Marker Identification of the Grade of Dysplasia of Intraductal Papillary Mucinous Neoplasm in Pancreatic Cyst Fluid by Quantitative Proteomic Profiling
Abstract
1. Introduction
2. Results
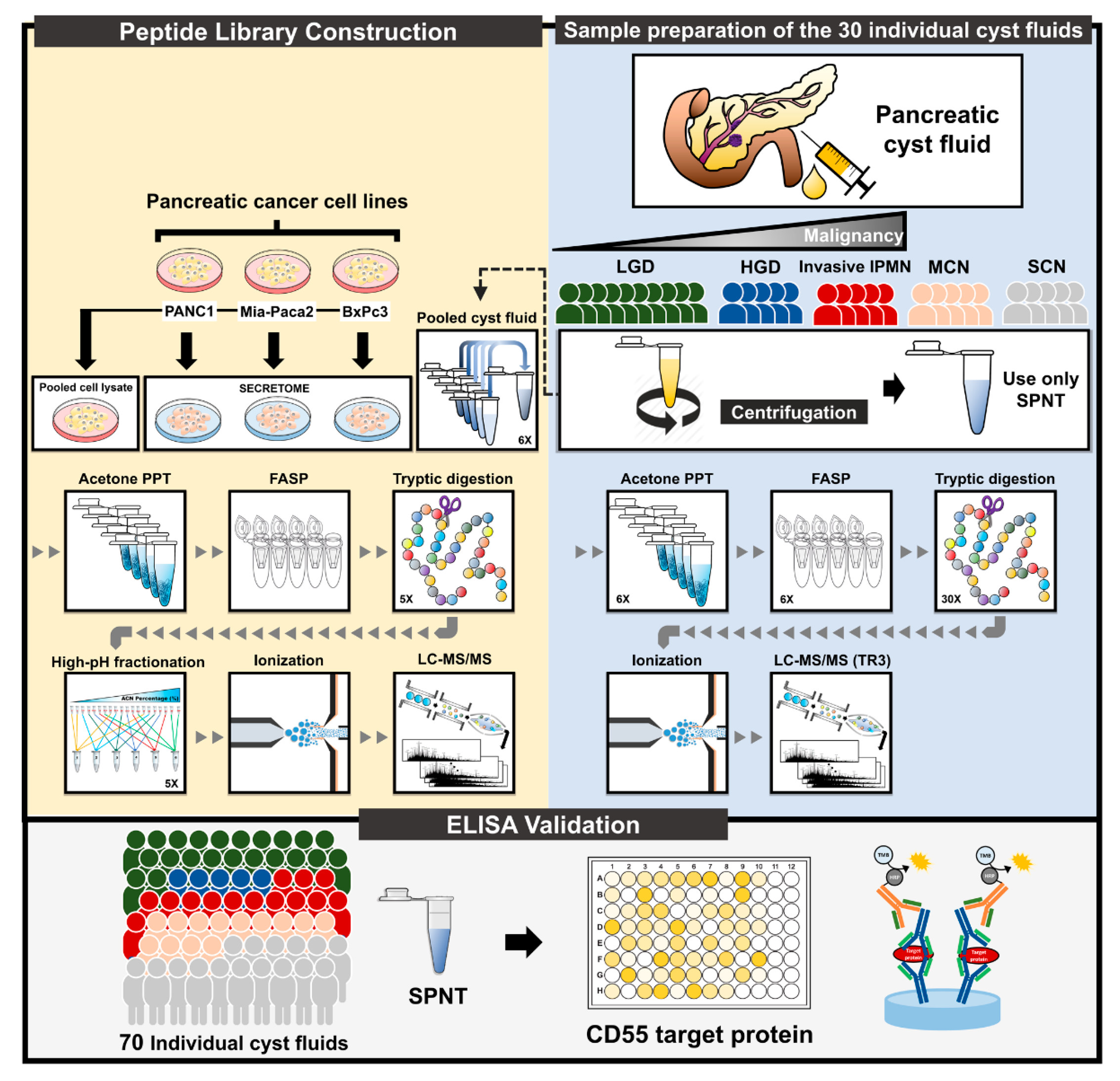
2.1. Results of Proteomic and Bioinformatic Analyses
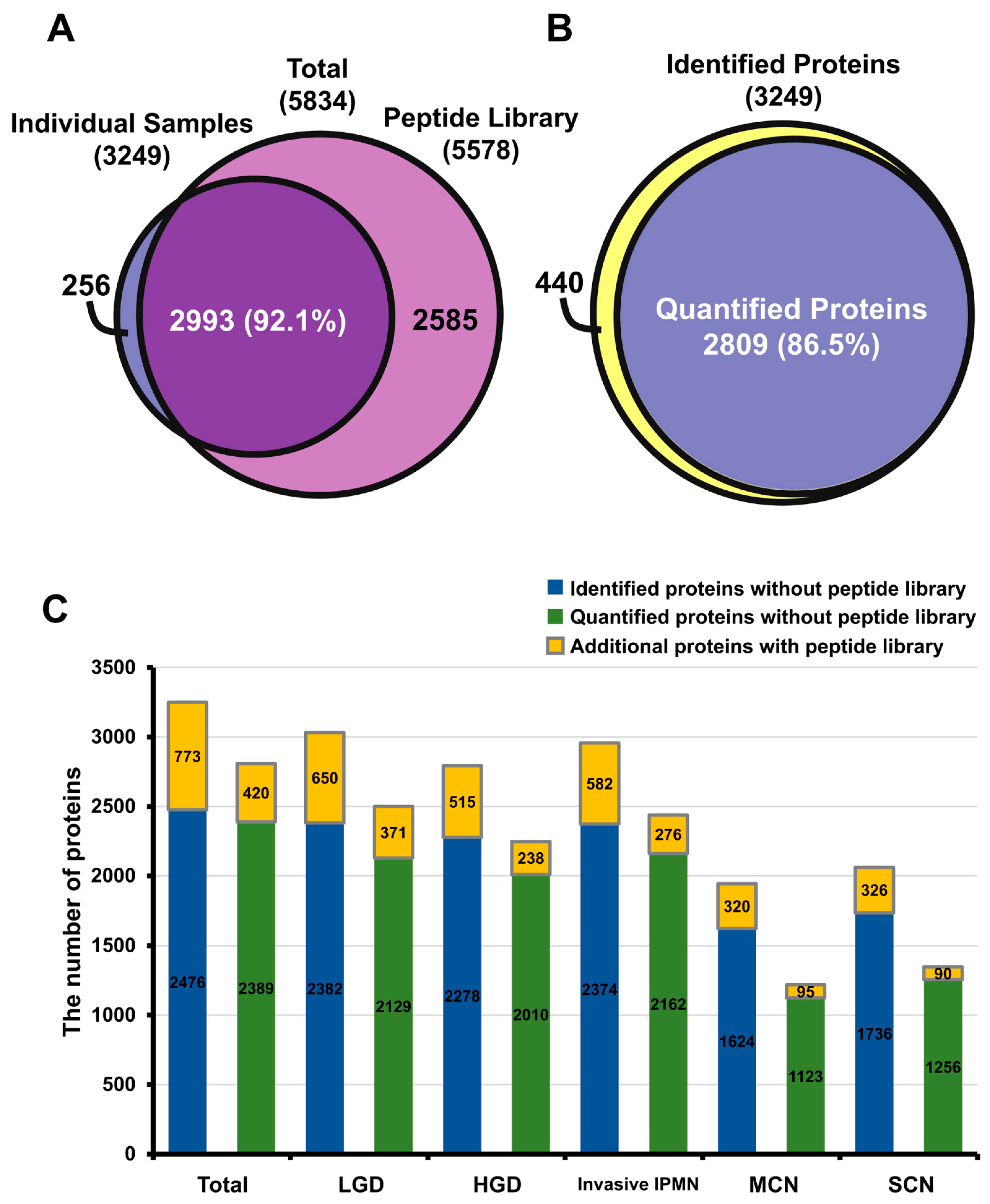
2.1.1. In-Depth Quantitative Proteomics of Pancreatic Cyst Fluid
2.1.2. Reproducibility of Data and Comparison with Other Proteome Databases
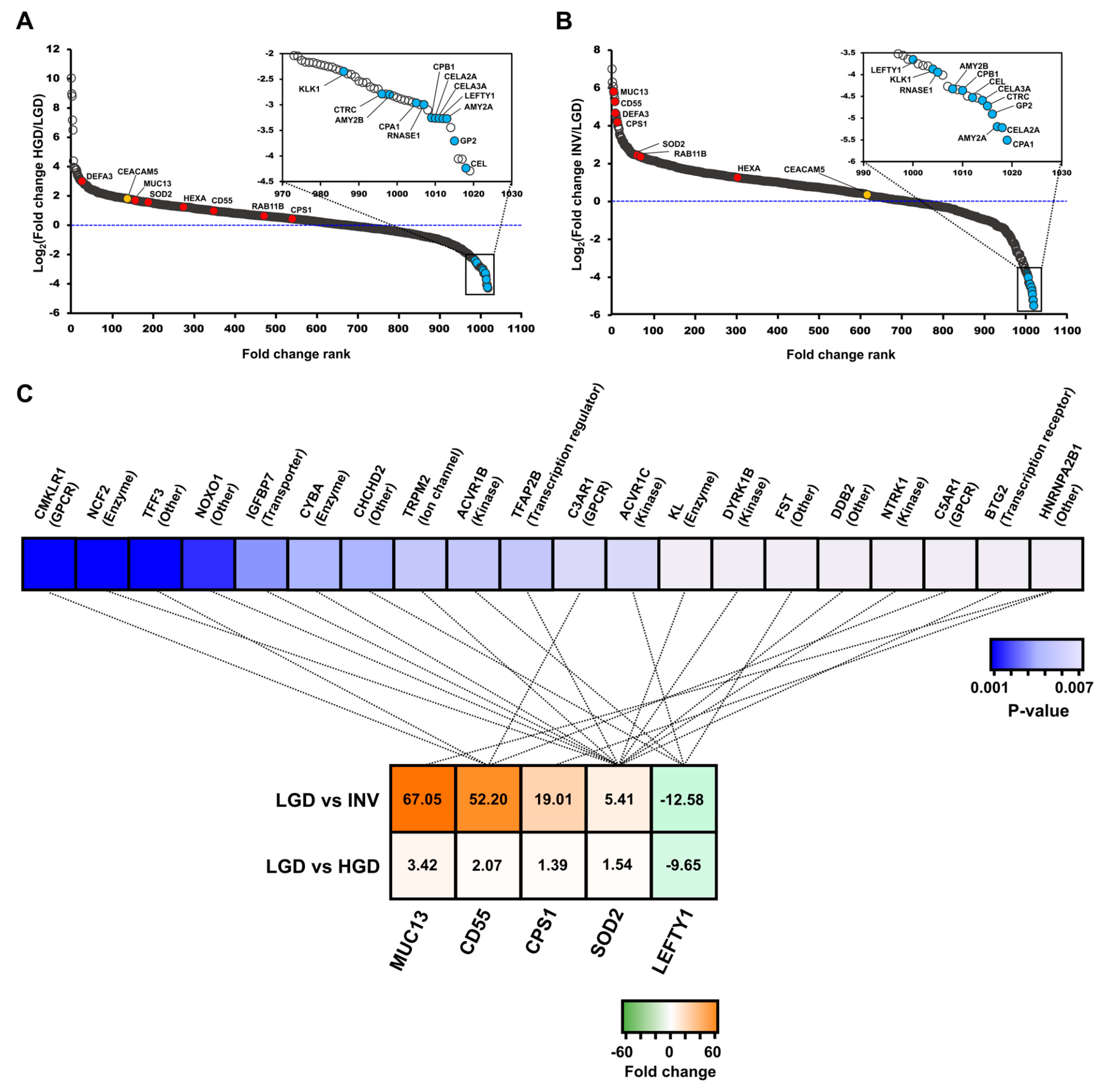
2.2. Discoverying Biomarker Candidates of IPMN Dysplasia
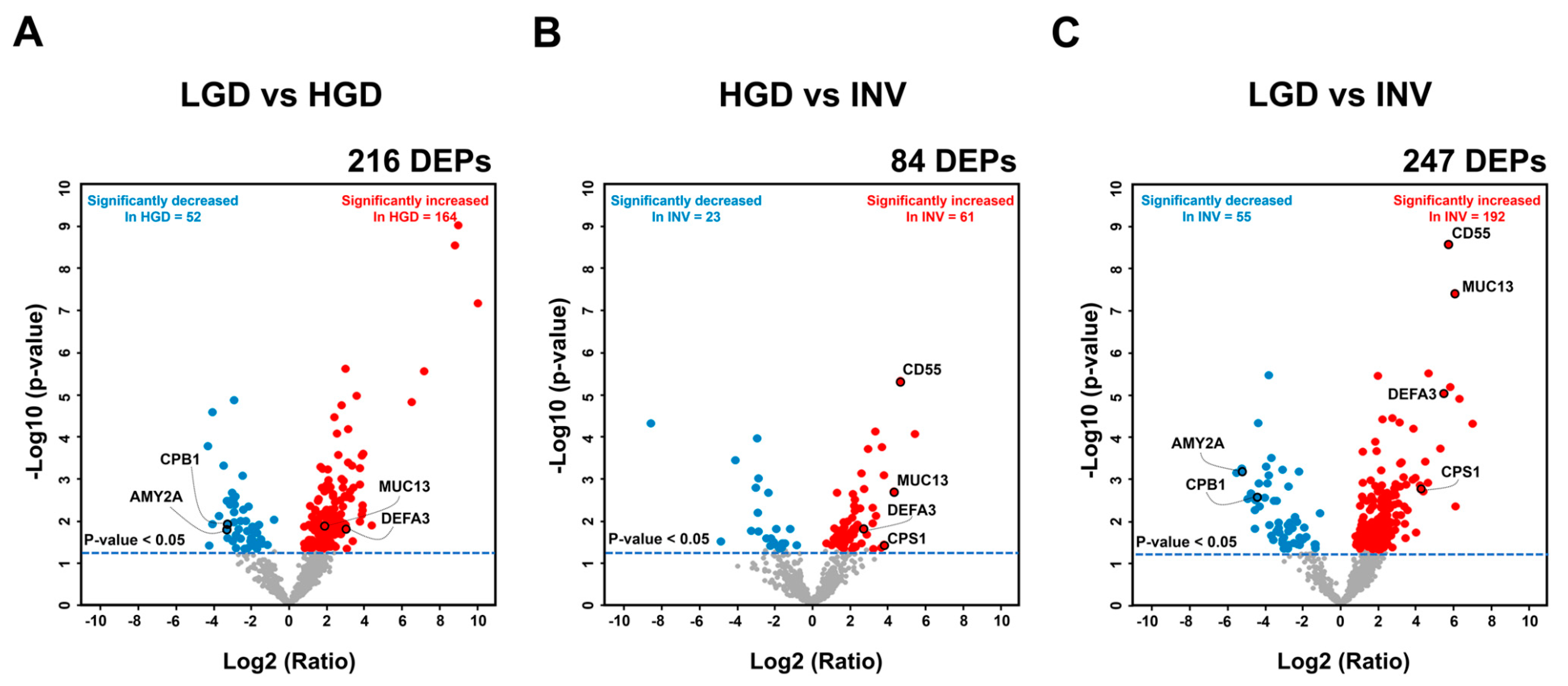
2.2.1. Differentially Expressed Proteins between IPMN Dysplasia
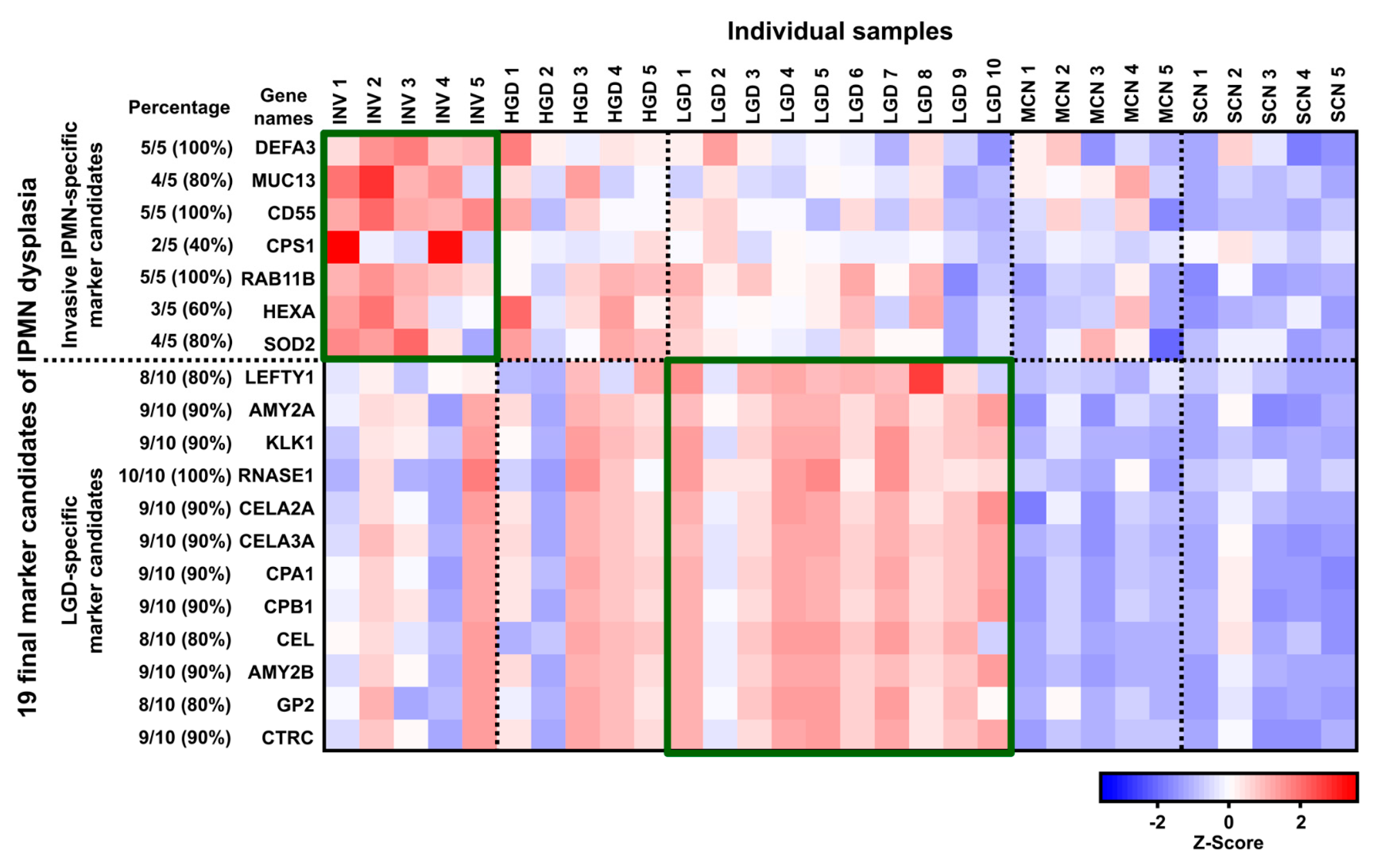
2.2.2. Biomarker Candidates of IPMN Dysplasia
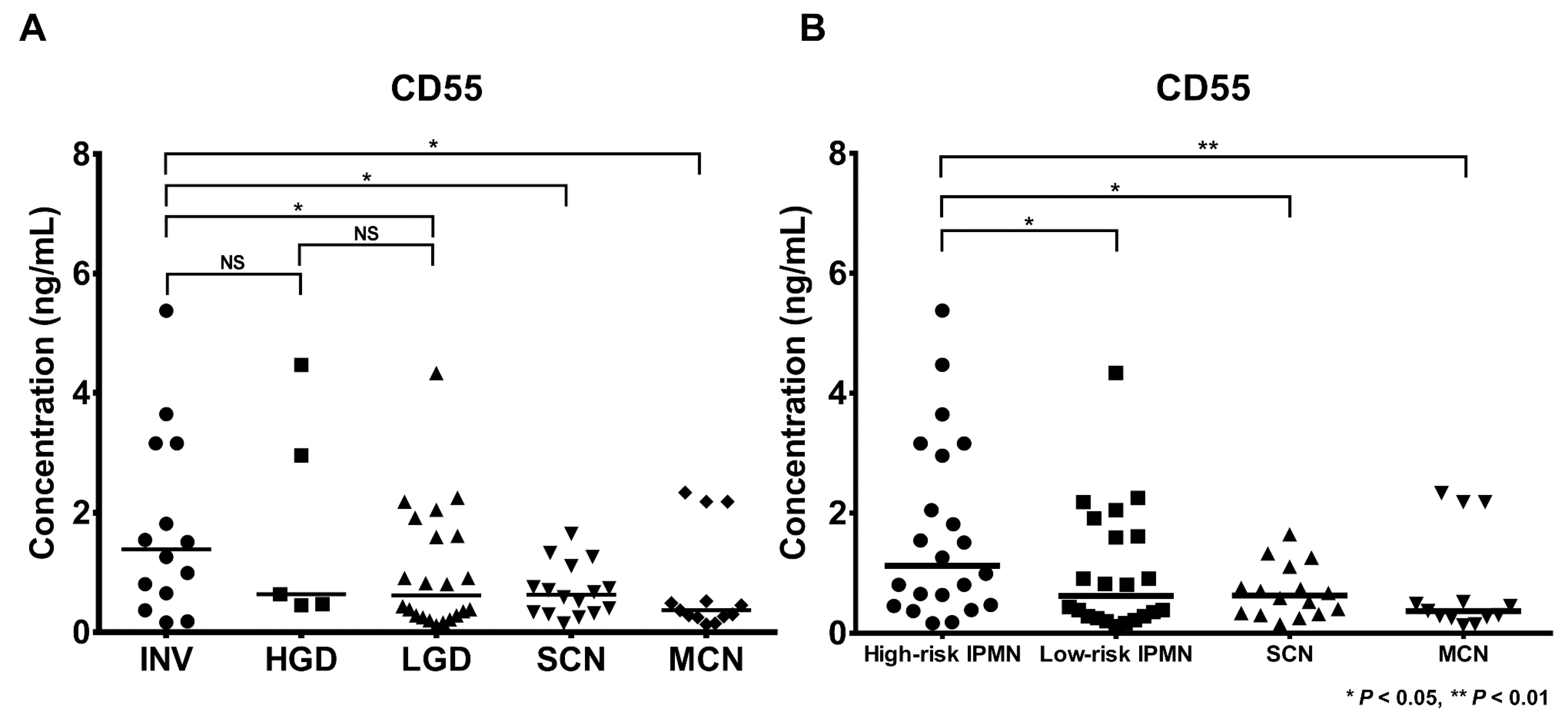
2.3. Determination of CD55 Levels by Antibody-Based Methods
2.3.1. Determination of CD55 by Enzyme-Linked Immunosorbent Assay (ELISA) and Western Blot
2.3.2. Immunohistochemistry (IHC) of CD55 and Myeloperoxidase (MPO)
3. Discussion
4. Materials and Methods
4.1. Patients and Cyst Fluid Samples and Preparation
4.2. Pancreatic Cyst Fluid Sample Preparation
4.3. LC-MS/MS and Statistical Analysis
4.4. Bioinformatics Analysis
4.5. Enzyme-Linked Immunosorbent Assay (ELISA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PCL | pancreatic cystic lesion |
| MRI | magnetic resonance imaging |
| CT | computed tomography |
| EUS | endoscopic ultrasound |
| IPMN | intraductal papillary mucinous neoplasm |
| LGD | low-grade dysplasia |
| HGD | high-grade dysplasia |
| CEA | carcinoembryonic antigen |
| CA19-9 | carbohydrate antigen 19-9 |
| EUS-FNA | endoscopic ultrasound-guided fine needle aspiration |
| DEP | differentially expressed protein |
| MCN | mucinous cystic neoplasm |
| SCN | serous cystic neoplasm |
| IRB | Institutional Review Board |
| BCA | bicinchoninic acid |
| SDS | sodium dodecyl sulfate |
| DTT | dithiothreitol |
| FASP | filter-aided sample preparation |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| GO | gene ontology |
| PPD | Plasma Proteome Database |
| IPA | Ingenuity Pathway Analysis |
| ELISA | enzyme-linked immunosorbent assay |
| MUC5AC | mucin-5AC |
| MUC2 | mucin-2 |
| PNLIP | pancreatic triacylglycerol lipase |
| CPA1 | carboxypeptidase A1 |
| CPB1 | carboxypeptidase B |
| PRSS1 | trypsin-1 |
| PRSS2 | trypsin-2 |
| CV | coefficient of variation |
| DEFA3 | neutrophil defensin 3 |
| MUC13 | mucin-13 |
| CD55 | complement decay-accelerating factor |
| CPS1 | carbamoyl-phosphate synthase [ammonia], mitochondrial |
| RAB11B | ras-related protein Rab-11B |
| HEXA | beta-hexosaminidase |
| SOD2 | superoxide dismutase [Mn], mitochondrial |
| LEFTY1 | left-right determination factor 1 |
| AMY2A | pancreatic alpha-amylase |
| KLK1 | kallikrein-1 |
| RNASE1 | ribonuclease pancreatic |
| CELA2A | chymotrypsin-like elastase family member 2A |
| CELA3A | chymotrypsin-like elastase family member 3A |
| CEL | bile salt-activated lipase |
| AMY2B | alpha-amylase 2B |
| GP2 | pancreatic secretory granule membrane major glycoprotein GP2 |
| CTRC | chymotrypsin-C |
| EPR | enhanced permeability and retention |
| IGFBP | insulin-like growth factor-binding protein 7 |
| KL | Klotho |
| BTG2 | protein BTG2 |
| IHC | immunohistochemistry |
| MPO | myeloperoxidase |
| FFPE | formalin-fixed paraffin-embedded |
References
- Klibansky, D.A.; Reid-Lombardo, K.M.; Gordon, S.R.; Gardner, T.B. The clinical relevance of the increasing incidence of intraductal papillary mucinous neoplasm. Clin. Gastroenterol. Hepatol. 2012, 10, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, M.; Uchinami, H.; Watanabe, G.; Sato, T.; Shibata, S.; Kume, M.; Ishiyama, K.; Takahashi, S.; Hashimoto, M.; Yamamoto, Y. F-18 fluorodeoxyglucose positron emission tomography for differential diagnosis of pancreatic tumors. Springerplus 2015, 4, 154. [Google Scholar] [CrossRef] [PubMed]
- Carbognin, G.; Zamboni, G.; Pinali, L.; Chiara, E.D.; Girardi, V.; Salvia, R.; Mucelli, R.P. Branch duct IPMTs: Value of cross-sectional imaging in the assessment of biological behavior and follow-up. Abdom. Imaging 2006, 31, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Bassi, C.; Sarr, M.G.; Lillemoe, K.D.; Reber, H.A. Natural history of intraductal papillary mucinous neoplasms (IPMN): Current evidence and implications for management. J. Gastrointest. Surg. 2008, 12, 645–650. [Google Scholar] [CrossRef]
- Chang, Y.R.; Park, J.K.; Jang, J.Y.; Kwon, W.; Yoon, J.H.; Kim, S.W. Incidental pancreatic cystic neoplasms in an asymptomatic healthy population of 21,745 individuals: Large-scale, single-center cohort study. Medicine 2016, 95, e5535. [Google Scholar] [CrossRef]
- Basturk, O.; Hong, S.M.; Wood, L.D.; Adsay, N.V.; Albores-Saavedra, J.; Biankin, A.V.; Brosens, L.A.; Fukushima, N.; Goggins, M.; Hruban, R.H.; et al. A Revised Classification System and Recommendations From the Baltimore Consensus Meeting for Neoplastic Precursor Lesions in the Pancreas. Am. J. Surg. Pathol. 2015, 39, 1730–1741. [Google Scholar] [CrossRef]
- Tanaka, M.; Fernandez-del Castillo, C.; Adsay, V.; Chari, S.; Falconi, M.; Jang, J.Y.; Kimura, W.; Levy, P.; Pitman, M.B.; Schmidt, C.M.; et al. International consensus guidelines 2012 for the management of IPMN and MCN of the pancreas. Pancreatology 2012, 12, 183–197. [Google Scholar] [CrossRef]
- Kang, M.J.; Lee, K.B.; Jang, J.Y.; Kwon, W.; Park, J.W.; Chang, Y.R.; Kim, S.W. Disease spectrum of intraductal papillary mucinous neoplasm with an associated invasive carcinoma invasive IPMN versus pancreatic ductal adenocarcinoma-associated IPMN. Pancreas 2013, 42, 1267–1274. [Google Scholar] [CrossRef]
- Scheiman, J.M.; Hwang, J.H.; Moayyedi, P. American gastroenterological association technical review on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology 2015, 148, 824–848. [Google Scholar] [CrossRef]
- European Study Group on Cystic Tumours of the Pancreas. European evidence-based guidelines on pancreatic cystic neoplasms. Gut 2018, 67, 789–804. [Google Scholar] [CrossRef]
- Tanaka, M.; Fernandez-Del Castillo, C.; Kamisawa, T.; Jang, J.Y.; Levy, P.; Ohtsuka, T.; Salvia, R.; Shimizu, Y.; Tada, M.; Wolfgang, C.L. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology 2017, 17, 738–753. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.M.; Yin, S.; Siddiqui, A.A.; Salem, R.R.; Schrope, B.; Sethi, A.; Poneros, J.M.; Gress, F.G.; Genkinger, J.M.; Do, C.; et al. Comparison of the diagnostic accuracy of three current guidelines for the evaluation of asymptomatic pancreatic cystic neoplasms. Medicine 2017, 96, e7900. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Park, T.; Lee, S.; Kim, Y.; Lee, S.Y.; Kim, S.W.; Kim, S.C.; Song, K.B.; Yamamoto, M.; Hatori, T.; et al. Proposed Nomogram Predicting the Individual Risk of Malignancy in the Patients With Branch Duct Type Intraductal Papillary Mucinous Neoplasms of the Pancreas. Ann. Surg. 2017, 266, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, M.A.; Fernandez-Del Castillo, C.; Al Efishat, M.; Eaton, A.A.; Gonen, M.; Batts, R.; Pergolini, I.; Rezaee, N.; Lillemoe, K.D.; Ferrone, C.R.; et al. Development and Validation of a Multi-institutional Preoperative Nomogram for Predicting Grade of Dysplasia in Intraductal Papillary Mucinous Neoplasms (IPMNs) of the Pancreas: A Report from The Pancreatic Surgery Consortium. Ann. Surg. 2018, 267, 157–163. [Google Scholar] [CrossRef]
- Bassi, C.; Salvia, R.; Molinari, E.; Biasutti, C.; Falconi, M.; Pederzoli, P. Management of 100 consecutive cases of pancreatic serous cystadenoma: Wait for symptoms and see at imaging or vice versa? World J. Surg. 2003, 27, 319–323. [Google Scholar] [CrossRef]
- Buscaglia, J.M.; Giday, S.A.; Kantsevoy, S.V.; Jagannath, S.B.; Magno, P.; Wolfgang, C.L.; Daniels, J.A.; Canto, M.I.; Okolo Iii, P.I. Patient- and cyst-related factors for improved prediction of malignancy within cystic lesions of the pancreas. Pancreatology 2009, 9, 631–638. [Google Scholar] [CrossRef]
- Park, W.G.; Mascarenhas, R.; Palaez-Luna, M.; Smyrk, T.C.; O’Kane, D.; Clain, J.E.; Levy, M.J.; Pearson, R.K.; Petersen, B.T.; Topazian, M.D.; et al. Diagnostic performance of cyst fluid carcinoembryonic antigen and amylase in histologically confirmed pancreatic cysts. Pancreas 2011, 40, 42–45. [Google Scholar] [CrossRef]
- Frossard, J.L.; Amouyal, P.; Amouyal, G.; Palazzo, L.; Amaris, J.; Soldan, M.; Giostra, E.; Spahr, L.; Hadengue, A.; Fabre, M. Performance of endosonography-guided fine needle aspiration and biopsy in the diagnosis of pancreatic cystic lesions. Am. J. Gastroenterol. 2003, 98, 1516–1524. [Google Scholar] [CrossRef]
- Singhi, A.D.; Nikiforova, M.N.; Fasanella, K.E.; McGrath, K.M.; Pai, R.K.; Ohori, N.P.; Bartholow, T.L.; Brand, R.E.; Chennat, J.S.; Lu, X.; et al. Preoperative GNAS and KRAS testing in the diagnosis of pancreatic mucinous cysts. Clin. Cancer Res. 2014, 20, 4381–4389. [Google Scholar] [CrossRef]
- Wu, J.; Matthaei, H.; Maitra, A.; Dal Molin, M.; Wood, L.D.; Eshleman, J.R.; Goggins, M.; Canto, M.I.; Schulick, R.D.; Edil, B.H.; et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci. Transl. Med. 2011, 3, 92ra66. [Google Scholar] [CrossRef]
- Thornton, G.D.; McPhail, M.J.; Nayagam, S.; Hewitt, M.J.; Vlavianos, P.; Monahan, K.J. Endoscopic ultrasound guided fine needle aspiration for the diagnosis of pancreatic cystic neoplasms: A meta-analysis. Pancreatology 2013, 13, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Woolf, K.M.; Liang, H.; Sletten, Z.J.; Russell, D.K.; Bonfiglio, T.A.; Zhou, Z. False-negative rate of endoscopic ultrasound-guided fine-needle aspiration for pancreatic solid and cystic lesions with matched surgical resections as the gold standard: One institution’s experience. Cancer Cytopathol. 2013, 121, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Ivry, S.L.; Sharib, J.M.; Dominguez, D.A.; Roy, N.; Hatcher, S.E.; Yip-Schneider, M.T.; Schmidt, C.M.; Brand, R.E.; Park, W.G.; Hebrok, M.; et al. Global Protease Activity Profiling Provides Differential Diagnosis of Pancreatic Cysts. Clin. Cancer Res. 2017, 23, 4865–4874. [Google Scholar] [CrossRef] [PubMed]
- Hata, T.; Dal Molin, M.; Hong, S.M.; Tamura, K.; Suenaga, M.; Yu, J.; Sedogawa, H.; Weiss, M.J.; Wolfgang, C.L.; Lennon, A.M.; et al. Predicting the Grade of Dysplasia of Pancreatic Cystic Neoplasms Using Cyst Fluid DNA Methylation Markers. Clin. Cancer Res. 2017, 23, 3935–3944. [Google Scholar] [CrossRef]
- Hata, T.; Dal Molin, M.; Suenaga, M.; Yu, J.; Pittman, M.; Weiss, M.; Canto, M.I.; Wolfgang, C.; Lennon, A.M.; Hruban, R.H.; et al. Cyst Fluid Telomerase Activity Predicts the Histologic Grade of Cystic Neoplasms of the Pancreas. Clin. Cancer Res. 2016, 22, 5141–5151. [Google Scholar] [CrossRef]
- Park, J.; Yun, H.S.; Lee, K.H.; Lee, K.T.; Lee, J.K.; Lee, S.Y. Discovery and Validation of Biomarkers That Distinguish Mucinous and Nonmucinous Pancreatic Cysts. Cancer Res. 2015, 75, 3227–3235. [Google Scholar] [CrossRef]
- Kristjansdottir, B.; Partheen, K.; Fung, E.T.; Marcickiewicz, J.; Yip, C.; Brannstrom, M.; Sundfeldt, K. Ovarian cyst fluid is a rich proteome resource for detection of new tumor biomarkers. Clin. Proteom. 2012, 9, 14. [Google Scholar] [CrossRef]
- Talebian, M.; von Bartheld, M.B.; Braun, J.; Versteegh, M.I.; Dekkers, O.M.; Rabe, K.F.; Annema, J.T. EUS-FNA in the preoperative staging of non-small cell lung cancer. Lung Cancer 2010, 69, 60–65. [Google Scholar] [CrossRef]
- Tanase, C.; Albulescu, R.; Neagu, M. Proteomic Approaches for Biomarker Panels in Cancer. J. Immunoass. Immunoch. 2016, 37, 1–15. [Google Scholar] [CrossRef]
- Thomas, S.; Hao, L.; Ricke, W.A.; Li, L. Biomarker discovery in mass spectrometry-based urinary proteomics. Proteom. Clin. Appl. 2016, 10, 358–370. [Google Scholar] [CrossRef]
- Jabbar, K.S.; Arike, L.; Verbeke, C.S.; Sadik, R.; Hansson, G.C. Highly Accurate Identification of Cystic Precursor Lesions of Pancreatic Cancer Through Targeted Mass Spectrometry: A Phase IIc Diagnostic Study. J. Clin. Oncol. 2017, 36, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Do, M.; Han, D.; Wang, J.I.; Kim, H.; Kwon, W.; Han, Y.; Jang, J.Y.; Kim, Y. Quantitative proteomic analysis of pancreatic cyst fluid proteins associated with malignancy in intraductal papillary mucinous neoplasms. Clin. Proteom. 2018, 15, 17. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Wong, H.P.; Wang, X.; Deasy, J.O. A bioinformatics filtering strategy for identifying radiation response biomarker candidates. PLoS ONE 2012, 7, e38870. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef]
- Ke, E.; Patel, B.B.; Liu, T.; Li, X.M.; Haluszka, O.; Hoffman, J.P.; Ehya, H.; Young, N.A.; Watson, J.C.; Weinberg, D.S.; et al. Proteomic analyses of pancreatic cyst fluids. Pancreas 2009, 38, e33–e42. [Google Scholar] [CrossRef]
- Schmidt, A.; Aebersold, R. High-accuracy proteome maps of human body fluids. Genome Biol. 2006, 7, 242. [Google Scholar] [CrossRef]
- Yan-Sanders, Y.; Hammons, G.J.; Lyn-Cook, B.D. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett. 2002, 183, 215–220. [Google Scholar] [CrossRef]
- Su, G.H.; Bansal, R.; Murphy, K.M.; Montgomery, E.; Yeo, C.J.; Hruban, R.H.; Kern, S.E. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc. Natl. Acad. Sci. USA 2001, 98, 3254–3257. [Google Scholar] [CrossRef]
- Sakamoto, H.; Kimura, H.; Sekijima, M.; Matsumoto, K.; Arao, T.; Chikugo, T.; Yamada, Y.; Kitano, M.; Ito, A.; Takeyama, Y.; et al. Plasma concentrations of angiogenesis-related molecules in patients with pancreatic cancer. Jpn. J. Clin. Oncol. 2012, 42, 105–112. [Google Scholar] [CrossRef]
- Perumalsamy, S.; Aqilah Mohd Zin, N.A.; Widodo, R.T.; Wan Ahmad, W.A.; Vethakkan, S.; Huri, H.Z. Chemokine Like Receptor-1 (CMKLR-1) Receptor: A Potential Therapeutic Target in Management of Chemerin Induced Type 2 Diabetes Mellitus and Cancer. Curr. Pharm. Des. 2017, 23, 3689–3698. [Google Scholar] [CrossRef]
- Murphy, K.M.; Brune, K.A.; Griffin, C.; Sollenberger, J.E.; Petersen, G.M.; Bansal, R.; Hruban, R.H.; Kern, S.E. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: Deleterious BRCA2 mutations in 17%. Cancer Res. 2002, 62, 3789–3793. [Google Scholar]
- Liu, C.; Yang, Z.; Li, D.; Liu, Z.; Miao, X.; Yang, L.; Zou, Q.; Yuan, Y. Overexpression of B2M and loss of ALK7 expression are associated with invasion, metastasis, and poor-prognosis of the pancreatic ductal adenocarcinoma. Cancer Biomark. 2015, 15, 735–743. [Google Scholar] [CrossRef]
- Jin, K.; Park, S.; Ewton, D.Z.; Friedman, E. The survival kinase Mirk/Dyrk1B is a downstream effector of oncogenic K-ras in pancreatic cancer. Cancer Res. 2007, 67, 7247–7255. [Google Scholar] [CrossRef] [PubMed]
- Hantute-Ghesquier, A.; Haustrate, A.; Prevarskaya, N.; Lehen’kyi, V. TRPM Family Channels in Cancer. Pharmaceuticals 2018, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Guppy, N.J.; El-Bahrawy, M.E.; Kocher, H.M.; Fritsch, K.; Qureshi, Y.A.; Poulsom, R.; Jeffery, R.E.; Wright, N.A.; Otto, W.R.; Alison, M.R. Trefoil factor family peptides in normal and diseased human pancreas. Pancreas 2012, 41, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Frampton, A.E.; Castellano, L.; Colombo, T.; Giovannetti, E.; Krell, J.; Jacob, J.; Pellegrino, L.; Roca-Alonso, L.; Funel, N.; Gall, T.M.; et al. MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology 2014, 146, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Diao, D.M.; Zhang, H.; Song, Y.C.; Dang, C.X. Proliferation enhanced by NGF-NTRK1 signaling makes pancreatic cancer cells more sensitive to 2DG-induced apoptosis. Int. J. Med. Sci. 2013, 10, 634–640. [Google Scholar] [CrossRef][Green Version]
- Barcelo, C.; Etchin, J.; Mansour, M.R.; Sanda, T.; Ginesta, M.M.; Sanchez-Arevalo Lobo, V.J.; Real, F.X.; Capella, G.; Estanyol, J.M.; Jaumot, M.; et al. Ribonucleoprotein HNRNPA2B1 interacts with and regulates oncogenic KRAS in pancreatic ductal adenocarcinoma cells. Gastroenterology 2014, 147, 882–892. [Google Scholar] [CrossRef]
- An, W.; Ben, Q.W.; Chen, H.T.; Zheng, J.M.; Huang, L.; Li, G.X.; Li, Z.S. Low expression of IGFBP7 is associated with poor outcome of pancreatic ductal adenocarcinoma. Ann. Surg. Oncol. 2012, 19, 3971–3978. [Google Scholar] [CrossRef]
- Ajona, D.; Ortiz-Espinosa, S.; Pio, R. Complement anaphylatoxins C3a and C5a: Emerging roles in cancer progression and treatment. Semin. Cell Dev. Biol. 2017, 85, 153–163. [Google Scholar] [CrossRef]
- Abramovitz, L.; Rubinek, T.; Ligumsky, H.; Bose, S.; Barshack, I.; Avivi, C.; Kaufman, B.; Wolf, I. KL1 internal repeat mediates klotho tumor suppressor activities and inhibits bFGF and IGF-I signaling in pancreatic cancer. Clin. Cancer Res. 2011, 17, 4254–4266. [Google Scholar] [CrossRef] [PubMed]
- Furniss, R.C.D.; Low, W.W.; Mavridou, D.A.I.; Dagley, L.F.; Webb, A.I.; Tate, E.W.; Clements, A. Plasma membrane profiling during enterohemorrhagic E. coli infection reveals that the metalloprotease StcE cleaves CD55 from host epithelial surfaces. J. Biol. Chem. 2018, 293, 17188–17199. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, N.; Park, W.G. Systematic Review of Pancreatic Cyst Fluid Biomarkers: The Path Forward. Clin. Transl. Gastroenterol. 2015, 6, e88. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Gbormittah, F.O.; Haab, B.B.; Partyka, K.; Garcia-Ott, C.; Hancapie, M.; Hancock, W.S. Characterization of glycoproteins in pancreatic cyst fluid using a high-performance multiple lectin affinity chromatography platform. J. Proteome Res. 2014, 13, 289–299. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cuoghi, A.; Farina, A.; Z’Graggen, K.; Dumonceau, J.M.; Tomasi, A.; Hochstrasser, D.F.; Genevay, M.; Lescuyer, P.; Frossard, J.L. Role of proteomics to differentiate between benign and potentially malignant pancreatic cysts. J. Proteome Res. 2011, 10, 2664–2670. [Google Scholar] [CrossRef] [PubMed]
- Greish, K. Enhanced permeability and retention (EPR) effect for anticancer nanomedicine drug targeting. Methods Mol. Biol. 2010, 624, 25–37. [Google Scholar]
- Argentiero, A.; De Summa, S.; Di Fonte, R.; Iacobazzi, R.M.; Porcelli, L.; Da Via, M.; Brunetti, O.; Azzariti, A.; Silvestris, N.; Solimando, A.G. Gene Expression Comparison between the Lymph Node-Positive and -Negative Reveals a Peculiar Immune Microenvironment Signature and a Theranostic Role for WNT Targeting in Pancreatic Ductal Adenocarcinoma: A Pilot Study. Cancers 2019, 11, 942. [Google Scholar] [CrossRef]
- Porcelli, L.; Iacobazzi, R.M.; Di Fonte, R.; Serrati, S.; Intini, A.; Solimando, A.G.; Brunetti, O.; Calabrese, A.; Leonetti, F.; Azzariti, A.; et al. CAFs and TGF-beta Signaling Activation by Mast Cells Contribute to Resistance to Gemcitabine/Nabpaclitaxel in Pancreatic Cancer. Cancers 2019, 11, 330. [Google Scholar] [CrossRef]
- Hiraoka, N.; Yamazaki-Itoh, R.; Ino, Y.; Mizuguchi, Y.; Yamada, T.; Hirohashi, S.; Kanai, Y. CXCL17 and ICAM2 Are Associated With a Potential Anti-Tumor Immune Response in Early Intraepithelial Stages of Human Pancreatic Carcinogenesis. Gastroenterology 2011, 140, 310–321. [Google Scholar] [CrossRef]
- Nasu, J.; Mizuno, M.; Uesu, T.; Takeuchi, K.; Inaba, T.; Ohya, S.; Kawada, M.; Shimo, K.; Okada, H.; Fujita, T.; et al. Cytokine-stimulated release of decay-accelerating factor (DAF;CD55) from HT-29 human intestinal epithelial cells. Clin. Exp. Immunol. 1998, 113, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Reis, E.S.; Mastellos, D.C.; Ricklin, D.; Mantovani, A.; Lambris, J.D. Complement in cancer: Untangling an intricate relationship. Nat. Rev. Immunol. 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Nanjappa, V.; Thomas, J.K.; Marimuthu, A.; Muthusamy, B.; Radhakrishnan, A.; Sharma, R.; Ahmad Khan, A.; Balakrishnan, L.; Sahasrabuddhe, N.A.; Kumar, S.; et al. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2014, 42, D959–D965. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, B.; Hanumanthu, G.; Suresh, S.; Rekha, B.; Srinivas, D.; Karthick, L.; Vrushabendra, B.M.; Sharma, S.; Mishra, G.; Chatterjee, P.; et al. Plasma Proteome Database as a resource for proteomics research. Proteomics 2005, 5, 3531–3536. [Google Scholar] [CrossRef]
- Wolf, K.; Wu, Y.I.; Liu, Y.; Geiger, J.; Tam, E.; Overall, C.; Stack, M.S.; Friedl, P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 2007, 9, 893–904. [Google Scholar] [CrossRef]
- Hornebeck, W.; Emonard, H.; Monboisse, J.C.; Bellon, G. Matrix-directed regulation of pericellular proteolysis and tumor progression. Semin. Cancer Biol. 2002, 12, 231–241. [Google Scholar] [CrossRef]
- Afshar-Kharghan, V. The role of the complement system in cancer. J. Clin. Investig. 2017, 127, 780–789. [Google Scholar] [CrossRef]
- Falanga, A.; Marchetti, M.; Vignoli, A. Coagulation and cancer: Biological and clinical aspects. J. Thromb. Haemost. 2013, 11, 223–233. [Google Scholar] [CrossRef]
- Bjorge, L.; Hakulinen, J.; Vintermyr, O.K.; Jarva, H.; Jensen, T.S.; Iversen, O.E.; Meri, S. Ascitic complement system in ovarian cancer. Br. J. Cancer 2005, 92, 895–905. [Google Scholar] [CrossRef]
- Ytting, H.; Jensenius, J.C.; Christensen, I.J.; Thiel, S.; Nielsen, H.J. Increased activity of the mannan-binding lectin complement activation pathway in patients with colorectal cancer. Scand. J. Gastroenterol. 2004, 39, 674–679. [Google Scholar] [CrossRef]
- Stiles, Z.E.; Khan, S.; Patton, K.T.; Jaggi, M.; Behrman, S.W.; Chauhan, S.C. Transmembrane mucin MUC13 distinguishes intraductal papillary mucinous neoplasms from non-mucinous cysts and is associated with high-risk lesions. HPB 2018, 21, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Mito, K.; Saito, M.; Morita, K.; Maetani, I.; Sata, N.; Mieno, M.; Fukushima, N. Clinicopathological and prognostic significance of MUC13 and AGR2 expression in intraductal papillary mucinous neoplasms of the pancreas. Pancreatology 2018, 18, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Khan, S.; Gupta, S.C.; Kashyap, V.K.; Yallapu, M.M.; Chauhan, S.C.; Jaggi, M. MUC13 contributes to rewiring of glucose metabolism in pancreatic cancer. Oncogenesis 2018, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Zafar, N.; Khan, S.S.; Setua, S.; Behrman, S.W.; Stiles, Z.E.; Yallapu, M.M.; Sahay, P.; Ghimire, H.; Ise, T.; et al. Clinical significance of MUC13 in pancreatic ductal adenocarcinoma. HPB 2018, 20, 563–572. [Google Scholar] [CrossRef]
- Khan, S.; Sikander, M.; Ebeling, M.C.; Ganju, A.; Kumari, S.; Yallapu, M.M.; Hafeez, B.B.; Ise, T.; Nagata, S.; Zafar, N.; et al. MUC13 interaction with receptor tyrosine kinase HER2 drives pancreatic ductal adenocarcinoma progression. Oncogene 2017, 36, 491–500. [Google Scholar] [CrossRef]
- Wobus, M.; Vogel, B.; Schmucking, E.; Hamann, J.; Aust, G. N-glycosylation of CD97 within the EGF domains is crucial for epitope accessibility in normal and malignant cells as well as CD55 ligand binding. Int. J. Cancer 2004, 112, 815–822. [Google Scholar] [CrossRef]
- Karpus, O.N.; Veninga, H.; Hoek, R.M.; Flierman, D.; van Buul, J.D.; Vandenakker, C.C.; vanBavel, E.; Medof, M.E.; van Lier, R.A.; Reedquist, K.A.; et al. Shear stress-dependent downregulation of the adhesion-G protein-coupled receptor CD97 on circulating leukocytes upon contact with its ligand CD55. J. Immunol. 2013, 190, 3740–3748. [Google Scholar] [CrossRef]
- Iacobuzio-Donahue, C.A.; Maitra, A.; Olsen, M.; Lowe, A.W.; Van Heek, N.T.; Rosty, C.; Walter, K.; Sato, N.; Parker, A.; Ashfaq, R.; et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am. J. Pathol. 2003, 162, 1151–1162. [Google Scholar] [CrossRef]
- Lowe, A.W.; Olsen, M.; Hao, Y.; Lee, S.P.; Taek Lee, K.; Chen, X.; van de Rijn, M.; Brown, P.O. Gene expression patterns in pancreatic tumors, cells and tissues. PLoS ONE 2007, 2, e323. [Google Scholar] [CrossRef]
- ong, Y.; Wang, Q.; Wang, D.; Junqiang, L.; Yang, J.; Li, H.; Wang, X.; Jin, X.; Jing, R.; Yang, J.H.; et al. Label-Free Quantitative Proteomics Unravels Carboxypeptidases as the Novel Biomarker in Pancreatic Ductal Adenocarcinoma. Transl. Oncol. 2018, 11, 691–699. [Google Scholar]
- Mikesch, J.H.; Schier, K.; Roetger, A.; Simon, R.; Buerger, H.; Brandt, B. The expression and action of decay-accelerating factor (CD55) in human malignancies and cancer therapy. Cell. Oncol. 2006, 28, 223–232. [Google Scholar] [PubMed]
- Saygin, C.; Wiechert, A.; Rao, V.S.; Alluri, R.; Connor, E.; Thiagarajan, P.S.; Hale, J.S.; Li, Y.; Chumakova, A.; Jarrar, A.; et al. CD55 regulates self-renewal and cisplatin resistance in endometrioid tumors. J. Exp. Med. 2017, 214, 2715–2732. [Google Scholar] [CrossRef] [PubMed]
- Macor, P.; Tripodo, C.; Zorzet, S.; Piovan, E.; Bossi, F.; Marzari, R.; Amadori, A.; Tedesco, F. In vivo targeting of human neutralizing antibodies against CD55 and CD59 to lymphoma cells increases the antitumor activity of rituximab. Cancer Res. 2007, 67, 10556–10563. [Google Scholar] [CrossRef] [PubMed]
- Mamidi, S.; Cinci, M.; Hasmann, M.; Fehring, V.; Kirschfink, M. Lipoplex mediated silencing of membrane regulators (CD46, CD55 and CD59) enhances complement-dependent anti-tumor activity of trastuzumab and pertuzumab. Mol. Oncol. 2013, 7, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.H.; Choi, Y.B.; Kim, J.H.; Weihl, C.C.; Ju, J.S. Comparisons of ELISA and Western blot assays for detection of autophagy flux. Data Brief. 2017, 13, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Gaugaz, F.Z. Fast and sensitive total protein and Peptide assays for proteomic analysis. Anal. Chem. 2015, 87, 4110–4116. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Park, J.; Han, D.; Do, M.; Woo, J.; Wang, J.I.; Han, Y.; Kwon, W.; Kim, S.W.; Jang, J.Y.; Kim, Y. Proteome characterization of human pancreatic cyst fluid from intraductal papillary mucinous neoplasm by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2017, 31, 1761–1772. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Vizcaino, J.A.; Cote, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 41, D1063–D1069. [Google Scholar] [CrossRef] [PubMed]
- Vizcaino, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Rios, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Jin, J.; Woo, J.; Min, H.; Kim, Y. Proteomic analysis of mouse astrocytes and their secretome by a combination of FASP and StageTip-based, high pH, reversed-phase fractionation. Proteomics 2014, 14, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; Von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | Pancreatic Cyst Fluids | ||||
|---|---|---|---|---|---|
| Low-Risk IPMN | High-Risk IPMN | Other Cystic Lesions | |||
| Group | LGD | HGD | Invasive IPMN | MCN | SCN |
| (n = 10) | (n = 5) | (n = 5) | (n = 5) | (n = 5) | |
| Age (years) | |||||
| 65.80 ± 5.55 | 67.80 ± 9.88 | 50.80 ± 14.45 | 49.00 ± 11.60 | 51.60 ± 17.08 | |
| Gender | |||||
| Male | 5 | 4 | 3 | 1 | 1 |
| Female | 5 | 1 | 2 | 4 | 4 |
| Gland Type | |||||
| Gastric | 9 | 2 (1) | 2 (1) | ||
| Intestinal | 0 | 1 (1) | 1 | ||
| Oncocytic | 0 | 0 | 1 | ||
| Pancreatobiliary | 0 | 1 | 0 | ||
| Pancreatic | 0 | 0 | (1) | ||
| Unknown | 1 | 0 | 0 | ||
| Duct Type | |||||
| Main | 0 | 0 | 1 | ||
| Branch | 4 | 4 | 1 | ||
| Mixed | 4 | 1 | 3 | ||
| Unknown | 2 | 0 | 0 | ||
| Cyst Focality | |||||
| Single | 8 | 5 | 4 | 2 | 0 |
| Multiple | 2 | 0 | 1 | 0 | 0 |
| Unknown | 0 | 0 | 0 | 3 | 5 |
| Mural Nodule | |||||
| Y | 2 | 3 | 5 | 0 | 0 |
| N | 8 | 2 | 0 | 2 | 0 |
| Unknown | 0 | 0 | 0 | 3 | 5 |
| Cyst Location | |||||
| Head | 4 | 3 | 3 | 0 | 1 |
| Body/Tail | 6 | 2 | 2 | 5 | 4 |
| Mixed | 0 | 0 | 0 | 0 | 0 |
| CEA Concentration (mg/L) | 1.44 ± 0.74 | 1.52 ± 1.04 | 5.48 ± 6.82 | 1.32 ± 0.96 | 1.44 ± 0.48 |
| CA 19-9 Concentration (mg/L) | 11.86 ± 8.69 | 22.80 ± 29.77 | 90.28 ± 129.71 | 20.20 ± 30.35 | 19.76 ± 17.81 |
| Cyst Size | |||||
| Cyst Size | 3.36 ± 1.33 | 3.56 ± 1.66 | 5.74 ± 3.69 | 7.50 ± 2.18 | 3.98 ± 1.93 |
| <3.0 cm | 4 | 3 | 2 | 0 | 1 |
| ≥3.0 cm | 6 | 2 | 3 | 5 | 4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Do, M.; Kim, H.; Shin, D.; Park, J.; Kim, H.; Han, Y.; Jang, J.-Y.; Kim, Y. Marker Identification of the Grade of Dysplasia of Intraductal Papillary Mucinous Neoplasm in Pancreatic Cyst Fluid by Quantitative Proteomic Profiling. Cancers 2020, 12, 2383. https://doi.org/10.3390/cancers12092383
Do M, Kim H, Shin D, Park J, Kim H, Han Y, Jang J-Y, Kim Y. Marker Identification of the Grade of Dysplasia of Intraductal Papillary Mucinous Neoplasm in Pancreatic Cyst Fluid by Quantitative Proteomic Profiling. Cancers. 2020; 12(9):2383. https://doi.org/10.3390/cancers12092383
Chicago/Turabian StyleDo, Misol, Hongbeom Kim, Dongyoon Shin, Joonho Park, Haeryoung Kim, Youngmin Han, Jin-Young Jang, and Youngsoo Kim. 2020. "Marker Identification of the Grade of Dysplasia of Intraductal Papillary Mucinous Neoplasm in Pancreatic Cyst Fluid by Quantitative Proteomic Profiling" Cancers 12, no. 9: 2383. https://doi.org/10.3390/cancers12092383
APA StyleDo, M., Kim, H., Shin, D., Park, J., Kim, H., Han, Y., Jang, J.-Y., & Kim, Y. (2020). Marker Identification of the Grade of Dysplasia of Intraductal Papillary Mucinous Neoplasm in Pancreatic Cyst Fluid by Quantitative Proteomic Profiling. Cancers, 12(9), 2383. https://doi.org/10.3390/cancers12092383





