. Cancers 2020, 12, 2455. https://doi.org/10.3390/cancers12092455
Tilley AMC, Howard CM, Sridharan S, Subramaniyan B, Bearss NR, Alkhalili S, Raman D. The CXCR4-Dependent LASP1-Ago2 Interaction in Triple-Negative Breast Cancer. Cancers. 2020; 12(9):2455. https://doi.org/10.3390/cancers12092455
Chicago/Turabian StyleTilley, Augustus M. C., Cory M. Howard, Sangita Sridharan, Boopathi Subramaniyan, Nicole R. Bearss, Sawsan Alkhalili, and Dayanidhi Raman. 2020. "The CXCR4-Dependent LASP1-Ago2 Interaction in Triple-Negative Breast Cancer" Cancers 12, no. 9: 2455. https://doi.org/10.3390/cancers12092455
APA StyleTilley, A. M. C., Howard, C. M., Sridharan, S., Subramaniyan, B., Bearss, N. R., Alkhalili, S., & Raman, D. (2020). The CXCR4-Dependent LASP1-Ago2 Interaction in Triple-Negative Breast Cancer. Cancers, 12(9), 2455. https://doi.org/10.3390/cancers12092455





