Metabolites as Prognostic Markers for Metastatic Non-Small Cell Lung Cancer (NSCLC) Patients Treated with First-Line Platinum-Doublet Chemotherapy
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
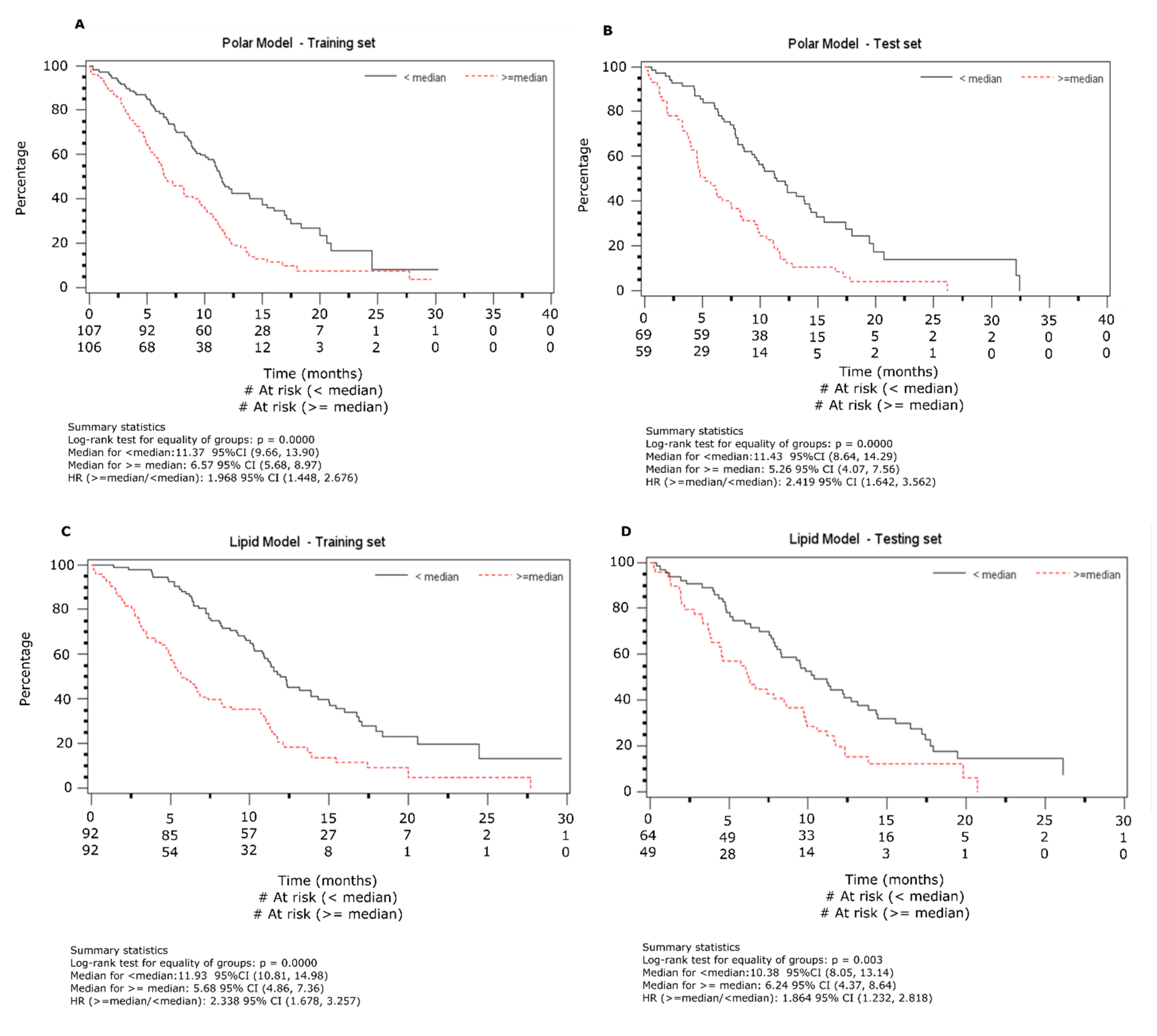
2.2. Overall Survival Analysis from NMR Data
2.3. Overall Survival Analysis from Lipid Data
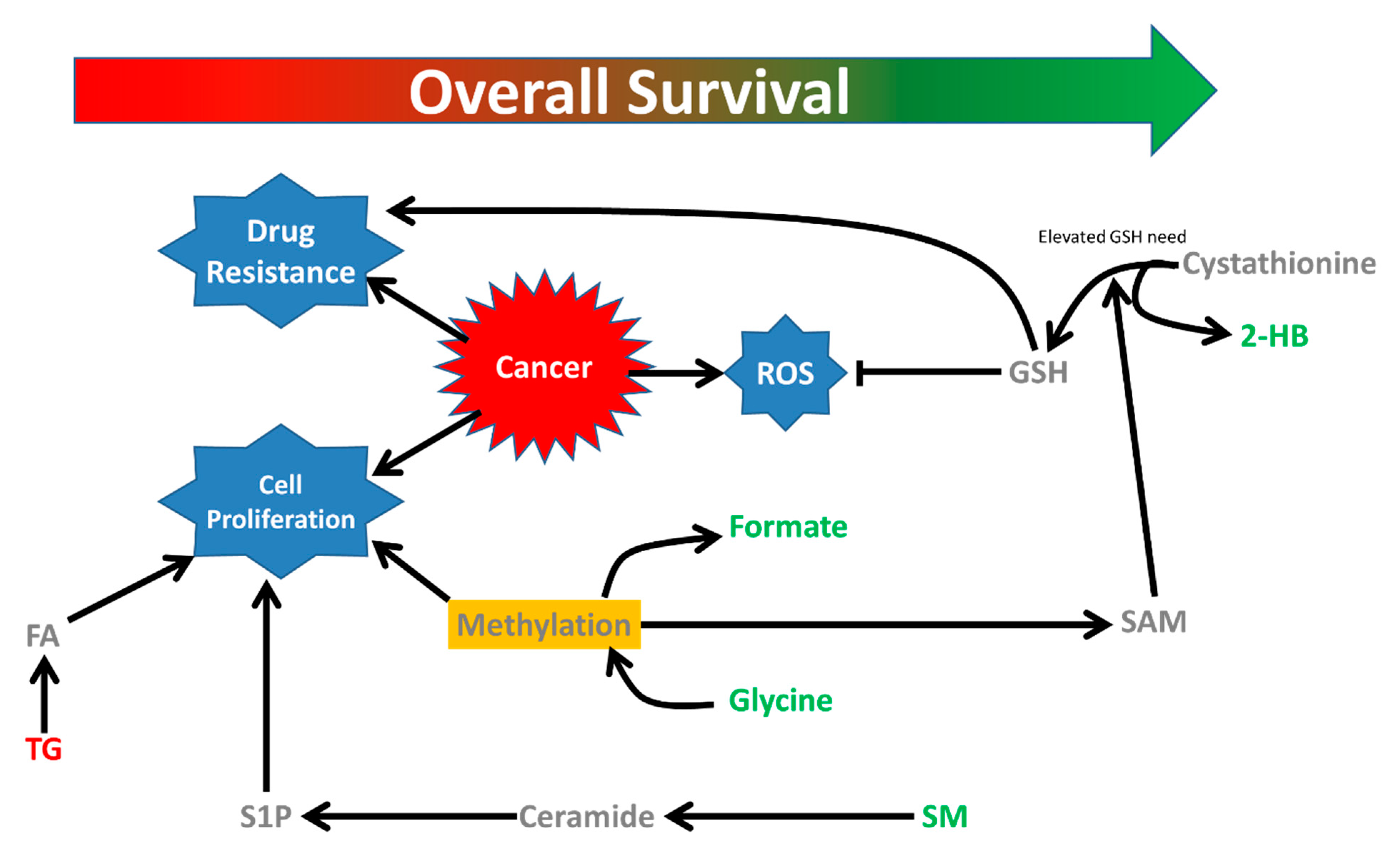
3. Discussion
4. Methods and Materials
4.1. Patients
4.2. Sample Preparation
4.3. NMR Spectroscopy
4.4. Lipidomics Assay by UPLC-Qtof-MS
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability
References
- Hori, S.; Nishiumi, S.; Kobayashi, K.; Shinohara, M.; Hatakeyama, Y.; Kotani, Y.; Hatano, N.; Maniwa, Y.; Nishio, W.; Bamba, T.; et al. A metabolomic approach to lung cancer. Lung Cancer 2011, 74, 284–292. [Google Scholar] [CrossRef]
- Jordan, K.W.; Adkins, C.B.; Su, L.; Halpern, E.F.; Mark, E.J.; Christiani, D.C.; Cheng, L.L. Comparison of squamous cell carcinoma and adenocarcinoma of the lung by metabolomic analysis of tissue-serum pairs. Lung Cancer 2010, 68, 44–50. [Google Scholar] [CrossRef]
- Deja, S.; Porebska, I.; Kowal, A.; Zabek, A.; Barg, W.; Pawelczyk, K.; Stanimirova, I.; Daszykowski, M.; Korzeniewska, A.; Jankowska, R.; et al. Metabolomics provide new insights on lung cancer staging and discrimination from chronic obstructive pulmonary disease. J. Pharm. Biomed. Anal. 2014, 100, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Sarfaraz, M.O.; Farshidfar, F.; Bebb, D.G.; Lee, C.Y.; Card, C.M.; David, M.; Weljie, A.M. Temporal characterization of serum metabolite signatures in lung cancer patients undergoing treatment. Metabolomics 2016, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Leighl, N.B.; Paz-Ares, L.; Douillard, J.-Y.; Peschel, C.; Arnold, A.; Depeirre, A.; Santoro, A.; Betticher, D.C.; Gatzemeier, U.; Jassem, J.; et al. Randomized Phase III Study of Matrix Metalloproteinase Inhibitor BMS-275291 in Combination With Paclitaxel and Carboplatin in Advanced Non-Small-Cell Lung Cancer: National Cancer Institute of Canada-Clinical Trials Group Study BR.18. J. Clin. Oncol. 2005, 23, 2831–2839. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wang, Z.; Liu, X.; Duan, J.; Feng, G.; Yin, Y.; Gu, J.; Chen, Z.; Gao, S.; Bai, H.; et al. Prediction of chemotherapeutic efficacy in non–small cell lung cancer by serum metabolomic profiling. Clin. Cancer Res. 2018, 24, 2100–2109. [Google Scholar] [CrossRef]
- Gall, W.E.; Beebe, K.; Lawton, K.A.; Adam, K.P.; Mitchell, M.W.; Nakhle, P.J.; Ryals, J.A.; Milburn, M.V.; Nannipieri, M.; Camastra, S.; et al. α-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS ONE 2010, 5, e10883. [Google Scholar] [CrossRef]
- Sengupta, R. Thioredoxin and glutaredoxin-mediated redox regulation of ribonucleotide reductase. World J. Biol. Chem. 2014, 5, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Harris, I.S.; Treloar, A.E.; Inoue, S.; Sasaki, M.; Gorrini, C.; Lee, K.C.; Yung, K.Y.; Brenner, D.; Knobbe-Thomsen, C.B.; Cox, M.A.; et al. Glutathione and Thioredoxin Antioxidant Pathways Synergize to Drive Cancer Initiation and Progression. Cancer Cell 2015, 27, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Ishida, T.; Sugio, K.; Maehara, Y.; Sugimachi, K. Glutathione S transferase Pi Is a powerful indicator in chemotherapy of human lung squamous-cell carcinoma. Respiration 1995, 62, 223–227. [Google Scholar] [CrossRef]
- Bai, F.; Nakanishi, Y.; Kawasaki, M.; Takayama, K.; Yatsunami, J.; Pei, X.H.; Tsuruta, N.; Wakamatsu, K.; Hara, N. Immunohistochemical expression of glutathione S-transferase-Pi can predict chemotherapy response in patients with nonsmall cell lung carcinoma. Cancer 1996, 78, 416–421. [Google Scholar] [CrossRef]
- Lu, S.C. Glutathione Synthesis. Biochim. Biophys. Acta 2014, 1830, 3143–3153. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.; Maddocks, O. One-carbon metabolism in cancer. Br. J. Cancer 2017, 116, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ye, Y.; Chang, D.W.; Huang, M.; Heymach, J.V.; Roth, J.A.; Wu, X.; Zhao, H. Circulating metabolite profiles to predict overall survival in advanced non-small cell lung cancer patients receiving first-line chemotherapy. Lung Cancer 2017, 114, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, P.; Pasman, W.J.; Van Vliet, T.; Urgert, R.; Katan, M.B. Contribution of caffeine to the homocysteine-raising effect of coffee: A randomized controlled trial in humans. Am. J. Clin. Nutr. 2002, 6, 1244–1248. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kulis, M.; Esteller, M. 2-DNA Methylation and Cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Maddocks, O.; Labuschagne, C.; Adams, P.; Vousden, K. Serine metabolism supports the methionine cycle and DNA/RNA Methylation through de novo ATP synthesis in cancer cells. Mol. Cell 2016, 61, 210–221. [Google Scholar] [CrossRef]
- Mullen, T.D.; Hannun, Y.A.; Obeid, L.M. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem. J. 2012, 441, 789–802. [Google Scholar] [CrossRef]
- Ponnusamy, S.; Meyers-Needham, M.; Senkal, C.E.; Saddoughi, S.A.; Sentelle, D.; Selvam, S.P.; Salas, A.; Ogretmen, B. Sphingolipids and cancer: Ceramide and sphingosine-1-phosphate in the regulation of cell death and drug resistance. Future Oncol. 2010, 6, 1603–1624. [Google Scholar] [CrossRef]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef] [PubMed]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Puchades-Carrasco, L.; Jantus-Lewintre, E.; Pérez-Rambla, C.; García-García, F.; Lucas, R.; Calabuig, S.; Blasco, A.; Dopazo, J.; Camps, C.; Pineda-Lucena, A. Serum metabolomic profiling facilitates the non-invasive identification of metabolic biomarkers associated with the onset and progression of non-small cell lung cancer. Oncotarget 2016, 7, 12904–12916. [Google Scholar] [CrossRef] [PubMed]
- Musharraf, S.G.; Mazhar, S.; Choudhary, M.I.; Rizi, N. Plasma metabolite profiling and chemometric analyses of lung cancer along with three controls through gas chromatography-mass spectrometry. Sci. Rep. 2015, 5, 8607. [Google Scholar] [CrossRef]
- Salvador, M.M.; de Cedrón, M.G.; Rubio, J.M.; Martínez, S.F.; Martínez, R.S.; Casado, E.; de Molina, A.R.; Sereno, M. Lipid metabolism and lung cancer. Crit. Rev. Oncol. Hematol. 2017, 112, 31–40. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Prot. 2007, 2, 2692–2703. [Google Scholar] [CrossRef]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, E.; Slupsky, C.M. Targeted Profiling: Quantitative Analysis of 1 H NMR Metabolomics Data. Anal. Chem. 2006, 78, 4430–4442. [Google Scholar] [CrossRef]

| A. Polar Metabolites Training Set | |||
| Factors | Hazard ratio | 95% C.I. of HR | p-Value |
| Risk group High Low | 2.16 1 | 1.57–2.97 | <0.0001 |
| ECOG Performance status 2, 3 0, 1 | 1.47 1 | 0.89–2.41 | 0.13 |
| Weight loss ≥5% <5% | 1.89 1 | 1.35–2.66 | 0.0004 |
| Sex Male Female | 1.30 1 | 0.87–1.94 | 0.20 |
| Disease Stage III IV | 1.20 1 | 0.81–1.78 | 0.35 |
| Histology Squamous Non-Squamous | 0.99 1 | 0.70–1.42 1 | 0.98 |
| Hemoglobin 1 Grade 1+ Grade 0 | 1.51 1 | 1.18–2.23 | 0.01 |
| B. Polar Metabolites Test Set | |||
| Factors | Hazard ratio | 95% C.I. of HR | p-Value |
| Risk group High Low | 2.42 1 | 1.61–3.65 | <0.0001 |
| ECOG Performance status 2, 3 0, 1 | 1.74 1 | 0.89–3.42 | 0.11 |
| Weight loss ≥5% <5% | 1.48 1 | 0.93–2.34 | 0.10 |
| Sex Male Female | 0.98 1 | 0.61–1.59 | 0.94 |
| Disease Stage III IV | 0.51 1 | 0.30–0.85 | 0.01 |
| Histology Squamous Non-Squamous | 0.97 1 | 0.59–1.59 1 | 0.90 |
| Hemoglobin 1 Grade 1+ Grade 0 | 1.59 | 1.06–2.68 | 0.04 |
| A. Lipids Training Set | |||
| Factors | Hazard ratio | 95% C.I. of HR | p-Value |
| Risk group High Low | 2.23 1 | 1.55–3.22 | <0.0001 |
| ECOG Performance status 2, 3 0, 1 | 1.51 1 | 0.87–2.64 | 0.14 |
| Weight loss ≥5% <5% | 1.93 1 | 1.31–2.85 | 0.0009 |
| Sex Male Female | 1.62 1 | 1.02–2.56 | 0.04 |
| Disease Stage III IV | 1.20 1 | 0.80–1.81 | 0.38 |
| Histology Squamous Non-Squamous | 0.95 1 | 0.60–1.49 1 | 0.81 |
| Hemoglobin 1 Grade 1+ Grade 0 | 1.52 1 | 1.12–2.34 | 0.03 |
| B. Lipids Test Set | |||
| Factors | Hazard ratio | 95% C.I. of HR | p-Value |
| Risk group High Low | 1.83 1 | 1.19–3.22 | 0.006 |
| ECOG Performance status 2, 3 0, 1 | 1.70 1 | 0.75–3.86 | 0.21 |
| Weight loss ≥5% <5% | 1.55 1 | 0.86–2.79 | 0.15 |
| Sex Male Female | 1.11 1 | 0.61–2.02 | 0.83 |
| Disease Stage III IV | 0.58 1 | 0.32–1.08 | 0.09 |
| Histology Squamous Non-Squamous | 0.85 1 | 0.45–1.61 | 0.62 |
| Hemoglobin 1 Grade 1+ Grade 0 | 1.60 1 | 1.03–2.68 | 0.04 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, D.; Sengupta, A.; Ding, K.; Ubeydullah, E.; Krishnaiah, S.; Leighl, N.B.; Shepherd, F.A.; Seymour, L.; Weljie, A. Metabolites as Prognostic Markers for Metastatic Non-Small Cell Lung Cancer (NSCLC) Patients Treated with First-Line Platinum-Doublet Chemotherapy. Cancers 2020, 12, 1926. https://doi.org/10.3390/cancers12071926
Hao D, Sengupta A, Ding K, Ubeydullah E, Krishnaiah S, Leighl NB, Shepherd FA, Seymour L, Weljie A. Metabolites as Prognostic Markers for Metastatic Non-Small Cell Lung Cancer (NSCLC) Patients Treated with First-Line Platinum-Doublet Chemotherapy. Cancers. 2020; 12(7):1926. https://doi.org/10.3390/cancers12071926
Chicago/Turabian StyleHao, Desirée, Arjun Sengupta, Keyue Ding, ER Ubeydullah, Saikumari Krishnaiah, Natasha B. Leighl, Frances A. Shepherd, Lesley Seymour, and Aalim Weljie. 2020. "Metabolites as Prognostic Markers for Metastatic Non-Small Cell Lung Cancer (NSCLC) Patients Treated with First-Line Platinum-Doublet Chemotherapy" Cancers 12, no. 7: 1926. https://doi.org/10.3390/cancers12071926
APA StyleHao, D., Sengupta, A., Ding, K., Ubeydullah, E., Krishnaiah, S., Leighl, N. B., Shepherd, F. A., Seymour, L., & Weljie, A. (2020). Metabolites as Prognostic Markers for Metastatic Non-Small Cell Lung Cancer (NSCLC) Patients Treated with First-Line Platinum-Doublet Chemotherapy. Cancers, 12(7), 1926. https://doi.org/10.3390/cancers12071926







