Emerging Cancer Epigenetic Mechanisms Regulated by All-Trans Retinoic Acid
Abstract
1. Introduction
2. RA and Annexins in Myelopoiesis and Acute Promyelocytic Leukemia
3. RA and Annexins in Mammary Morphogenesis and Breast Cancer
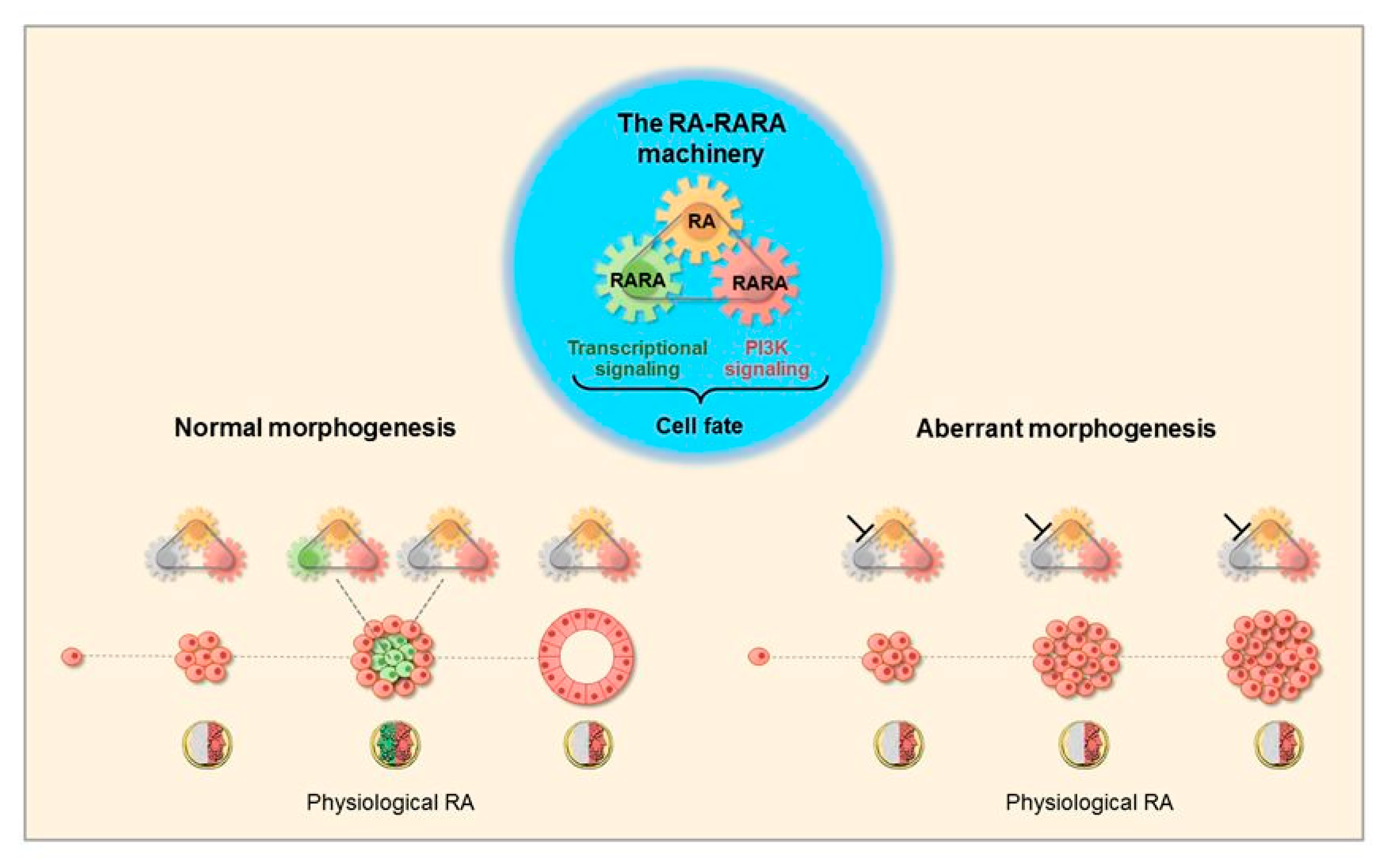
3.1. Morphogenetic Effects of RA
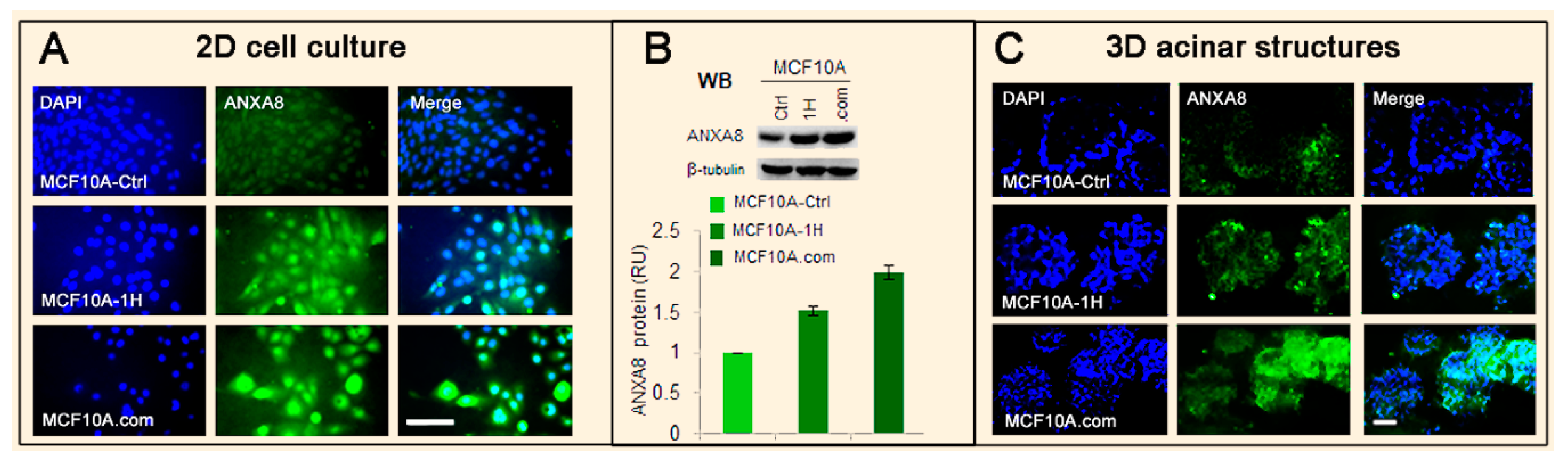
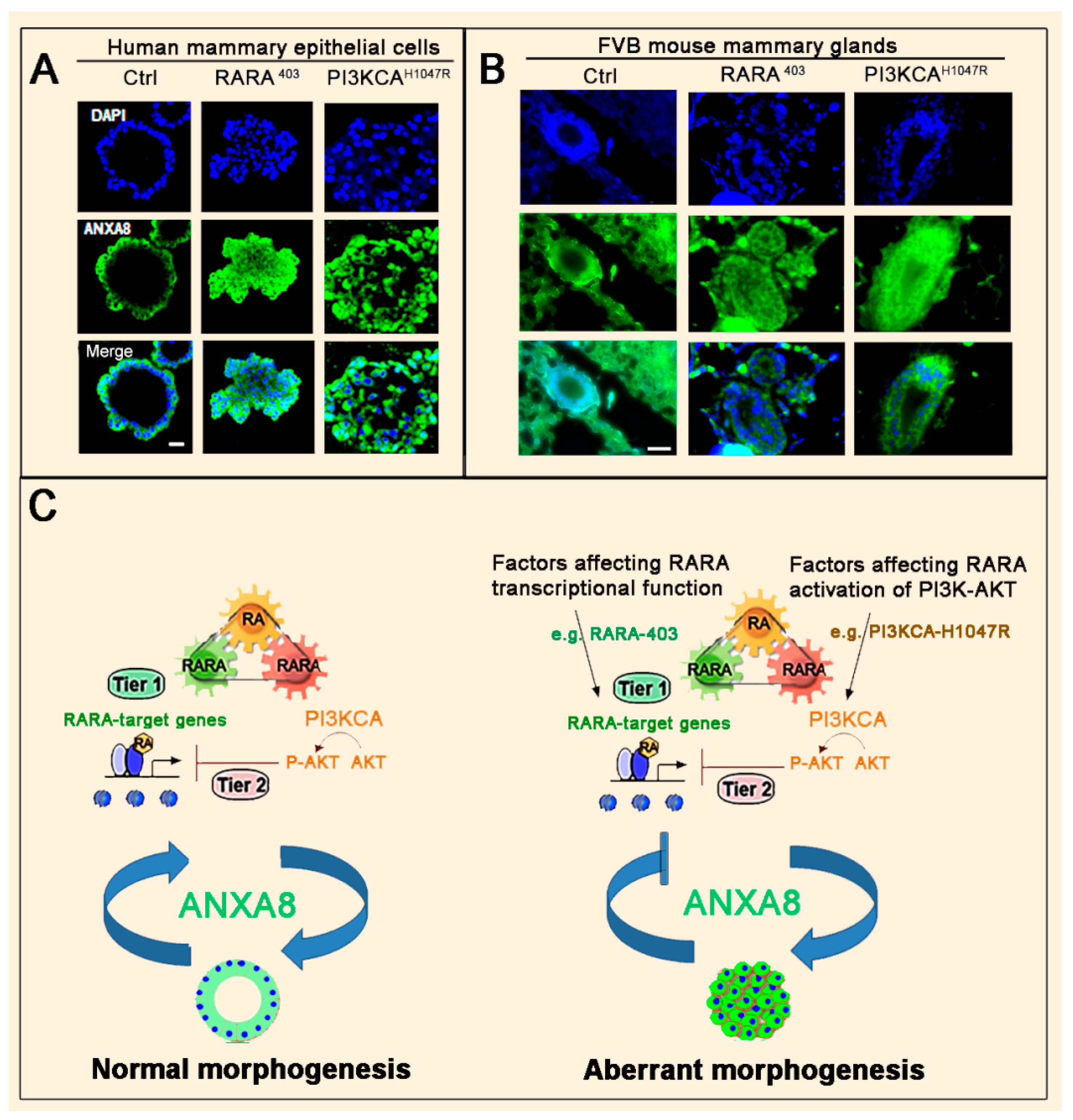
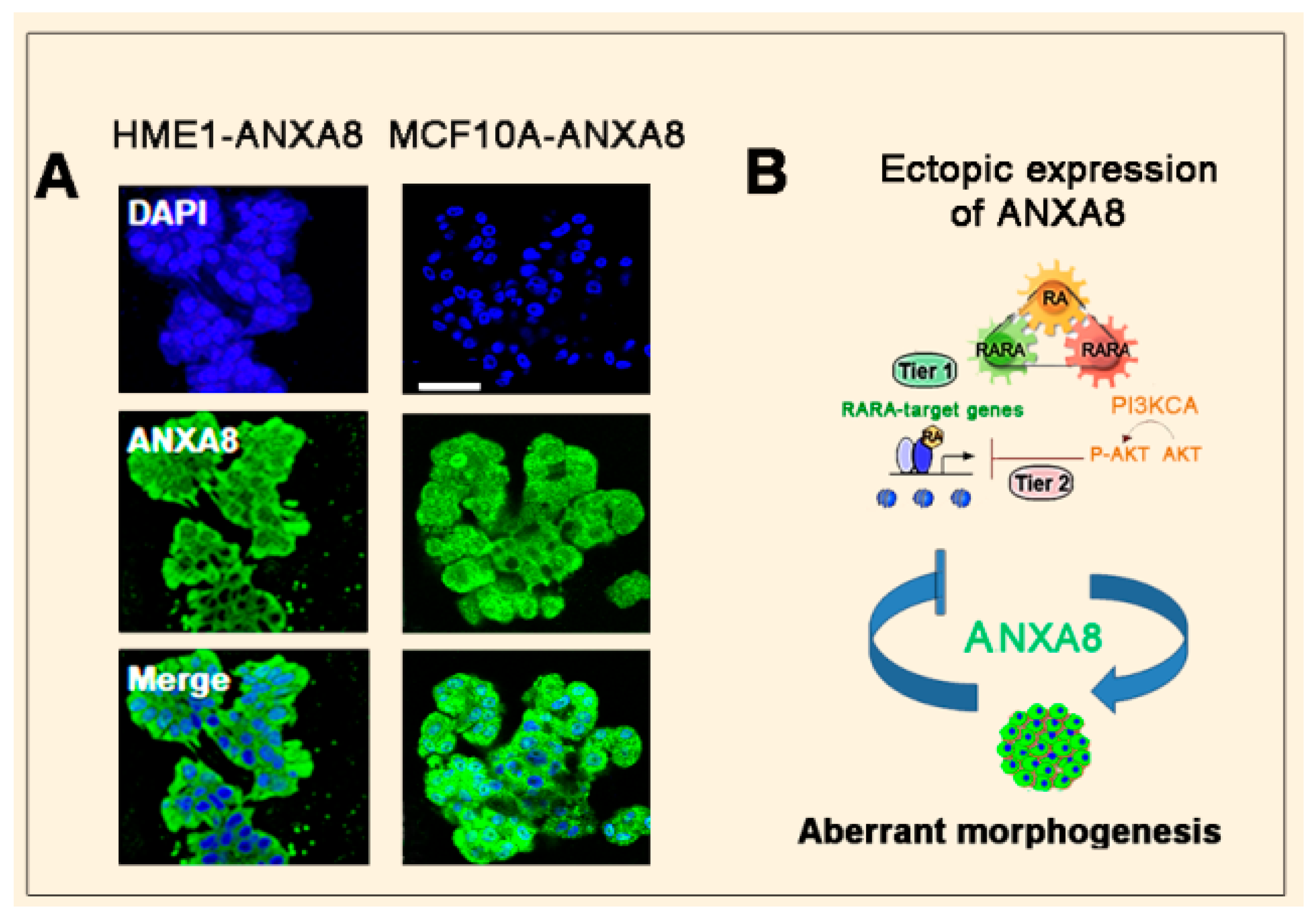
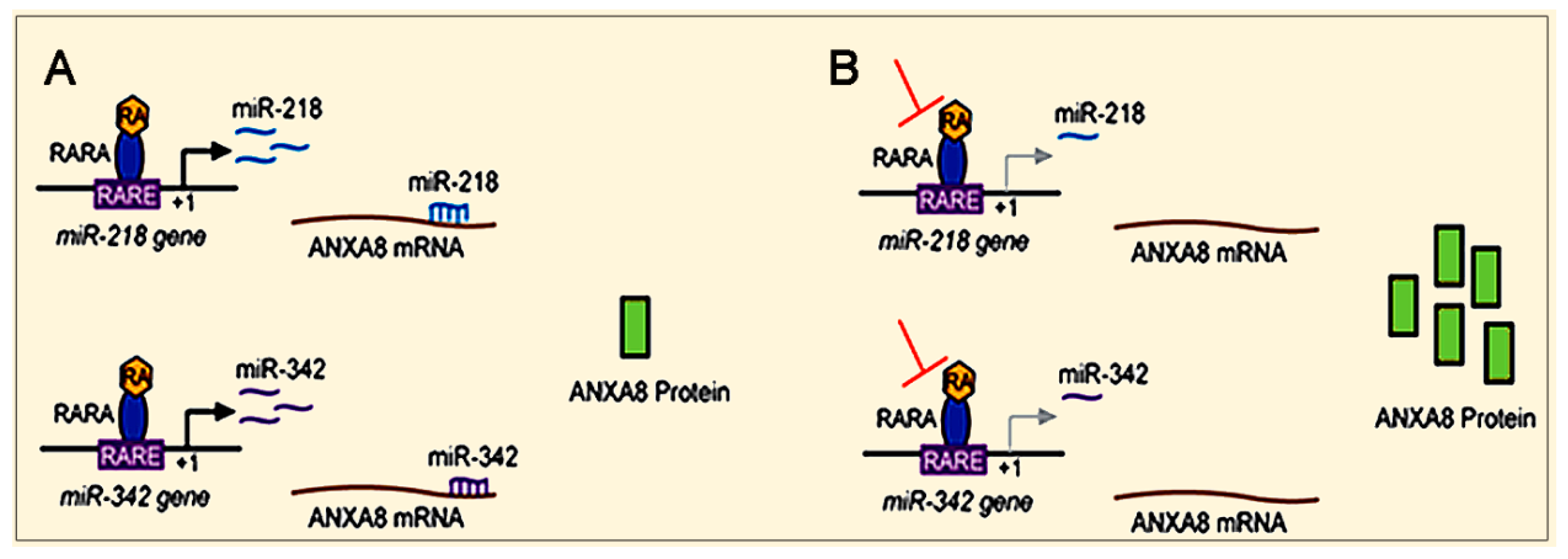
3.2. The RA-RARA and ANXA8 Connection
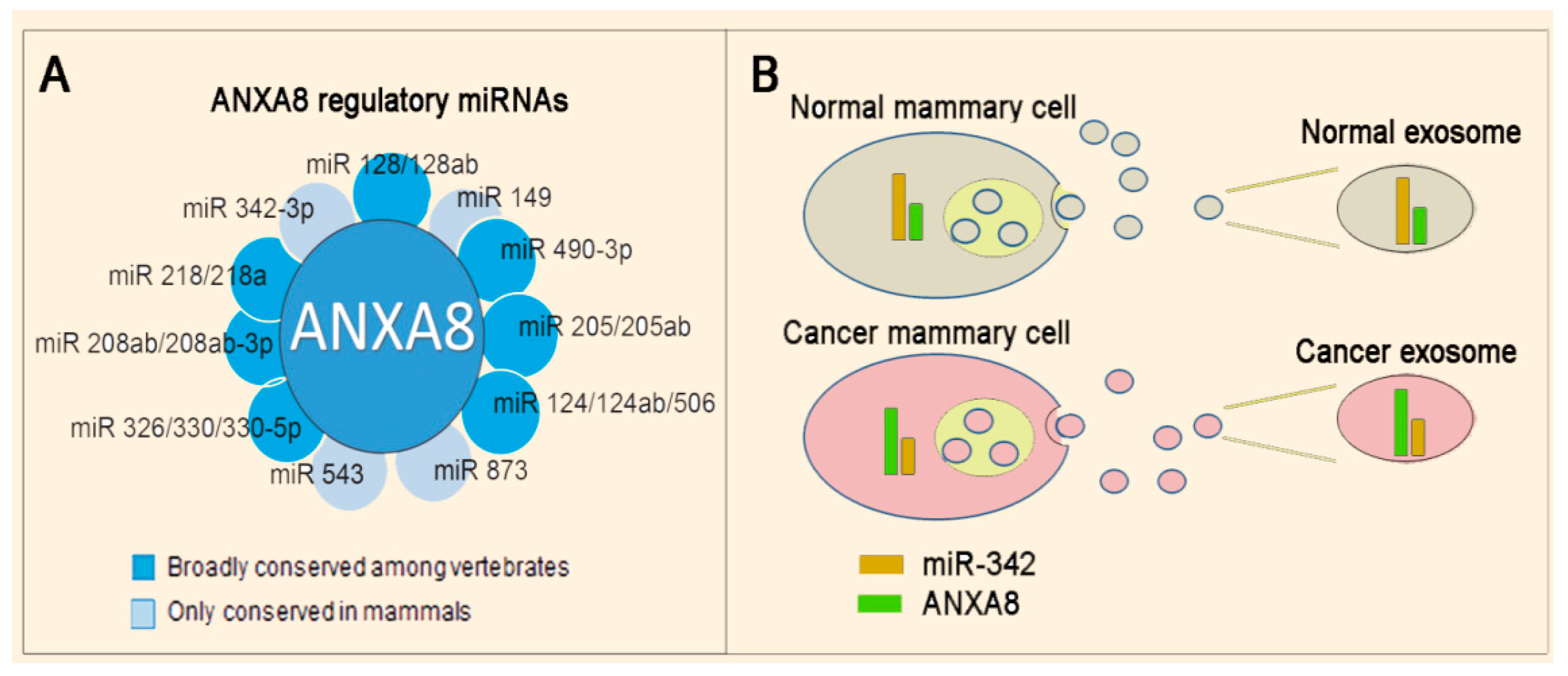
3.3. Mechanisms and Biomarkers Involving Regulatory microRNAs (miRNAs)
3.3.1. Regulatory miRNAs of ANXA2 and ANXA8
3.3.2. Long Non-Coding RNAs (LncRNAs)
4. Emerging Mechanisms Involving ANXA2 and/or ANXA8 in Different Cancers
4.1. Prostate Cancer
4.2. Ovarian Cancer
4.3. Pancreatic Cancer
4.4. Gastric Carcinoma
4.5. Cholangiocarcinoma
4.6. Oral Squamous Cell Carcinoma
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bushue, N.; Wan, Y.J. Retinoid pathway and cancer therapeutics. Adv. Drug Deliv. Rev. 2010, 62, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.J.; Russell, R.M. Beta-carotene. In Encyclopedia of Dietary Supplements, 2nd ed.; Coates, P.M., Betz, J.M., Blackman, M.R., Cragg, G.M., Levine, M., Moss, J., White, J.D., Eds.; Informa Healthcare: London, UK; New York, NY, USA, 2010; pp. 115–120. [Google Scholar]
- Ross, A.; Vitamin, A. Carotenoids. In Modern Nutrition in Health and Disease, 10th ed.; Shils, M.S.M., Ross, A., Caballero, B., Cousins, R., Eds.; Lippincott Williams & Wilkins: Baltimore, MD, USA, 2006; pp. 351–375. [Google Scholar]
- Ross, C.A.; Vitamin, A. Encyclopedia of Dietary Supplements, 2nd ed.; Coates, P.M., Betz, J.M., Blackman, M.R., Cragg, G.M., Levine, M., Moss, J., White, J.D., Eds.; Informa Healthcare: London, UK; New York, NY, USA, 2010; pp. 778–791. [Google Scholar]
- Solomons, N.W.; Vitamin, A. Present Knowledge in Nutrition, 9th ed.; Bowman, B., Russell, R., Eds.; International Life Sciences Institute: Washington, DC, USA, 2006; pp. 157–183. [Google Scholar]
- Institute of Medicine. Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc; The National Academies Press: Washington, DC, USA, 2001. [Google Scholar]
- Cabezuelo, M.T.; Zaragoza, R.; Barber, T.; Vina, J.R. Role of vitamin a in mammary gland development and lactation. Nutrients 2019, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Prevalence of Vitamin A Deficiency in Populations at Risk 1995–2005; World Health Organization: Geneva, Switzerland, 2009. [Google Scholar]
- Mayo-Wilson, E.; Imdad, A.; Herzer, K.; Yakoob, M.Y.; Bhutta, Z.A. Vitamin A supplements for preventing mortality, illness, and blindness in children aged under 5: Systematic review and meta-analysis. BMJ 2011, 343, d5094. [Google Scholar] [CrossRef] [PubMed]
- Sommer, A. Vitamin a deficiency and clinical disease: An historical overview. J. Nutr. 2008, 138, 1835–1839. [Google Scholar] [CrossRef] [PubMed]
- Darlow, B.A.; Graham, P.J. Vitamin A supplementation to prevent mortality and short and long-term morbidity in very low birthweight infants. Cochrane Database Syst. Rev. 2016. [Google Scholar] [CrossRef]
- Mactier, H.; Weaver, L.T. Vitamin A and preterm infants: What we know, what we don’t know, and what we need to know. Arch. Dis. Child. Fetal Neonatal Ed. 2005, 90, F103–F108. [Google Scholar] [CrossRef]
- Oliveira-Menegozzo, J.M.; Bergamaschi, D.P.; Middleton, P.; East, C.E. Vitamin A supplementation for postpartum women. Cochrane Database Syst. Rev. 2010. [Google Scholar] [CrossRef]
- van den Broek, N.; Dou, L.; Othman, M.; Neilson, J.P.; Gates, S.; Gulmezoglu, A.M. Vitamin A supplementation during pregnancy for maternal and newborn outcomes. Cochrane Database Syst. Rev. 2010. [Google Scholar] [CrossRef]
- Cohlan, S.Q. Excessive intake of vitamin A as a cause of congenital anomalies in the rat. Science 1953, 117, 535–536. [Google Scholar] [CrossRef]
- Ribaya-Mercado, J.D.; Blumberg, J.B. Vitamin A: Is it a risk factor for osteoporosis and bone fracture? Nutr. Rev. 2007, 65, 425–438. [Google Scholar] [CrossRef]
- Niederreither, K.; Fraulob, V.; Garnier, J.M.; Chambon, P.; Dolle, P. Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech. Dev. 2002, 110, 165–171. [Google Scholar] [CrossRef]
- Ross, A.C.; Zolfaghari, R. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annu. Rev. Nutr. 2011, 31, 65–87. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.M.; Leung, C.Y.; Tang, W.W.; Choi, H.L.; Leung, Y.C.; McCaffery, P.J.; Wang, C.C.; Woolf, A.S.; Shum, A.S. A paradoxical teratogenic mechanism for retinoic acid. Proc. Natl. Acad. Sci. USA 2012, 109, 13668–13673. [Google Scholar] [CrossRef] [PubMed]
- Napoli, J.L. Effects of ethanol on physiological retinoic acid levels. IUBMB Life 2011, 63, 701–706. [Google Scholar] [CrossRef]
- Kot-Leibovich, H.; Fainsod, A. Ethanol induces embryonic malformations by competing for retinaldehyde dehydrogenase activity during vertebrate gastrulation. Dis. Model Mech. 2009, 2, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Freemantle, S.J.; Dragnev, K.H.; Dmitrovsky, E. The retinoic acid paradox in cancer chemoprevention. J. Natl. Cancer Inst. 2006, 98, 426–427. [Google Scholar] [CrossRef]
- Omenn, G.S.; Goodman, G.E.; Thornquist, M.D.; Balmes, J.; Cullen, M.R.; Glass, A.; Keogh, J.P.; Meyskens, F.L.; Valanis, B.; Williams, J.H.; et al. Effects of a combination of beta carotene and vitamin A on lung cancer and cardiovascular disease. N. Engl. J. Med. 1996, 334, 1150–1155. [Google Scholar] [CrossRef]
- Hennekens, C.H.; Buring, J.E.; Manson, J.E.; Stampfer, M.; Rosner, B.; Cook, N.R.; Belanger, C.; LaMotte, F.; Gaziano, J.M.; Ridker, P.M.; et al. Lack of effect of long-term supplementation with beta carotene on the incidence of malignant neoplasms and cardiovascular disease. N. Engl. J. Med. 1996, 334, 1145–1149. [Google Scholar] [CrossRef]
- The Alpha-Tocopherol Beta-Carotene Cancer Prevention Study Group. The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. N. Engl. J. Med. 1994, 330, 1029–1035. [Google Scholar] [CrossRef]
- Khuri, F.R.; Lee, J.J.; Lippman, S.M.; Kim, E.S.; Cooper, J.S.; Benner, S.E.; Winn, R.; Pajak, T.F.; Williams, B.; Shenouda, G.; et al. Randomized phase III trial of low-dose isotretinoin for prevention of second primary tumors in stage I and II head and neck cancer patients. J. Natl. Cancer Inst. 2006, 98, 441–450. [Google Scholar] [CrossRef]
- Lippman, S.M.; Lee, J.J.; Karp, D.D.; Vokes, E.E.; Benner, S.E.; Goodman, G.E.; Khuri, F.R.; Marks, R.; Winn, R.J.; Fry, W.; et al. Randomized phase III intergroup trial of isotretinoin to prevent second primary tumors in stage I non-small-cell lung cancer. J. Natl. Cancer Inst. 2001, 93, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Blaner, W.S. Vitamin A signaling and homeostasis in obesity, diabetes, and metabolic disorders. Pharmacol. Ther. 2019, 197, 153–178. [Google Scholar] [CrossRef] [PubMed]
- Ecker, B.L.; Lee, J.Y.; Sterner, C.J.; Solomon, A.C.; Pant, D.K.; Shen, F.; Peraza, J.; Vaught, L.; Mahendra, S.; Belka, G.K.; et al. Impact of obesity on breast cancer recurrence and minimal residual disease. Breast Cancer Res. 2019, 21, 41. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.C. Vitamin A and retinoic acid in T cell-related immunity. Am. J. Clin. Nutr. 2012, 96, 1166S–1172S. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.C.; Esterhazy, D.; Sarde, A.; London, M.; Pullabhatla, V.; Osma-Garcia, I.; Al-Bader, R.; Ortiz, C.; Elgueta, R.; Arno, M.; et al. Retinoic acid is essential for Th1 cell lineage stability and prevents transition to a Th17 cell program. Immunity 2015, 42, 499–511. [Google Scholar] [CrossRef]
- LaBarge, M.A.; Mora-Blanco, E.L.; Samson, S.; Miyano, M. Breast cancer beyond the age of mutation. Gerontology 2016, 62, 434–442. [Google Scholar] [CrossRef]
- Todhunter, M.E.; LaBarge, M.A. Cell and tissue biology paves a path to breast cancer prevention. Trends Cancer 2017, 3, 313–315. [Google Scholar] [CrossRef]
- Hendrickson, S.J.; Lindstrom, S.; Eliassen, A.H.; Rosner, B.A.; Chen, C.; Barrdahl, M.; Brinton, L.; Buring, J.; Canzian, F.; Chanock, S.; et al. Plasma carotenoid- and retinol-weighted multi-SNP scores and risk of breast cancer in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Cancer Epidemiol. Biomark. Prev. 2013, 22, 927–936. [Google Scholar] [CrossRef][Green Version]
- Eliassen, A.H.; Hendrickson, S.J.; Brinton, L.A.; Buring, J.E.; Campos, H.; Dai, Q.; Dorgan, J.F.; Franke, A.A.; Gao, Y.T.; Goodman, M.T.; et al. Circulating carotenoids and risk of breast cancer: Pooled analysis of eight prospective studies. J. Natl. Cancer Inst. 2012, 104, 1905–1916. [Google Scholar] [CrossRef]
- Eliassen, A.H.; Liao, X.; Rosner, B.; Tamimi, R.M.; Tworoger, S.S.; Hankinson, S.E. Plasma carotenoids and risk of breast cancer over 20 y of follow-up. Am. J. Clin. Nutr. 2015, 101, 1197–1205. [Google Scholar] [CrossRef]
- Nagel, G.; Linseisen, J.; van Gils, C.H.; Peeters, P.H.; Boutron-Ruault, M.C.; Clavel-Chapelon, F.; Romieu, I.; Tjonneland, A.; Olsen, A.; Roswall, N.; et al. Dietary beta-carotene, vitamin C and E intake and breast cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC). Breast Cancer Res. Treat 2010, 119, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.F.; Peeters, P.H.; Klaasen, V.M.; Bueno-de-Mesquita, H.B.; Jansen, E.H.; Ros, M.M.; Travier, N.; Olsen, A.; Tjonneland, A.; Overvad, K.; et al. Plasma carotenoids, vitamin C, tocopherols, and retinol and the risk of breast cancer in the European Prospective Investigation into Cancer and Nutrition cohort. Am. J. Clin. Nutr. 2016, 103, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Sachs, L. The differentiation of myeloid leukaemia cells: New possibilities for therapy. Br. J. Haematol. 1978, 40, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.S.; Wang, G.; Freireich, E.J.; Daly, M.; Naylor, S.L.; Trujillo, J.M.; Stass, S.A. Specific expression of the annexin VIII gene in acute promyelocytic leukemia. Blood 1992, 79, 1802–1810. [Google Scholar] [CrossRef]
- Stavem, P. Acute hypergranular promyelocytic leukaemia. Priority of discovery (Jean Bernard and Leif Hillestad). Scand J. Haematol. 1978, 20, 287–288. [Google Scholar]
- Collins, S.J.; Gallo, R.C.; Gallagher, R.E. Continuous growth and differentiation of human myeloid leukaemic cells in suspension culture. Nature 1977, 270, 347–349. [Google Scholar] [CrossRef]
- Breitman, T.R.; Selonick, S.E.; Collins, S.J. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc. Natl. Acad. Sci. USA 1980, 77, 2936–2940. [Google Scholar] [CrossRef]
- Koeffler, H.P. Induction of differentiation of human acute myelogenous leukemia cells: Therapeutic implications. Blood 1983, 62, 709–721. [Google Scholar] [CrossRef]
- Chomienne, C.; Ballerini, P.; Balitrand, N.; Amar, M.; Bernard, J.F.; Boivin, P.; Daniel, M.T.; Berger, R.; Castaigne, S.; Degos, L. Retinoic acid therapy for promyelocytic leukaemia. Lancet 1989, 2, 746–747. [Google Scholar] [CrossRef]
- Chen, Z.X.; Xue, Y.Q.; Zhang, R.; Tao, R.F.; Xia, X.M.; Li, C.; Wang, W.; Zu, W.Y.; Yao, X.Z.; Ling, B.J. A clinical and experimental study on all-trans retinoic acid-treated acute promyelocytic leukemia patients. Blood 1991, 78, 1413–1419. [Google Scholar] [CrossRef]
- Rao, Y.; Li, R.; Zhang, D. A drug from poison: How the therapeutic effect of arsenic trioxide on acute promyelocytic leukemia was discovered. Sci. China Life Sci. 2013, 56, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.D.; Golomb, H.M.; Dougherty, C. 15/17 translocation, a consistent chromosomal change in acute promyelocytic leukaemia. Lancet 1977, 1, 549–550. [Google Scholar] [CrossRef]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Lallemand-Breitenbach, V.; de The, H. PML nuclear bodies: Regulation, function and therapeutic perspectives. J. Pathol. 2014, 234, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.E.; Ye, Y.C.; Chen, S.R.; Chai, J.R.; Lu, J.X.; Zhoa, L.; Gu, L.J.; Wang, Z.Y. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 1988, 72, 567–572. [Google Scholar] [CrossRef]
- Kakizuka, A.; Miller, W.H., Jr.; Umesono, K.; Warrell, R.P., Jr.; Frankel, S.R.; Murty, V.V.; Dmitrovsky, E.; Evans, R.M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 1991, 66, 663–674. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Chen, Z. Acute promyelocytic leukemia: From highly fatal to highly curable. Blood 2008, 111, 2505–2515. [Google Scholar] [CrossRef]
- Carbone, R.; Botrugno, O.A.; Ronzoni, S.; Insinga, A.; di Croce, L.; Pelicci, P.G.; Minucci, S. Recruitment of the histone methyltransferase SUV39H1 and its role in the oncogenic properties of the leukemia-associated PML-retinoic acid receptor fusion protein. Mol. Cell Biol. 2006, 26, 1288–1296. [Google Scholar] [CrossRef]
- Villa, R.; Pasini, D.; Gutierrez, A.; Morey, L.; Occhionorelli, M.; Vire, E.; Nomdedeu, J.F.; Jenuwein, T.; Pelicci, P.G.; Minucci, S.; et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell 2007, 11, 513–525. [Google Scholar] [CrossRef]
- Grignani, F.; de Matteis, S.; Nervi, C.; Tomassoni, L.; Gelmetti, V.; Cioce, M.; Fanelli, M.; Ruthardt, M.; Ferrara, F.F.; Zamir, I.; et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 1998, 391, 815–818. [Google Scholar] [CrossRef]
- Nowak, D.; Stewart, D.; Koeffler, H.P. Differentiation therapy of leukemia: 3 decades of development. Blood 2009, 113, 3655–3665. [Google Scholar] [CrossRef] [PubMed]
- de The, H.; Pandolfi, P.P.; Chen, Z. Acute Promyelocytic leukemia: A Paradigm for oncoprotein-targeted cure. Cancer Cell 2017, 32, 552–560. [Google Scholar] [CrossRef] [PubMed]
- He, L.Z.; Bhaumik, M.; Tribioli, C.; Rego, E.M.; Ivins, S.; Zelent, A.; Pandolfi, P.P. Two critical hits for promyelocytic leukemia. Mol. Cell 2000, 6, 1131–1141. [Google Scholar] [CrossRef]
- Guidez, F.; Parks, S.; Wong, H.; Jovanovic, J.V.; Mays, A.; Gilkes, A.F.; Mills, K.I.; Guillemin, M.C.; Hobbs, R.M.; Pandolfi, P.P.; et al. RARalpha-PLZF overcomes PLZF-mediated repression of CRABPI, contributing to retinoid resistance in t(11;17) acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 2007, 104, 18694–18699. [Google Scholar] [CrossRef] [PubMed]
- Lehmann-Che, J.; Bally, C.; de The, H. Resistance to therapy in acute promyelocytic leukemia. N. Engl. J. Med. 2014, 371, 1170–1172. [Google Scholar] [CrossRef]
- Sainty, D.; Liso, V.; Cantu-Rajnoldi, A.; Head, D.; Mozziconacci, M.J.; Arnoulet, C.; Benattar, L.; Fenu, S.; Mancini, M.; Duchayne, E.; et al. A new morphologic classification system for acute promyelocytic leukemia distinguishes cases with underlying PLZF/RARA gene rearrangements. Blood 2000, 96, 1287–1296. [Google Scholar] [PubMed]
- Wells, R.A.; Catzavelos, C.; Kamel-Reid, S. Fusion of retinoic acid receptor alpha to NuMA, the nuclear mitotic apparatus protein, by a variant translocation in acute promyelocytic leukaemia. Nat. Genet. 1997, 17, 109–113. [Google Scholar] [CrossRef]
- Arnould, C.; Philippe, C.; Bourdon, V.; goire, M.J.G.; Berger, R.; Jonveaux, P. The signal transducer and activator of transcription STAT5b gene is a new partner of retinoic acid receptor alpha in acute promyelocytic-like leukaemia. Hum Mol. Genet. 1999, 8, 1741–1749. [Google Scholar] [CrossRef]
- Noguera, N.I.; Catalano, G.; Banella, C.; Divona, M.; Faraoni, I.; Ottone, T.; Arcese, W.; Voso, M.T. Acute promyelocytic leukemia: Update on the mechanisms of leukemogenesis, resistance and on innovative treatment strategies. Cancers 2019, 11, 1591. [Google Scholar] [CrossRef]
- Geoffroy, M.C.; de The, H. Classic and variants APLs, as viewed from a therapy response. Cancers 2020, 12, 967. [Google Scholar] [CrossRef]
- Monastyrskaya, K. Functional association between regulatory RNAs and the annexins. Int. J. Mol. Sci. 2018, 19, 591. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Moss, S.E. Annexins: From structure to function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Creutz, C.E.; Moss, S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Raynal, P.; Pollard, H.B. Annexins: The problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim. Biophys. Acta 1994, 1197, 63–93. [Google Scholar] [CrossRef]
- Bandorowicz, J.; Pikula, S. Annexins--multifunctional, calcium-dependent, phospholipid-binding proteins. Acta Biochim. Pol. 1993, 40, 281–293. [Google Scholar] [CrossRef]
- Satoh, A.; Miwa, H.E.; Kojima, K.; Hirabayashi, J.; Matsumoto, I. Ligand-binding properties of annexin from Caenorhabditis elegans (annexin XVI, Nex-1). J. Biochem. 2000, 128, 377–381. [Google Scholar] [CrossRef]
- Rescher, U.; Gerke, V. Annexins—Unique membrane binding proteins with diverse functions. J. Cell Sci. 2004, 117, 2631–2639. [Google Scholar] [CrossRef]
- Rosengarth, A.; Luecke, H. A calcium-driven conformational switch of the N-terminal and core domains of annexin A1. J. Mol. Biol. 2003, 326, 1317–1325. [Google Scholar] [CrossRef]
- Faure, A.V.; Migne, C.; Devilliers, G.; Ayala-Sanmartin, J. Annexin 2 “secretion” accompanying exocytosis of chromaffin cells: Possible mechanisms of annexin release. Exp. Cell Res. 2002, 276, 79–89. [Google Scholar] [CrossRef]
- Benaud, C.; Gentil, B.J.; Assard, N.; Court, M.; Garin, J.; Delphin, C.; Baudier, J. AHNAK interaction with the annexin 2/S100A10 complex regulates cell membrane cytoarchitecture. J. Cell Biol. 2004, 164, 133–144. [Google Scholar] [CrossRef]
- Jacob, R.; Heine, M.; Eikemeyer, J.; Frerker, N.; Zimmer, K.P.; Rescher, U.; Gerke, V.; Naim, H.Y. Annexin II is required for apical transport in polarized epithelial cells. J. Biol. Chem. 2004, 279, 3680–3684. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Yang, P.; Fan, Y.H.; Mu, Z.M.; Hauptmann, R.; Adolf, G.R.; Stass, S.A.; Chang, K.S. Regulation of the expression of annexin VIII in acute promyelocytic leukemia. Blood 1994, 84, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Stass, S.A.; Chang, K.S. Expression of the annexin VIII gene in acute promyelocytic leukemia. Leuk. Lymphoma 1994, 13, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Olwill, S.A.; McGlynn, H.; Gilmore, W.S.; Alexander, H.D. All-trans retinoic acid-induced downregulation of annexin II expression in myeloid leukaemia cell lines is not confined to acute promyelocytic leukaemia. Br. J. Haematol. 2005, 131, 258–264. [Google Scholar] [CrossRef]
- Kwaan, H.C.; Weiss, I.; Tallman, M.S. The role of abnormal hemostasis and fibrinolysis in morbidity and mortality of acute promyelocytic leukemia. Semin. Thromb. Hemost. 2019, 45, 612–621. [Google Scholar] [CrossRef]
- Huang, D.; Yang, Y.; Sun, J.; Dong, X.; Wang, J.; Liu, H.; Lu, C.; Chen, X.; Shao, J.; Yan, J. Annexin A2-S100A10 heterotetramer is upregulated by PML/RARalpha fusion protein and promotes plasminogen-dependent fibrinolysis and matrix invasion in acute promyelocytic leukemia. Front. Med. 2017, 11, 410–422. [Google Scholar] [CrossRef]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef]
- Garzon, R.; Pichiorri, F.; Palumbo, T.; Visentini, M.; Aqeilan, R.; Cimmino, A.; Wang, H.; Sun, H.; Volinia, S.; Alder, H.; et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene 2007, 26, 4148–4157. [Google Scholar] [CrossRef]
- Careccia, S.; Mainardi, S.; Pelosi, A.; Gurtner, A.; Diverio, D.; Riccioni, R.; Testa, U.; Pelosi, E.; Piaggio, G.; Sacchi, A.; et al. A restricted signature of miRNAs distinguishes APL blasts from normal promyelocytes. Oncogene 2009, 28, 4034–4040. [Google Scholar] [CrossRef]
- de Marchis, M.L.; Ballarino, M.; Salvatori, B.; Puzzolo, M.C.; Bozzoni, I.; Fatica, A. A new molecular network comprising PU.1, interferon regulatory factor proteins and miR-342 stimulates ATRA-mediated granulocytic differentiation of acute promyelocytic leukemia cells. Leukemia 2009, 23, 856–862. [Google Scholar] [CrossRef]
- Inman, J.L.; Robertson, C.; Mott, J.D.; Bissell, M.J. Mammary gland development: Cell fate specification, stem cells and the microenvironment. Development 2015, 142, 1028–1042. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, S.; Ren, M.; Visconti, N.; Corlazzoli, F.; Gagliostro, V.; Somenzi, G.; Yao, J.; Sun, Y.; Sacchi, N. Tracing anti-cancer and cancer-promoting actions of all-trans retinoic acid in breast cancer to a RARalpha epigenetic mechanism of mammary epithelial cell fate. Oncotarget 2016, 7, 87064–87080. [Google Scholar] [CrossRef] [PubMed]
- Montesano, R.; Soulie, P. Retinoids induce lumen morphogenesis in mammary epithelial cells. J. Cell Sci. 2002, 115, 4419–4431. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.A.; Shen, K.; Wang, Y.; Brooks, S.C. Retinoic acid signaling is required for proper morphogenesis of mammary gland. Dev. Dyn. 2005, 234, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.W.; Kwon, H.J.; Shin, J.O.; Lee, J.M.; Cho, S.W.; Tickle, C.; Jung, H.S. Retinoic acid signaling and the initiation of mammary gland development. Dev. Biol. 2012, 365, 259–266. [Google Scholar] [CrossRef]
- Cohn, E.; Ossowski, L.; Bertran, S.; Marzan, C.; Farias, E.F. RARalpha1 control of mammary gland ductal morphogenesis and wnt1-tumorigenesis. Breast Cancer Res. 2010, 12, R79. [Google Scholar] [CrossRef]
- Centritto, F.; Paroni, G.; Bolis, M.; Garattini, S.K.; Kurosaki, M.; Barzago, M.M.; Zanetti, A.; Fisher, J.N.; Scott, M.F.; Pattini, L.; et al. Cellular and molecular determinants of all-trans retinoic acid sensitivity in breast cancer: Luminal phenotype and RARalpha expression. EMBO Mol. Med. 2015, 7, 950–972. [Google Scholar] [CrossRef]
- Somenzi, G.; Sala, G.; Rossetti, S.; Ren, M.; Ghidoni, R.; Sacchi, N. Disruption of retinoic acid receptor alpha reveals the growth promoter face of retinoic acid. PLoS ONE 2007, 2, e836. [Google Scholar] [CrossRef]
- Schug, T.T.; Berry, D.C.; Shaw, N.S.; Travis, S.N.; Noy, N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 2007, 129, 723–733. [Google Scholar] [CrossRef]
- Schug, T.T.; Berry, D.C.; Toshkov, I.A.; Cheng, L.; Nikitin, A.Y.; Noy, N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc. Natl. Acad. Sci. USA 2008, 105, 7546–7551. [Google Scholar] [CrossRef]
- Bosch, A.; Bertran, S.P.; Lu, Y.; Garcia, A.; Jones, A.M.; Dawson, M.I.; Farias, E.F. Reversal by RARalpha agonist Am580 of c-Myc-induced imbalance in RARalpha/RARgamma expression during MMTV-Myc tumorigenesis. Breast Cancer Res. 2012, 14, R121. [Google Scholar] [CrossRef] [PubMed]
- Garattini, E.; Bolis, M.; Garattini, S.K.; Fratelli, M.; Centritto, F.; Paroni, G.; Gianni, M.; Zanetti, A.; Pagani, A.; Fisher, J.N.; et al. Retinoids and breast cancer: From basic studies to the clinic and back again. Cancer Treat. Rev. 2014, 40, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Garattini, E.; Paroni, G.; Terao, M. Retinoids and breast cancer: New clues to increase their activity and selectivity. Breast Cancer Res. 2012, 14, 111. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.J.; Wolman, S.R.; Tait, L.; Heppner, G.H.; Miller, F.R. MCF10AT: A model for the evolution of cancer from proliferative breast disease. Am. J. Pathol. 1996, 148, 313–319. [Google Scholar]
- Miller, F.R.; Santner, S.J.; Tait, L.; Dawson, P.J. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J. Natl. Cancer Inst. 2000, 92, 1185–1186. [Google Scholar] [CrossRef] [PubMed]
- Miller, F.R.; Soule, H.D.; Tait, L.; Pauley, R.J.; Wolman, S.R.; Dawson, P.J.; Heppner, G.H. Xenograft model of progressive human proliferative breast disease. J. Natl. Cancer Inst. 1993, 85, 1725–1732. [Google Scholar] [CrossRef]
- Behbod, F.; Kittrell, F.S.; LaMarca, H.; Edwards, D.; Kerbawy, S.; Heestand, J.C.; Young, E.; Mukhopadhyay, P.; Yeh, H.W.; Allred, D.C.; et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009, 11, R66. [Google Scholar] [CrossRef]
- Imbalzano, K.M.; Tatarkova, I.; Imbalzano, A.N.; Nickerson, J.A. Increasingly transformed MCF-10A cells have a progressively tumor-like phenotype in three-dimensional basement membrane culture. Cancer Cell Int. 2009, 9, 7. [Google Scholar] [CrossRef]
- Bistulfi, G.; Pozzi, S.; Ren, M.; Rossetti, S.; Sacchi, N. A repressive epigenetic domino effect confers susceptibility to breast epithelial cell transformation: Implications for predicting breast cancer risk. Cancer Res. 2006, 66, 10308–10314. [Google Scholar] [CrossRef]
- Corlazzoli, F.; Rossetti, S.; Bistulfi, G.; Ren, M.; Sacchi, N. Derangement of a factor upstream of RARalpha triggers the repression of a pleiotropic epigenetic network. PLoS ONE 2009, 4, e4305. [Google Scholar] [CrossRef]
- Rossetti, S.; Hoogeveen, A.T.; Esposito, J.; Sacchi, N. Loss of MTG16a (CBFA2T3), a novel rDNA repressor, leads to increased ribogenesis and disruption of breast acinar morphogenesis. J. Cell Mol. Med. 2010, 14, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, S.; Corlazzoli, F.; Gregorski, A.; Azmi, N.H.; Sacchi, N. Identification of an estrogen-regulated circadian mechanism necessary for breast acinar morphogenesis. Cell Cycle 2012, 11, 3691–3700. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, S.; Bshara, W.; Reiners, J.A.; Corlazzoli, F.; Miller, A.; Sacchi, N. Harnessing 3D models of mammary epithelial morphogenesis: An off the beaten path approach to identify candidate biomarkers of early stage breast cancer. Cancer Lett. 2016, 380, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, S.; Sacchi, N. 3D mammary epithelial cell models: A goldmine of DCIS Biomarkers and morphogenetic mechanisms. Cancers 2019, 11, 130. [Google Scholar] [CrossRef] [PubMed]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef]
- Ma, I.; Allan, A.L. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. Rep. 2011, 7, 292–306. [Google Scholar] [CrossRef]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef]
- Badve, S.S.; Gokmen-Polar, Y. Ductal carcinoma in situ of breast: Update 2019. Pathology 2019, 51, 563–569. [Google Scholar] [CrossRef]
- Peng, X.; Yun, D.; Christov, K. Breast cancer progression in MCF10A series of cell lines is associated with alterations in retinoic acid and retinoid X receptors and with differential response to retinoids. Int. J. Oncol. 2004, 25, 961–971. [Google Scholar]
- Stein, T.; Morris, J.S.; Davies, C.R.; Weber-Hall, S.J.; Duffy, M.A.; Heath, V.J.; Bell, A.K.; Ferrier, R.K.; Sandilands, G.P.; Gusterson, B.A. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004, 6, R75–R91. [Google Scholar] [CrossRef]
- Stein, T.; Price, K.N.; Morris, J.S.; Heath, V.J.; Ferrier, R.K.; Bell, A.K.; Pringle, M.A.; Villadsen, R.; Petersen, O.W.; Sauter, G.; et al. Annexin A8 is up-regulated during mouse mammary gland involution and predicts poor survival in breast cancer. Clin. Cancer Res. 2005, 11, 6872–6879. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, J.M.; Cairney, C.J.; Ferrier, R.K.; McDonald, L.; Soady, K.; Kendrick, H.; Pringle, M.A.; Morgan, R.O.; Martin, F.; Smalley, M.J.; et al. Annexin A8 identifies a subpopulation of transiently quiescent c-kit positive luminal progenitor cells of the ductal mammary epithelium. PLoS ONE 2015, 10, e0119718. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Gonzalez, I.; Bobien, A.; Molnar, C.; Schmid, S.; Strotbek, M.; Boerries, M.; Busch, H.; Olayioye, M.A. miR-149 suppresses breast cancer metastasis by blocking paracrine interactions with macrophages. Cancer Res. 2020, 80, 1330–1341. [Google Scholar] [CrossRef] [PubMed]
- Romero-Cordoba, S.L.; Rodriguez-Cuevas, S.; Bautista-Pina, V.; Maffuz-Aziz, A.; D’Ippolito, E.; Cosentino, G.; Baroni, S.; Iorio, M.V.; Hidalgo-Miranda, A. Loss of function of miR-342-3p results in MCT1 over-expression and contributes to oncogenic metabolic reprogramming in triple negative breast cancer. Sci. Rep. 2018, 8, 12252. [Google Scholar] [CrossRef] [PubMed]
- Young, J.; Kawaguchi, T.; Yan, L.; Qi, Q.; Liu, S.; Takabe, K. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget 2017, 8, 99978–99989. [Google Scholar] [CrossRef]
- Halvaei, S.; Daryani, S.; Eslami, S.Z.; Samadi, T.; Jafarbeik-Iravani, N.; Bakhshayesh, T.O.; Majidzadeh, A.K.; Esmaeili, R. Exosomes in cancer liquid biopsy: A focus on breast cancer. Mol. Ther. Nucleic Acids 2018, 10, 131–141. [Google Scholar] [CrossRef]
- Alimirzaie, S.; Bagherzadeh, M.; Akbari, M.R. Liquid biopsy in breast cancer: A comprehensive review. Clin. Genet. 2019, 95, 643–660. [Google Scholar] [CrossRef]
- Lennon, A.M.; Buchanan, A.H.; Kinde, I.; Warren, A.; Honushefsky, A.; Cohain, A.T.; Ledbetter, D.H.; Sanfilippo, F.; Sheridan, K.; Rosica, D.; et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 2020. [Google Scholar] [CrossRef]
- Chen, S.L.; Chen, C.Y.; Hsieh, J.C.; Yu, Z.Y.; Cheng, S.J.; Hsieh, K.Y.; Yang, J.W.; Kumar, P.V.; Lin, S.F.; Chen, G.Y. Graphene oxide-based biosensors for liquid biopsies in cancer diagnosis. Nanomaterials 2019, 9, 1725. [Google Scholar] [CrossRef]
- Yang, Y.; Kannisto, E.; Yu, G.; Reid, M.E.; Patnaik, S.K.; Wu, Y. An immuno-biochip selectively captures tumor-derived exosomes and detects exosomal RNAs for cancer diagnosis. ACS Appl. Mater. Interfaces 2018, 10, 43375–43386. [Google Scholar] [CrossRef]
- Ren, M.; Pozzi, S.; Bistulfi, G.; Somenzi, G.; Rossetti, S.; Sacchi, N. Impaired retinoic acid (RA) signal leads to RARbeta2 epigenetic silencing and RA resistance. Mol. Cell Biol. 2005, 25, 10591–10603. [Google Scholar] [CrossRef] [PubMed]
- Sirchia, S.M.; Ferguson, A.T.; Sironi, E.; Subramanyan, S.; Orlandi, R.; Sukumar, S.; Sacchi, N. Evidence of epigenetic changes affecting the chromatin state of the retinoic acid receptor beta2 promoter in breast cancer cells. Oncogene 2000, 19, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Sirchia, S.M.; Ren, M.; Pili, R.; Sironi, E.; Somenzi, G.; Ghidoni, R.; Toma, S.; Nicolo, G.; Sacchi, N. Endogenous reactivation of the RARbeta2 tumor suppressor gene epigenetically silenced in breast cancer. Cancer Res. 2002, 62, 2455–2461. [Google Scholar] [PubMed]
- Evron, E.; Dooley, W.C.; Umbricht, C.B.; Rosenthal, D.; Sacchi, N.; Gabrielson, E.; Soito, A.B.; Hung, D.T.; Ljung, B.; Davidson, N.E.; et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet 2001, 357, 1335–1336. [Google Scholar] [CrossRef]
- Bean, G.R.; Scott, V.; Yee, L.; Ratliff-Daniel, B.; Troch, M.M.; Seo, P.; Bowie, M.L.; Marcom, P.K.; Slade, J.; Kimler, B.F.; et al. Retinoic acid receptor-beta2 promoter methylation in random periareolar fine needle aspiration. Cancer Epidemiol Biomark. Prev. 2005, 14, 790–798. [Google Scholar] [CrossRef][Green Version]
- Fang, C.; Jian, Z.Y.; Shen, X.F.; Wei, X.M.; Yu, G.Z.; Zeng, X.T. Promoter methylation of the retinoic acid receptor beta2 (RARbeta2) Is associated with increased risk of breast cancer: A PRISMA compliant meta-analysis. PLoS ONE 2015, 10, e0140329. [Google Scholar] [CrossRef]
- Kim, J.; Piao, H.L.; Kim, B.J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef]
- Tang, J.; Zhong, G.; Zhang, H.; Yu, B.; Wei, F.; Luo, L.; Kang, Y.; Wu, J.; Jiang, J.; Li, Y.; et al. LncRNA DANCR upregulates PI3K/AKT signaling through activating serine phosphorylation of RXRA. Cell Death Dis. 2018, 9, 1167. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, H.; Yan, G.; Wu, T.; Liu, S.; Chen, W.; Ning, Y.; Lu, Z. Long non-coding RNA and breast cancer. Technol. Cancer Res. Treat. 2019, 18, 860–863. [Google Scholar] [CrossRef]
- Tang, T.; Guo, C.; Xia, T.; Zhang, R.; Zen, K.; Pan, Y.; Jin, L. LncCCAT1 promotes breast cancer stem cell function through activating WNT/beta-catenin signaling. Theranostics 2019, 9, 7384–7402. [Google Scholar] [CrossRef]
- Lueck, K.; Carr, A.F.; Yu, L.; Greenwood, J.; Moss, S.E. Annexin A8 regulates wnt signaling to maintain the phenotypic plasticity of retinal pigment epithelial cells. Sci. Rep. 2020, 10, 1256. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.V.; Hogdall, C.K.; Jochumsen, K.M.; Hogdall, E.V.S. Annexin A2 and cancer: A systematic review. Int. J. Oncol. 2018, 52, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Rhodes, D.R.; Ingold, C.; Chinnaiyan, A.M.; Rubin, M.A. Dysregulation of the annexin family protein family is associated with prostate cancer progression. Am. J. Pathol. 2003, 162, 255–261. [Google Scholar] [CrossRef]
- Liu, J.W.; Shen, J.J.; Tanzillo-Swarts, A.; Bhatia, B.; Maldonado, C.M.; Person, M.D.; Lau, S.S.; Tang, D.G. Annexin II expression is reduced or lost in prostate cancer cells and its re-expression inhibits prostate cancer cell migration. Oncogene 2003, 22, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Ardura, J.A.; Alvarez-Carrion, L.; Gutierrez-Rojas, I.; Alonso, V. Role of calcium signaling in prostate cancer progression: Effects on cancer hallmarks and bone metastatic mechanisms. Cancers 2020, 12, 1071. [Google Scholar] [CrossRef] [PubMed]
- Labrecque, M.P.; Coleman, I.M.; Brown, L.G.; True, L.D.; Kollath, L.; Lakely, B.; Nguyen, H.M.; Yang, Y.C.; da Costa, R.M.G.; Kaipainen, A.; et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Investig. 2019, 129, 4492–4505. [Google Scholar] [CrossRef]
- Gou, R.; Zhu, L.; Zheng, M.; Guo, Q.; Hu, Y.; Li, X.; Liu, J.; Lin, B. Annexin A8 can serve as potential prognostic biomarker and therapeutic target for ovarian cancer: Based on the comprehensive analysis of Annexins. J. Transl. Med. 2019, 17, 275. [Google Scholar] [CrossRef]
- Lokman, N.A.; Ho, R.; Gunasegaran, K.; Bonner, W.M.; Oehler, M.K.; Ricciardelli, C. Anti-tumour effects of all-trans retinoid acid on serous ovarian cancer. J. Exp. Clin. Cancer Res. 2019, 38, 10. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef]
- Gress, T.M.; Wallrapp, C.; Frohme, M.; Muller-Pillasch, F.; Lacher, U.; Friess, H.; Buchler, M.; Adler, G.; Hoheisel, J.D. Identification of genes with specific expression in pancreatic cancer by cDNA representational difference analysis. Genes Chromosomes Cancer 1997, 19, 97–103. [Google Scholar] [CrossRef]
- Logsdon, C.D.; Simeone, D.M.; Binkley, C.; Arumugam, T.; Greenson, J.K.; Giordano, T.J.; Misek, D.E.; Kuick, R.; Hanash, S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003, 63, 2649–2657. [Google Scholar] [PubMed]
- Karanjawala, Z.E.; Illei, P.B.; Ashfaq, R.; Infante, J.R.; Murphy, K.; Pandey, A.; Schulick, R.; Winter, J.; Sharma, R.; Maitra, A.; et al. New markers of pancreatic cancer identified through differential gene expression analyses: Claudin 18 and annexin A8. Am. J. Surg. Pathol. 2008, 32, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Hata, H.; Tatemichi, M.; Nakadate, T. Involvement of annexin A8 in the properties of pancreatic cancer. Mol. Carcinog. 2014, 53, 181–191. [Google Scholar] [CrossRef]
- Pimiento, J.M.; Chen, D.T.; Centeno, B.A.; Davis-Yadley, A.H.; Husain, K.; Fulp, W.J.; Wang, C.; Zhang, A.; Malafa, M.P. Annexin A8 is a prognostic marker and potential therapeutic target for pancreatic cancer. Pancreas 2015, 44, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Inaguma, S.; Ito, H.; Riku, M.; Ikeda, H.; Kasai, K. Addiction of pancreatic cancer cells to zinc-finger transcription factor ZIC2. Oncotarget 2015, 6, 28257–28268. [Google Scholar] [CrossRef]
- Inaguma, S.; Riku, M.; Ito, H.; Tsunoda, T.; Ikeda, H.; Kasai, K. GLI1 orchestrates CXCR4/CXCR7 signaling to enhance migration and metastasis of breast cancer cells. Oncotarget 2015, 6, 33648–33657. [Google Scholar] [CrossRef]
- Deng, P.C.; Chen, W.B.; Cai, H.H.; An, Y.; Wu, X.Q.; Chen, X.M.; Sun, D.L.; Yang, Y.; Shi, L.Q. LncRNA SNHG14 potentiates pancreatic cancer progression via modulation of annexin A2 expression by acting as a competing endogenous RNA for miR-613. J. Cell Mol. Med. 2019, 23, 7222–7232. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- Ma, F.; Li, X.; Fang, H.; Jin, Y.; Sun, Q. Prognostic value of ANXA8 in gastric carcinoma. J. Cancer 2020, 11, 3551–3558. [Google Scholar] [CrossRef]
- Yoo, H.J.; Yun, B.R.; Kwon, J.H.; Ahn, H.S.; Seol, M.A.; Lee, M.J.; Yu, G.R.; Yu, H.C.; Hong, B.; Choi, K.; et al. Genetic and expression alterations in association with the sarcomatous change of cholangiocarcinoma cells. Exp. Mol. Med. 2009, 41, 102–115. [Google Scholar] [CrossRef]
- Lee, M.J.; Yu, G.R.; Yoo, H.J.; Kim, J.H.; Yoon, B.I.; Choi, Y.K.; Kim, D.G. ANXA8 down-regulation by EGF-FOXO4 signaling is involved in cell scattering and tumor metastasis of cholangiocarcinoma. Gastroenterology 2009, 137, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Oka, R.; Nakashiro, K.; Goda, H.; Iwamoto, K.; Tokuzen, N.; Hamakawa, H. Annexin A8 is a novel molecular marker for detecting lymph node metastasis in oral squamous cell carcinoma. Oncotarget 2016, 7, 4882–4889. [Google Scholar] [CrossRef] [PubMed]
- Staquicini, D.I.; Rangel, R.; Guzman-Rojas, L.; Staquicini, F.I.; Dobroff, A.S.; Tarleton, C.A.; Ozbun, M.A.; Kolonin, M.G.; Gelovani, J.G.; Marchio, S.; et al. Intracellular targeting of annexin A2 inhibits tumor cell adhesion, migration, and in vivo grafting. Sci. Rep. 2017, 7, 4243. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossetti, S.; Sacchi, N. Emerging Cancer Epigenetic Mechanisms Regulated by All-Trans Retinoic Acid. Cancers 2020, 12, 2275. https://doi.org/10.3390/cancers12082275
Rossetti S, Sacchi N. Emerging Cancer Epigenetic Mechanisms Regulated by All-Trans Retinoic Acid. Cancers. 2020; 12(8):2275. https://doi.org/10.3390/cancers12082275
Chicago/Turabian StyleRossetti, Stefano, and Nicoletta Sacchi. 2020. "Emerging Cancer Epigenetic Mechanisms Regulated by All-Trans Retinoic Acid" Cancers 12, no. 8: 2275. https://doi.org/10.3390/cancers12082275
APA StyleRossetti, S., & Sacchi, N. (2020). Emerging Cancer Epigenetic Mechanisms Regulated by All-Trans Retinoic Acid. Cancers, 12(8), 2275. https://doi.org/10.3390/cancers12082275



