Diversity in the Extracellular Vesicle-Derived Microbiome of Tissues According to Tumor Progression in Pancreatic Cancer
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
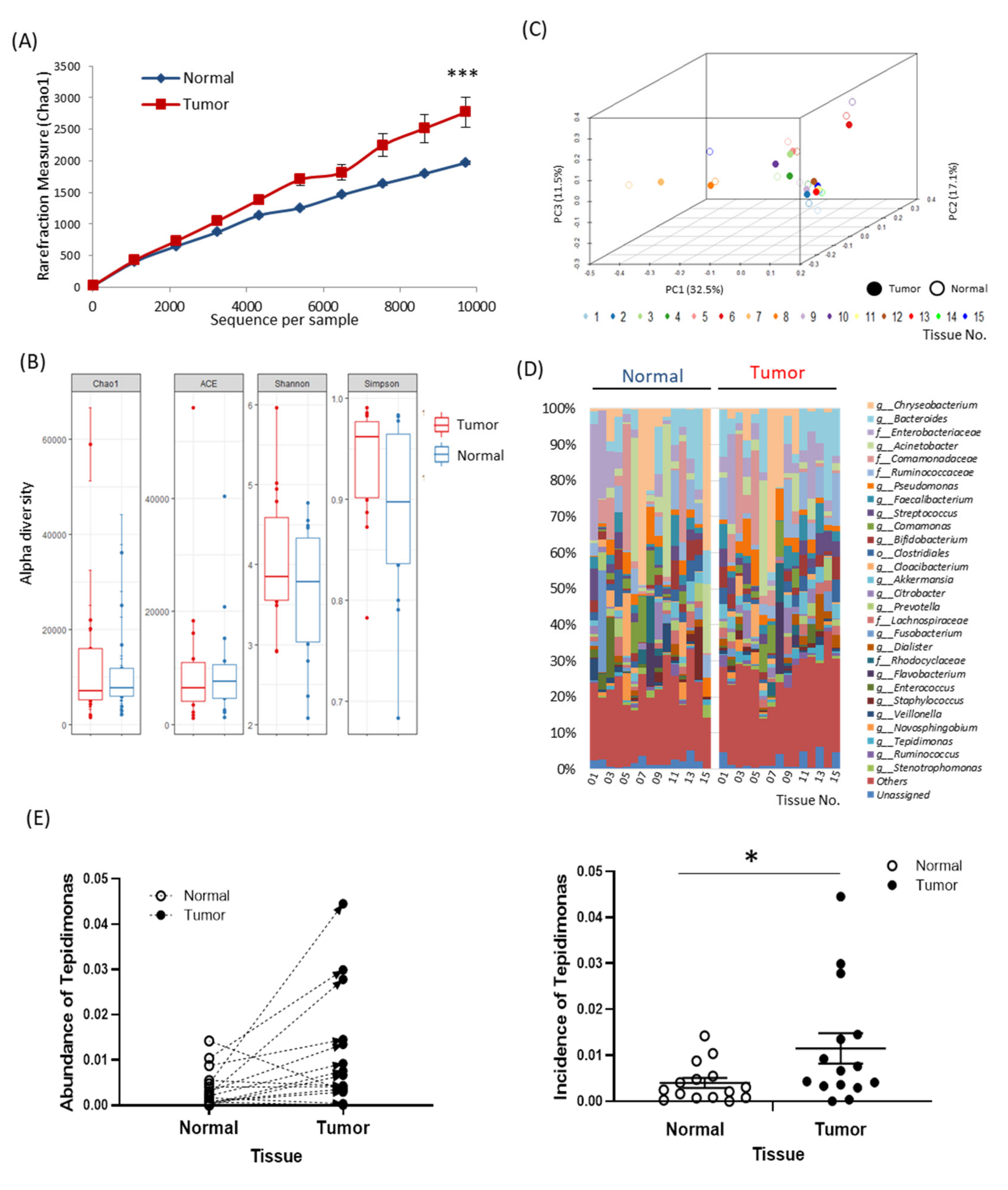
2.2. Tissue-Specific EV-Derived Microbiomes
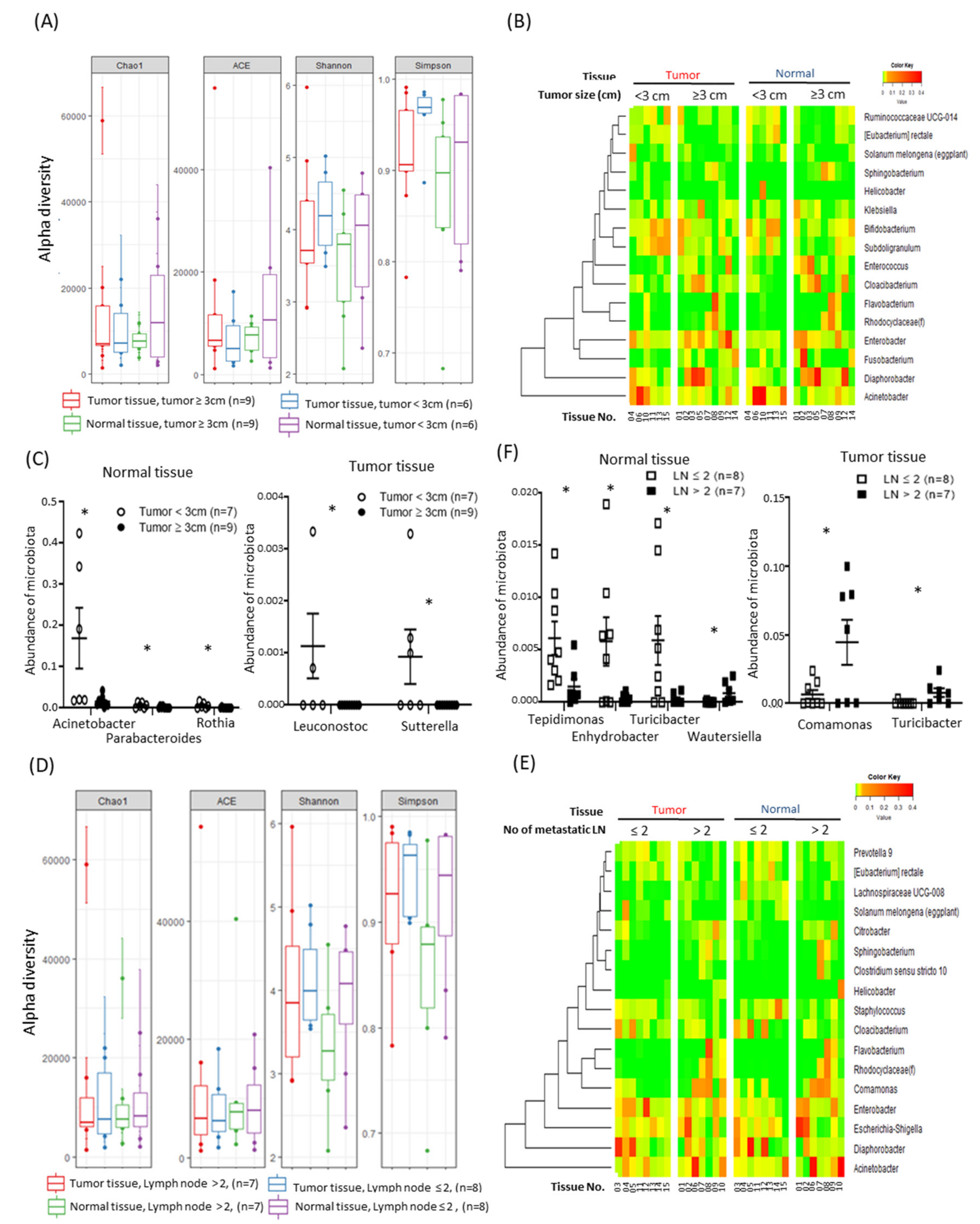
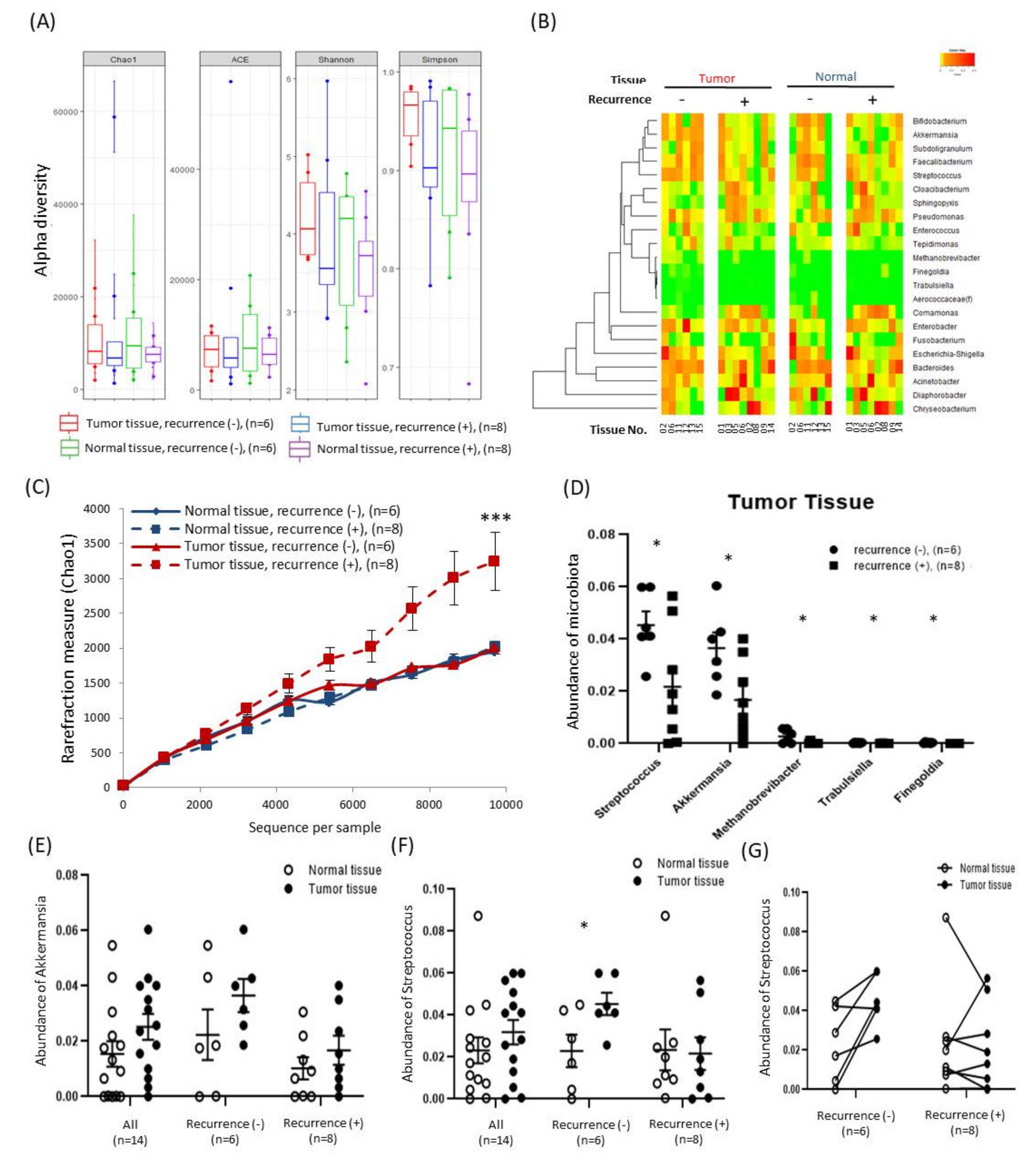
2.3. Alterations in Tissue-Specific EV-Derived Microbiomes According to Tumor Progression
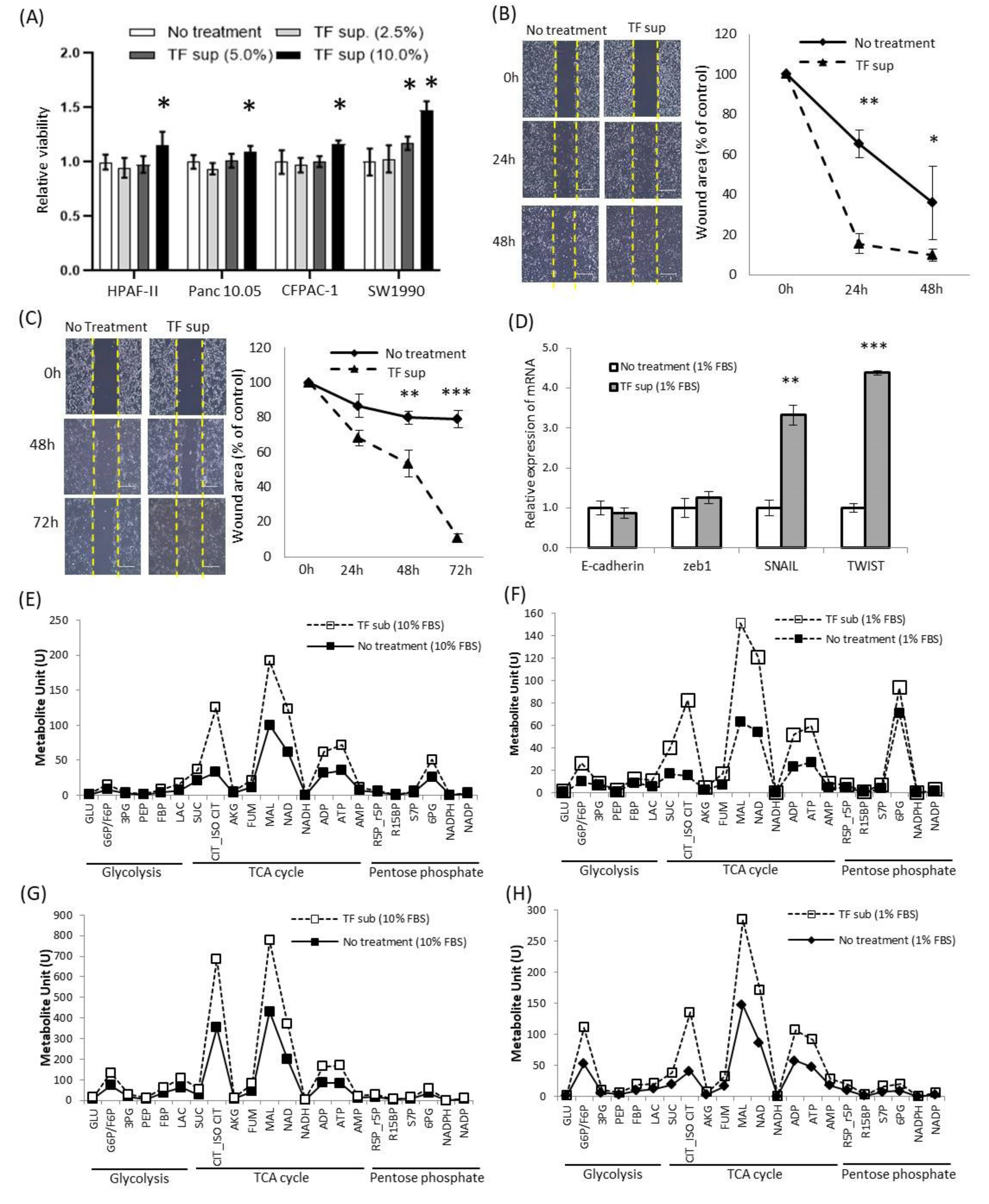
2.4. Alteration of Oncological Function of Tumor Cell by Tepidimoas Fonticaldi
3. Discussion
4. Materials and Methods
4.1. Clinicopathological Characteristics of Patients
4.2. Isolation of EVs and DNA Extraction
4.3. PCR Amplification and Sequencing of 16S Rrna Genes
4.4. Taxonomic Assignment and Analysis of the Bacterial Composition of the Microbiota
4.5. Comparative Analysis for Microbiome Diversity
4.6. Culture of Tepidimonas Fonticaldi and Cancer Cells
4.7. In Vitro Assay of Cancer Cells
4.8. Metabolite Analysis
4.9. Statistics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gillen, S.; Schuster, T.; Meyer Zum Buschenfelde, C.; Friess, H.; Kleeff, J. Preoperative/neoadjuvant therapy in pancreatic cancer: A systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010, 7, e1000267. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Lin, C.C.; Mariotto, A.B.; Siegel, R.L.; Stein, K.D.; Kramer, J.L.; Alteri, R.; Robbins, A.S.; Jemal, A. Cancer treatment and survivorship statistics, 2014. CA Cancer J. Clin. 2014, 64, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Sethi, V.; Vitiello, G.A.; Saxena, D.; Miller, G.; Dudeja, V. The Role of the Microbiome in Immunologic Development and its Implication For Pancreatic Cancer Immunotherapy. Gastroenterology 2019, 156, 2097–2115.e2092. [Google Scholar] [CrossRef]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e712. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 2011, 77, 3219–3226. [Google Scholar] [CrossRef]
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 2018, 67, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Heintz-Buschart, A.; Wilmes, P. Human Gut Microbiome: Function Matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Sheflin, A.M.; Whitney, A.K.; Weir, T.L. Cancer-promoting effects of microbial dysbiosis. Curr. Oncol. Rep. 2014, 16, 406. [Google Scholar] [CrossRef] [PubMed]
- Owyang, C.; Wu, G.D. The gut microbiome in health and disease. Gastroenterology 2014, 146, 1433–1436. [Google Scholar] [CrossRef] [PubMed]
- Rajilic-Stojanovic, M.; Figueiredo, C.; Smet, A.; Hansen, R.; Kupcinskas, J.; Rokkas, T.; Andersen, L.; Machado, J.C.; Ianiro, G.; Gasbarrini, A.; et al. Systematic review: Gastric microbiota in health and disease. Aliment. Pharmacol. Ther. 2020, 51, 582–602. [Google Scholar] [CrossRef] [PubMed]
- Hold, G.L. Gastrointestinal Microbiota and Colon Cancer. Dig. Dis. 2016, 34, 244–250. [Google Scholar] [CrossRef]
- Yu, L.X.; Schwabe, R.F. The gut microbiome and liver cancer: Mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 527–539. [Google Scholar] [CrossRef]
- Schulte, A.; Pandeya, N.; Fawcett, J.; Fritschi, L.; Risch, H.A.; Webb, P.M.; Whiteman, D.C.; Neale, R.E. Association between Helicobacter pylori and pancreatic cancer risk: A meta-analysis. Cancer Causes Control 2015, 26, 1027–1035. [Google Scholar] [CrossRef]
- Trikudanathan, G.; Philip, A.; Dasanu, C.A.; Baker, W.L. Association between Helicobacter pylori infection and pancreatic cancer. A cumulative meta-analysis. JOP 2011, 12, 26–31. [Google Scholar]
- Sethi, V.; Kurtom, S.; Tarique, M.; Lavania, S.; Malchiodi, Z.; Hellmund, L.; Zhang, L.; Sharma, U.; Giri, B.; Garg, B.; et al. Gut Microbiota Promotes Tumor Growth in Mice by Modulating Immune Response. Gastroenterology 2018, 155, 33–37.e36. [Google Scholar] [CrossRef]
- Moya, A.; Ferrer, M. Functional Redundancy-Induced Stability of Gut Microbiota Subjected to Disturbance. Trends Microbiol. 2016, 24, 402–413. [Google Scholar] [CrossRef]
- Macia, L.; Nanan, R.; Hosseini-Beheshti, E.; Grau, G.E. Host- and Microbiota-Derived Extracellular Vesicles, Immune Function, and Disease Development. Int. J. Mol. Sci. 2019, 21, 107. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kim, E.K.; McDowell, A.; Kim, Y.K. Microbe-derived extracellular vesicles as a smart drug delivery system. Transl. Clin. Pharmacol. 2018, 26, 103–110. [Google Scholar] [CrossRef]
- Choi, Y.; Kwon, Y.; Kim, D.K.; Jeon, J.; Jang, S.C.; Wang, T.; Ban, M.; Kim, M.H.; Jeon, S.G.; Kim, M.S.; et al. Gut microbe-derived extracellular vesicles induce insulin resistance, thereby impairing glucose metabolism in skeletal muscle. Sci. Rep. 2015, 5, 15878. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, Y.S.; Kim, K.H.; Choi, J.P.; Kim, Y.K.; Yun, S.; Sharma, L.; Dela Cruz, C.S.; Lee, J.S.; Oh, Y.M.; et al. The microbiome of the lung and its extracellular vesicles in nonsmokers, healthy smokers and COPD patients. Exp. Mol. Med. 2017, 49, e316. [Google Scholar] [CrossRef] [PubMed]
- Komesu, Y.M.; Dinwiddie, D.L.; Richter, H.E.; Lukacz, E.S.; Sung, V.W.; Siddiqui, N.Y.; Zyczynski, H.M.; Ridgeway, B.; Rogers, R.G.; Arya, L.A.; et al. Defining the relationship between vaginal and urinary microbiomes. Am. J. Obstet. Gynecol. 2020, 222, 154.e1–154.e10. [Google Scholar] [CrossRef]
- Chen, W.M.; Huang, H.W.; Chang, J.S.; Han, Y.L.; Guo, T.R.; Sheu, S.Y. Tepidimonas fonticaldi sp. nov., a slightly thermophilic betaproteobacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 1810–1816. [Google Scholar] [CrossRef]
- Kanokratana, P.; Wongwilaiwalin, S.; Mhuantong, W.; Tangphatsornruang, S.; Eurwilaichitr, L.; Champreda, V. Characterization of cellulolytic microbial consortium enriched on Napier grass using metagenomic approaches. J. Biosci. Bioeng. 2018, 125, 439–447. [Google Scholar] [CrossRef]
- Ling, Z.; Shao, L.; Liu, X.; Cheng, Y.; Yan, C.; Mei, Y.; Ji, F.; Liu, X. Regulatory T Cells and Plasmacytoid Dendritic Cells Within the Tumor Microenvironment in Gastric Cancer Are Correlated With Gastric Microbiota Dysbiosis: A Preliminary Study. Front. Immunol. 2019, 10, 533. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H.K.; Min, S.K.; Lee, W.H. 16S rDNA microbiome composition pattern analysis as a diagnostic biomarker for biliary tract cancer. World J. Surg. Oncol. 2020, 18, 19. [Google Scholar] [CrossRef]
- Li, D.; Chen, H.; Mao, B.; Yang, Q.; Zhao, J.; Gu, Z.; Zhang, H.; Chen, Y.Q.; Chen, W. Microbial Biogeography and Core Microbiota of the Rat Digestive Tract. Sci. Rep. 2017, 8, 45840. [Google Scholar] [CrossRef] [PubMed]
- Gavazza, A.; Rossi, G.; Lubas, G.; Cerquetella, M.; Minamoto, Y.; Suchodolski, J.S. Faecal microbiota in dogs with multicentric lymphoma. Vet. Comp. Oncol. 2018, 16, E169–E175. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Li, J.; An, Y.; Li, P.; Xiong, W.; Li, J.; Yan, D.; Wang, M.; Zhong, G. Chitooligosaccharides Prevents the Development of Colitis-Associated Colorectal Cancer by Modulating the Intestinal Microbiota and Mycobiota. Front. Microbiol. 2019, 10, 2101. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef]
- Cruz-Aguliar, R.M.; Wantia, N.; Clavel, T.; Vehreschild, M.; Buch, T.; Bajbouj, M.; Haller, D.; Busch, D.; Schmid, R.M.; Stein-Thoeringer, C.K. An Open-Labeled Study on Fecal Microbiota Transfer in Irritable Bowel Syndrome Patients Reveals Improvement in Abdominal Pain Associated with the Relative Abundance of Akkermansia Muciniphila. Digestion 2019, 100, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Dingemanse, C.; Belzer, C.; van Hijum, S.A.; Gunthel, M.; Salvatori, D.; den Dunnen, J.T.; Kuijper, E.J.; Devilee, P.; de Vos, W.M.; van Ommen, G.B.; et al. Akkermansia muciniphila and Helicobacter typhlonius modulate intestinal tumor development in mice. Carcinogenesis 2015, 36, 1388–1396. [Google Scholar] [CrossRef]
- Wang, L.; Tang, L.; Feng, Y.; Zhao, S.; Han, M.; Zhang, C.; Yuan, G.; Zhu, J.; Cao, S.; Wu, Q.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8(+) T cells in mice. Gut 2020. [Google Scholar] [CrossRef]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef]
- Mager, D.L.; Haffajee, A.D.; Devlin, P.M.; Norris, C.M.; Posner, M.R.; Goodson, J.M. The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J. Transl. Med. 2005, 3, 27. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, T.M. Role of Oral Microbiota in Cancer Development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.J.; Zhang, L.; Zhou, H.; Chia, D.; Elashoff, D.; Akin, D.; Paster, B.J.; Joshipura, K.; Wong, D.T. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012, 61, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.J.; Fletcher, E.M.; Gibbons, S.M.; Bouvet, M.; Doran, K.S.; Kelley, S.T. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ 2015, 3, e1373. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Choi, J.; Lee, Y.; Lee, J.E.; Lee, E.H.; Kwon, H.J.; Yang, J.; Jeong, B.R.; Kim, Y.K.; Han, P.L. Metagenome Analysis of Bodily Microbiota in a Mouse Model of Alzheimer Disease Using Bacteria-derived Membrane Vesicles in Blood. Exp. Neurobiol. 2017, 26, 369–379. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Wang, X.Q.; Zhang, A.H.; Miao, J.H.; Sun, H.; Yan, G.L.; Wu, F.F.; Wang, X.J. Gut microbiota as important modulator of metabolism in health and disease. Rsc. Adv. 2018, 8, 42380–42389. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, S.; Yoo, S.; Rho, J.K.; Jun, E.S.; Chang, S.; Kim, K.K.; Kim, S.C.; Kim, I. Downregulation of X-linked inhibitor of apoptosis protein by ’7-Benzylidenenaltrexone maleate’ sensitizes pancreatic cancer cells to TRAIL-induced apoptosis. Oncotarget 2017, 8, 61057–61071. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Jang, J.; Lee, E.J.; Kim, S.J.; Yoo, H.J.; Lee, S.; Kang, M.J. Potential involvement of Drosophila flightless-1 in carbohydrate metabolism. BMB Rep. 2018, 51, 462–467. [Google Scholar] [CrossRef] [PubMed]
| Factors | Values | |
|---|---|---|
| Age, y | Mean ± SD, (range) | 65.0 ± 8.2 (52.0–79.0) |
| Sex (Female/Male) | N (%) | 4/11 (26.7/73.3) |
| Preoperative laboratory | Mean ± SD, (range) | |
| WBC (*103/μL) | 6.5 ± 1.9 (2.1–9.1 ) | |
| Neutrophil (%) | 55.6 ± 10.0 (32.3–71.2) | |
| Lymphocyte (%) | 34.5 ± 9.4 (21.8–58.0) | |
| Monocyte (%) | 6.7 ± 2.0 (2.4–11.8) | |
| Hb (g/dL) | 13.6 ± 1.2 (11.7–15.7) | |
| Plt (*103/μL) | 210.6 ± 39.0 (144.0–268.0) | |
| AST (IU/L) | 27.3 ± 8.9 (20.0–49.0) | |
| ALT (IU/L) | 28.7 ± 18.1 (11.0–74.0) | |
| ALP (IU/L)) | 100.7 ± 48.0 (38.0–211.0) | |
| TP (g/dL) | 6.6 ± 0.5 (5.7–7.5) | |
| Alb (g/dL) | 3.8 ± 0.5 (2.8–4.6) | |
| TB (mg/dL) | 1.1 ± 1.7 (0.2–7.1) | |
| BUN (mg/dL) | 12.8 ± 3.1 (9.0–19.0) | |
| Crea (mg/dL) | 0.8 ± 0.1 (0.5–1.1) | |
| CA 19-9 (U/mL) | 190.3 ± 337.4 (3.4–1290.0) | |
| CEA (U/mL) | 2.2 ± 1.1 (0.9–5.5) | |
| Pathologic finding | ||
| Tumor location (head + uncinate/body + tail) | N (%) | 5/10 (33.3/66.7) |
| Operation (PD + TPS/DPS) | N (%) | 5/10 (33.3/66.7) |
| Tumor size (cm) | Mean ± SD, (range) | 3.3 ± 1.9 (1.4–9.2) |
| Tumor differentiation (wel/mod/por) | N (%) | 1/13/1 (6.7/86.7/6.7) |
| Lymphovascular invasion (absent/present) | N (%) | 10/5 (66.7/33.3) |
| Perineural invasion (absent/present) | N (%) | 1/14 (6.7/93.3) |
| metastatic LN (absent/present) | N (%) | 5/10 (33.3/66.7) |
| Neoadjuvant CTx ( non-received/received) | N (%) | 14/1 (6.7/93.3) |
| Recurrence within 4 y (absent/present/n.i) | N (%) | 6/8/1 (40.0/53.3/6.7) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, J.-Y.; Kim, T.-B.; Kim, J.; Choi, H.W.; Kim, E.J.; Yoo, H.J.; Lee, S.; Jun, H.R.; Yoo, W.; Kim, S.; et al. Diversity in the Extracellular Vesicle-Derived Microbiome of Tissues According to Tumor Progression in Pancreatic Cancer. Cancers 2020, 12, 2346. https://doi.org/10.3390/cancers12092346
Jeong J-Y, Kim T-B, Kim J, Choi HW, Kim EJ, Yoo HJ, Lee S, Jun HR, Yoo W, Kim S, et al. Diversity in the Extracellular Vesicle-Derived Microbiome of Tissues According to Tumor Progression in Pancreatic Cancer. Cancers. 2020; 12(9):2346. https://doi.org/10.3390/cancers12092346
Chicago/Turabian StyleJeong, Jin-Yong, Tae-Bum Kim, Jinju Kim, Hwi Wan Choi, Eo Jin Kim, Hyun Ju Yoo, Song Lee, Hye Ryeong Jun, Wonbeak Yoo, Seokho Kim, and et al. 2020. "Diversity in the Extracellular Vesicle-Derived Microbiome of Tissues According to Tumor Progression in Pancreatic Cancer" Cancers 12, no. 9: 2346. https://doi.org/10.3390/cancers12092346
APA StyleJeong, J.-Y., Kim, T.-B., Kim, J., Choi, H. W., Kim, E. J., Yoo, H. J., Lee, S., Jun, H. R., Yoo, W., Kim, S., Kim, S. C., & Jun, E. (2020). Diversity in the Extracellular Vesicle-Derived Microbiome of Tissues According to Tumor Progression in Pancreatic Cancer. Cancers, 12(9), 2346. https://doi.org/10.3390/cancers12092346






