Cylindromatosis Lysine 63 Deubiquitinase (CYLD) Regulates NF-kB Signaling Pathway and Modulates Fibroblast and Endothelial Cells Recruitment in Nasopharyngeal Carcinoma
Abstract
1. Introduction
2. Results
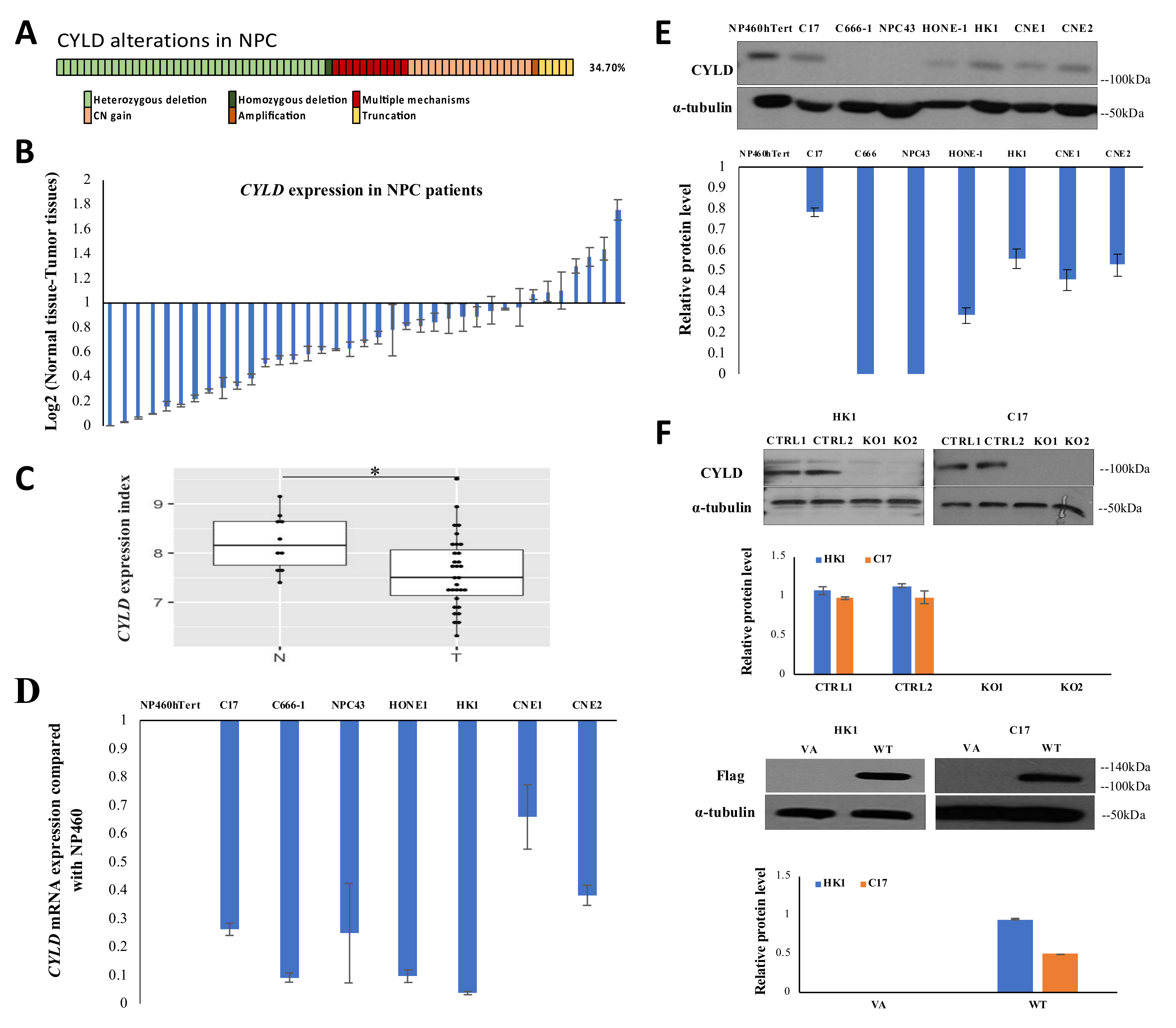
2.1. Integrative Genomic Analysis Reveals Multiple Mechanisms Leading to CYLD Inactivation and Down-Regulation in NPC
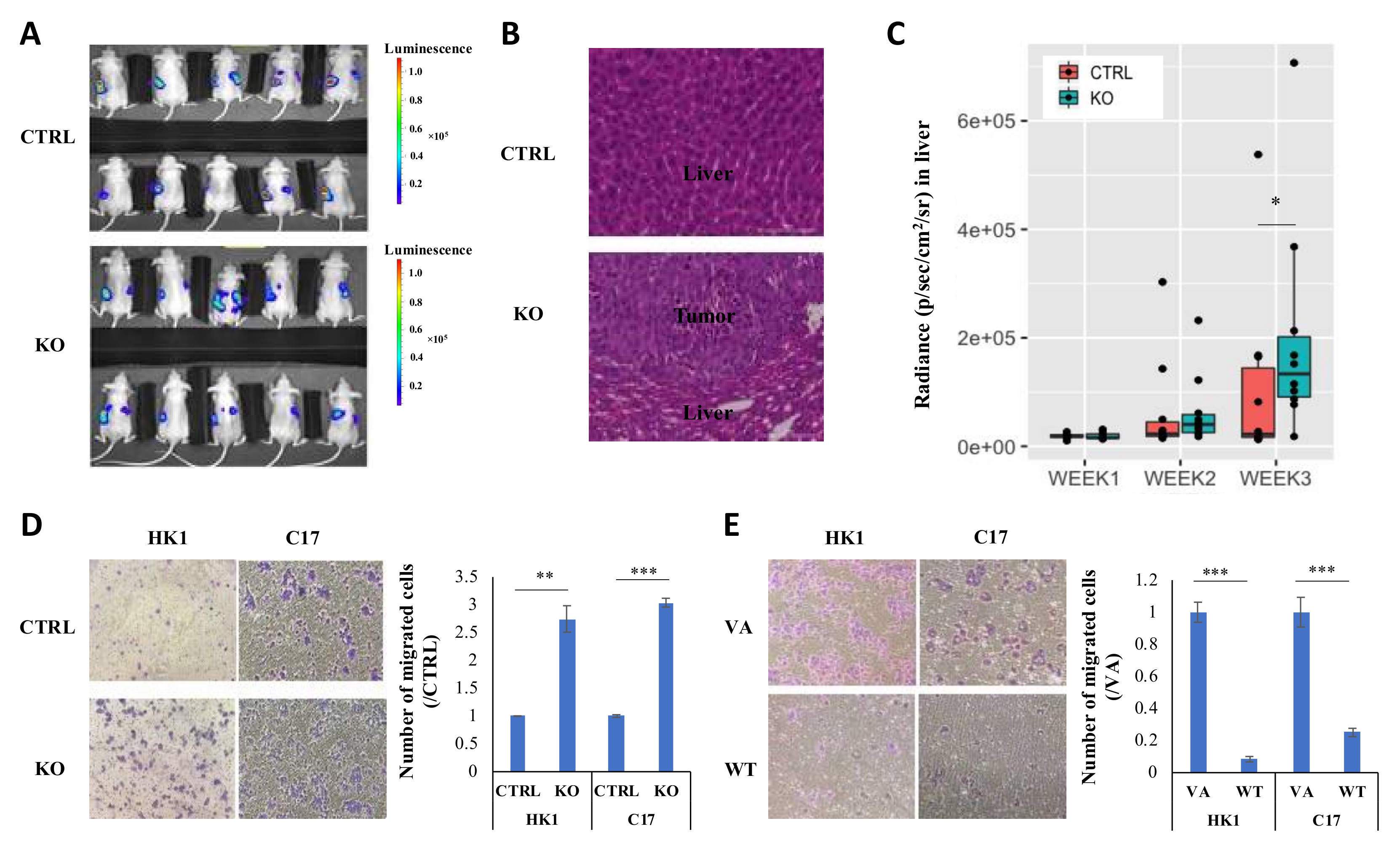
2.2. CYLD Expression Level Affects NPC Tumor Growth In Vivo and Cell Proliferation In Vitro
2.3. CYLD Expression Level Affects NPC Metastasis In Vivo and Migration In Vitro
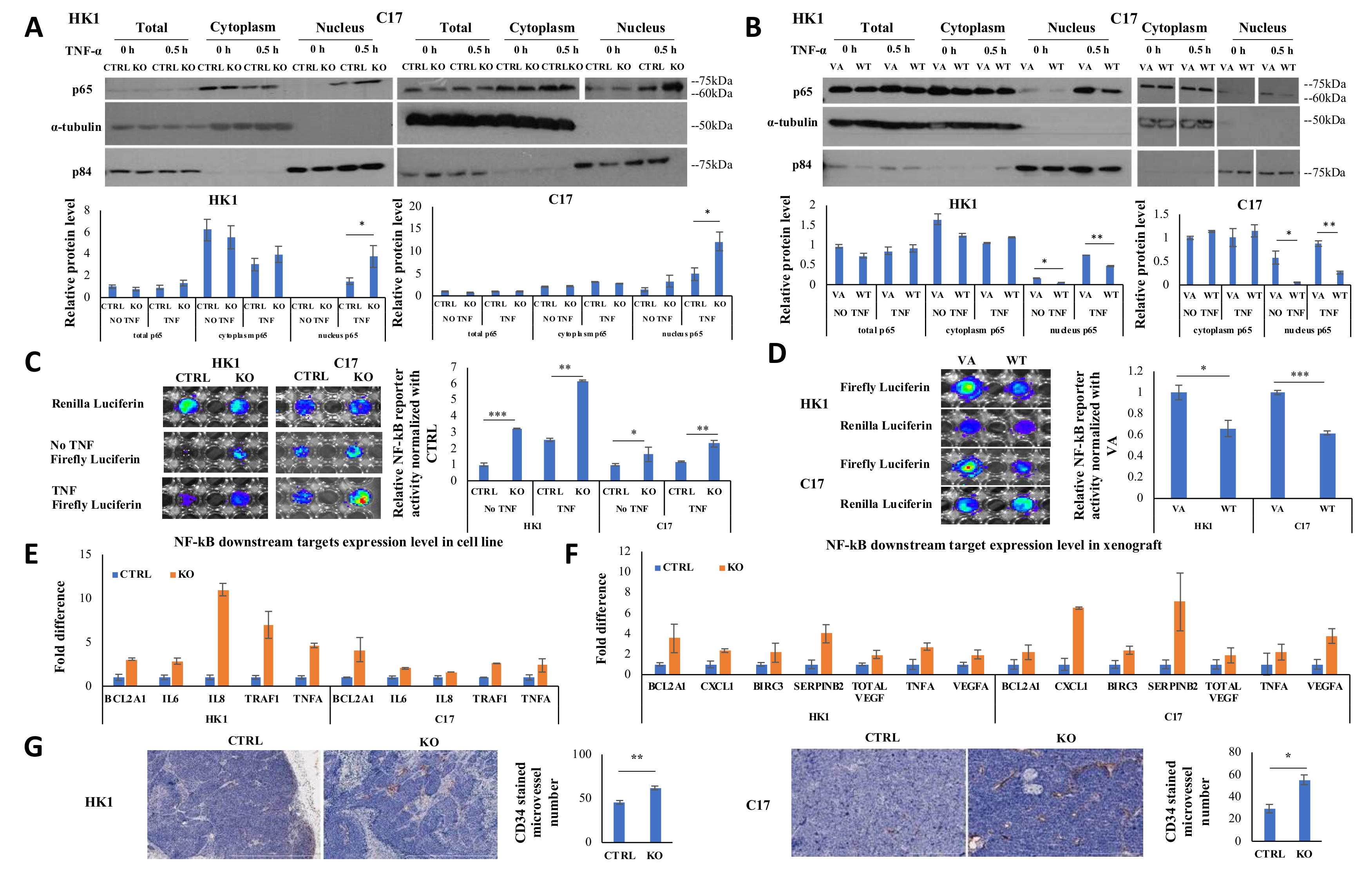
2.4. CYLD Suppresses NF-kB Signaling Pathway via Inhibiting p65 Nuclear Translocation
2.5. CYLD Expression Affects NF-kB Transcriptional Function and CYLD Knockout Increases Angiogenesis
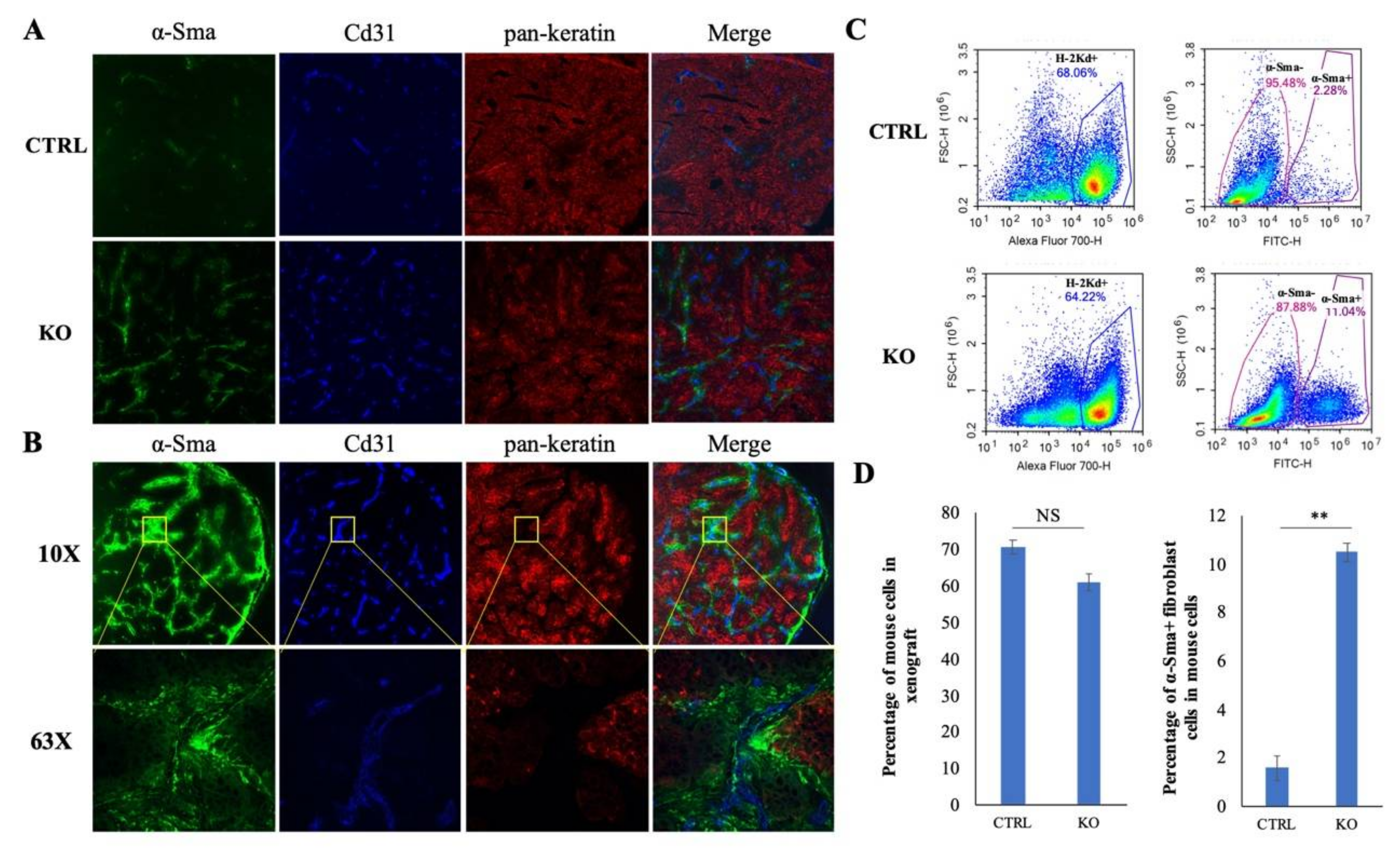
2.6. Single Cell Analysis Shows CYLD Suppresses Specific Stromal Cell Infiltration in the TME
2.7. CYLD Suppresses Fibroblast and Endothelial Cell Infiltration in NPC TME
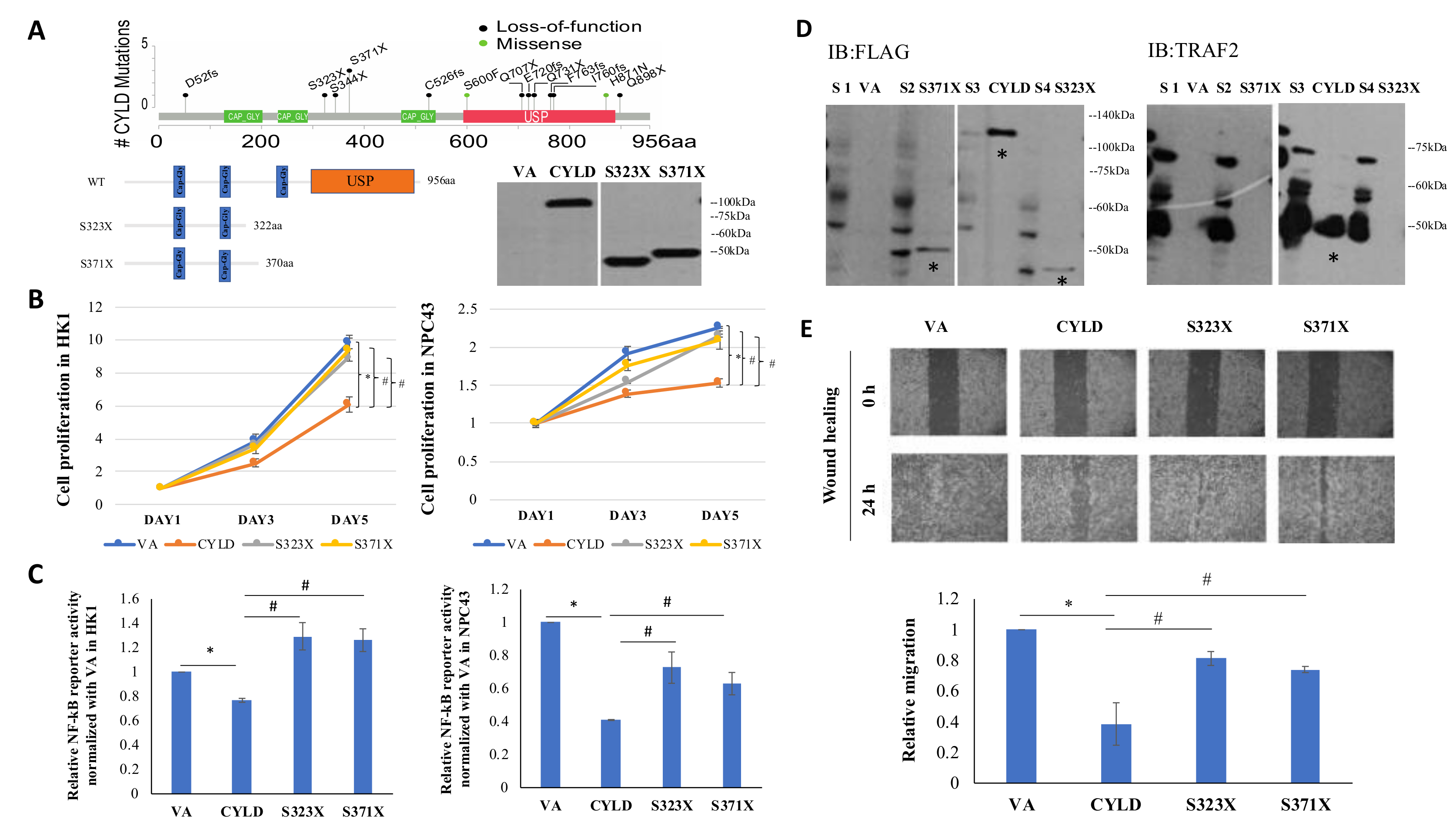
2.8. CYLD Truncation Mutations (S323X, S371X) Abolish Wild-Type (WT) Function as a NF-kB Inhibitor
3. Discussion
4. Materials and Methods
4.1. NPC Patient Specimens
4.2. Genomic Analysis in NPC
4.3. Cell Lines
4.4. Plasmids and Lentivirus Preparation and Infection
4.5. Real-Time Quantitative PCR
4.6. Tumorigenicity in Nude Mice and Intrasplenic Metastasis Assay
4.7. Immunohistochemistry Staining
4.8. Cell Proliferation Assay and 3D Colony Formation
4.9. Western Blot and Antibodies
4.10. Subcellular Fractionation
4.11. Co-Immunoprecipitation Assay
4.12. Wound Healing and Chamber Migration Assay
4.13. NF-kB-Specific Single Plasmid Dual Reporter Assay
4.14. 10× Genomics Single Cell RNA Sequencing and Data Analysis
4.15. Immunofluorescence Staining of Cryosected Tissue Sections
4.16. Flow Cytometry of Single Cells Dissociated from Xenograft
4.17. Statistical Analysis
4.18. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lin, J.-H.; Jiang, C.-Q.; Ho, S.Y.; Zhang, W.S.; Mai, Z.-M.; Xu, L.; Lo, C.-M.; Lam, T.H. Smoking and nasopharyngeal carcinoma mortality: A cohort study of 101,823 adults in Guangzhou, China. BMC Cancer 2015, 15, 906. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M.; Muir, C.S. Cancer Incidence in Five Continents. Comparability and quality of data. IARC Sci. Publ. 1992, 120, 45–173. [Google Scholar]
- Biggs, P.J.; Wooster, R.; Ford, D.; Chapman, P.; Mangion, J.; Quirk, Y.; Easton, D.F.; Burn, J.; Stratton, M.R. Familial cylindromatosis (turban tumour syndrome) gene localised to chromosome 16q12–16q13: Evidence for its role as a tumour suppressor gene. Nat. Genet. 1995, 11, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Massoumi, R. CYLD: A deubiquitination enzyme with multiple roles in cancer. Futur. Oncol. 2011, 7, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Dai, W.; Cheung, A.K.; Ko, J.M.; Kan, R.; Wong, B.W.; Leong, M.M.; Deng, M.; Kwok, T.C.; Chan, J.Y.; et al. Whole-exome sequencing identifies multiple loss-of-function mutations of NF-kappaB pathway regulators in nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA 2016, 113, 11283–11288. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Kigawa, T.; Koshiba, S.; Sato, K.; Matsuo, Y.; Sakamoto, A.; Takagi, T.; Shirouzu, M.; Yabuki, T.; Nunokawa, E.; et al. The CAP-Gly Domain of CYLD Associates with the Proline-Rich Sequence in NEMO/IKKγ. Structure 2004, 12, 1719–1728. [Google Scholar] [CrossRef]
- Brummelkamp, T.R.; Nijman, S.M.B.; Dirac, A.M.G.; Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 2003, 424, 797–801. [Google Scholar] [CrossRef]
- Trompouki, E.; Hatzivassiliou, E.; Tsichritzis, T.; Farmer, H.; Ashworth, A.; Mosialos, G. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature 2003, 424, 793–796. [Google Scholar] [CrossRef]
- Wright, A.; Reiley, W.W.; Chang, M.; Jin, W.; Lee, A.J.; Zhang, M.; Sun, S.-C. Regulation of Early Wave of Germ Cell Apoptosis and Spermatogenesis by Deubiquitinating Enzyme CYLD. Dev. Cell 2007, 13, 705–716. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.; Karin, M. Regulation and Function of NF-κB Transcription Factors in the Immune System. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef]
- Hoffmann, A.; Baltimore, D. Circuitry of nuclear factor kappaB signaling. Immunol. Rev. 2006, 210, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.K.F.; Lo, K.W.; Tsao, S.W.; Wong, H.L.; Hui, J.W.Y.; To, K.F.; Hayward, S.D.; Chui, Y.L.; Lau, Y.L.; Takada, K.; et al. Epstein-Barr Virus Infection Alters Cellular Signal Cascades in Human Nasopharyngeal Epithelial Cells. Neoplasia 2006, 8, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Vokes, E.E.; Liebowitz, D.N.; Weichselbaum, R.R. Nasopharyngeal carcinoma. Lancet 1997, 350, 1087–1091. [Google Scholar] [CrossRef]
- Young, L.S.; Rickinson, A.B. Epstein–Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.-C.; Meng, X.; Hazawa, M.; Nagata, Y.; Varela, A.M.; Xu, L.; Sato, Y.; Liu, L.-Z.; Ding, L.-W.; Sharma, A.; et al. The genomic landscape of nasopharyngeal carcinoma. Nat. Genet. 2014, 46, 866–871. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chung, G.T.; Lui, V.W.; To, K.F.; Ma, B.B.; Chow, C.; Woo, J.K.; Yip, K.Y.; Seo, J.; Hui, E.P.; et al. Exome and genome sequencing of nasopharynx cancer identifies NF-kappaB pathway activating mutations. Nat. Commun. 2017, 8, 14121. [Google Scholar] [CrossRef]
- Alameda, J.P.; Fernández-Aceñero, M.J.; Quintana, R.M.; Page, A.; Ramírez, Á.; Navarro, M.; Casanova, M.L. Functional Inactivation of CYLD Promotes the Metastatic Potential of Tumor Epidermal Cells. J. Investig. Dermatol. 2013, 133, 1870–1878. [Google Scholar] [CrossRef]
- Sun, S.-C. CYLD: A tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010, 17, 25–34. [Google Scholar] [CrossRef]
- Gringhuis, S.I.; Kaptein, T.M.; Wevers, B.A.; Mesman, A.W.; Geijtenbeek, T.B. Fucose-specific DC-SIGN signalling directs T helper cell type-2 responses via IKKepsilon- and CYLD-dependent Bcl3 activation. Nat. Commun. 2014, 5, 3898. [Google Scholar] [CrossRef]
- Tabruyn, S.P.; Memet, S.; Ave, P.; Verhaeghe, C.; Mayo, K.H.; Struman, I.; Martial, J.A.; Griffioen, A.W. NF-kappaB activation in endothelial cells is critical for the activity of angiostatic agents. Mol. Cancer Ther. 2009, 8, 2645–2654. [Google Scholar] [CrossRef]
- Erez, N.; Truitt, M.; Olson, P.; Hanahan, D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-κB-Dependent Manner. Cancer Cell 2010, 17, 135–147. [Google Scholar] [CrossRef]
- Wang, S.; Ma, N.; Kawanishi, S.; Hiraku, Y.; Oikawa, S.; Xie, Y.; Zhang, Z.; Huang, G.; Murata, M. Relationships of Alpha-SMA-Positive Fibroblasts and SDF-1-Positive Tumor Cells with Neoangiogenesis in Nasopharyngeal Carcinoma. Biomed. Res. Int. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Park, Y.M.; Lee, S.J.; Cho, H.-J.; Kim, D.-H.; Lee, J.-I.; Kang, M.-S.; Seol, H.J.; Shim, Y.M.; Nam, H.; et al. Alpha-Smooth Muscle Actin (ACTA2) Is Required for Metastatic Potential of Human Lung Adenocarcinoma. Clin. Cancer Res. 2013, 19, 5879–5889. [Google Scholar] [CrossRef] [PubMed]
- Anggorowati, N.; Kurniasari, C.R.; Damayanti, K.; Cahyanti, T.; Widodo, I.; Ghozali, A.; Romi, M.M.; Sari, D.C.R.; Arfian, N. Histochemical and Immunohistochemical Study of α-SMA, Collagen, and PCNA in Epithelial Ovarian Neoplasm. Asian Pac. J. Cancer Prev. 2017, 18, 667–671. [Google Scholar] [PubMed]
- Hellerbrand, C.; Bumes, E.; Bataille, F.; Dietmaier, W.; Massoumi, R.; Bosserhoff, A.K. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis 2007, 28, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Massoumi, R.; Kuphal, S.; Hellerbrand, C.; Haas, B.; Wild, P.; Spruss, T.; Pfeifer, A.; Fassler, R.; Bosserhoff, A.K. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J. Exp. Med. 2009, 206, 221–232. [Google Scholar] [CrossRef]
- Hayashi, M.; Jono, H.; Shinriki, S.; Nakamura, T.; Guo, J.; Sueta, A.; Tomiguchi, M.; Fujiwara, S.; Yamamoto-Ibusuki, M.; Murakami, K.-I.; et al. Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Res. Treat. 2014, 143, 447–457. [Google Scholar] [CrossRef]
- Shinriki, S.; Jono, H.; Maeshiro, M.; Nakamura, T.; Guo, J.; Li, J.-D.; Ueda, M.; Yoshida, R.; Shinohara, M.; Nakayama, H.; et al. Loss of CYLD promotes cell invasion via ALK5 stabilization in oral squamous cell carcinoma. J. Pathol. 2018, 244, 367–379. [Google Scholar] [CrossRef]
- Zhao, C.; Qian, G.-Q.; Xiaoyin, C.; Chen, C. Retrograde Analysis of Clinical Characteristics of Bone Metastasis in 1,031 Cases of Preliminarily Diagnosed Nasopharyngeal Carcinoma. Asian Pac. J. Cancer Prev. 2014, 15, 3785–3788. [Google Scholar] [CrossRef]
- Cao, X.; He, L.; Xie, F.; Chen, Y.-F.; Wen, Z. Factors determining the survival of nasopharyngeal carcinoma with lung metastasis alone: Does combined modality treatment benefit? BMC Cancer 2011, 11, 370. [Google Scholar] [CrossRef]
- Ong, Y.; Heng, D.; Chung, B.; Leong, S.; Wee, J.; Fong, K.; Tan, T.; Tan, E.H. Design of a prognostic index score for metastatic nasopharyngeal carcinoma. Eur. J. Cancer 2003, 39, 1535–1541. [Google Scholar] [CrossRef]
- Lu, J.J.; Cooper, J.S.; Lee, A.W. Nasopharyngeal Cancer: Multidisciplinary Management; Springer Science & Business Media: Berlin, Germany, 2010. [Google Scholar]
- Chung, G.T.; Lou, W.P.; Chow, C.; To, K.F.; Choy, K.W.; Leung, A.W.; Tong, C.Y.; Yuen, J.W.; Ko, C.W.; Yip, T.T.; et al. Constitutive activation of distinct NF-kappaB signals in EBV-associated nasopharyngeal carcinoma. J. Pathol. 2013, 231, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, X.-M.; Huang, H.-R.; Zhao, F.-P.; Wang, F.; Liu, X.; Li, X. Detailed analysis of inflammatory cell infiltration and the prognostic impact on nasopharyngeal carcinoma. Head Neck 2018, 40, 1245–1253. [Google Scholar] [CrossRef]
- Huang, D.; Du, X. Crosstalk between tumor cells and microenvironment via Wnt pathway in colorectal cancer dissemination. World J. Gastroenterol. 2008, 14, 1823–1827. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2016, 36, 1461–1473. [Google Scholar] [CrossRef]
- Lim, J.H.; Jono, H.; Komatsu, K.; Woo, C.H.; Lee, J.; Miyata, M.; Matsuno, T.; Xu, X.; Huang, Y.; Zhang, W.; et al. CYLD negatively regulates transforming growth factor-beta-signalling via deubiquitinating Akt. Nat. Commun. 2012, 3, 771. [Google Scholar] [CrossRef]
- Baud, V.; Karin, M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 2009, 8, 33–40. [Google Scholar] [CrossRef]
- Roman-Blas, J.A.; A Jimenez, S. NF-κB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthr. Cartil. 2006, 14, 839–848. [Google Scholar] [CrossRef]
- Zhao, W.; Ma, N.; Wang, S.; Mo, Y.; Zhang, Z.; Huang, G.; Midorikawa, K.; Hiraku, Y.; Olikawa, S.; Murata, M.; et al. RERG suppresses cell proliferation, migration and angiogenesis through ERK/NF-kappaB signaling pathway in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 88. [Google Scholar] [CrossRef]
- Jia, C.-C.; Wang, T.-T.; Liu, W.; Fu, B.-S.; Hua, X.; Wang, G.-Y.; Li, T.-J.; Li, X.; Wu, X.-Y.; Tai, Y.; et al. Cancer-Associated Fibroblasts from Hepatocellular Carcinoma Promote Malignant Cell Proliferation by HGF Secretion. PLoS ONE 2013, 8, e63243. [Google Scholar] [CrossRef]
- Nemeth, Z.; Li, M.; Csizmadia, E.; Dome, B.; Johansson, M.; Persson, J.L.; Seth, P.; Otterbein, L.; Wegiel, B. Heme oxygenase-1 in macrophages controls prostate cancer progression. Oncotarget 2015, 6, 33675–33688. [Google Scholar] [CrossRef]
- Yu, Q.; Qiu, Y.; Chen, X.; Wang, X.; Mei, L.; Wu, H.; Liu, K.; Liu, Y.; Li, M.; Zhang, Z.; et al. Chemotherapy priming of the Pancreatic Tumor Microenvironment Promotes Delivery and Anti-Metastasis Efficacy of Intravenous Low-Molecular-Weight Heparin-Coated Lipid-siRNA Complex. Theranostics 2019, 9, 355–368. [Google Scholar] [CrossRef]
- Goldman, J.; Blundell, M.P.; Lopes, L.; Kinnon, C.; Di Santo, J.P.; Thrasher, A.J. Enhanced human cell engraftment in mice deficient in RAG2 and the common cytokine receptor gamma chain. Br. J. Haematol. 1998, 103, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Alameda, J.P.; Fernández-Aceñero, M.J.; Moreno-Maldonado, R.; Navarro, M.; Quintana, R.; Page, A.; Ramírez, Á.; Bravo, A.; Casanova, M.L. CYLD regulates keratinocyte differentiation and skin cancer progression in humans. Cell Death Dis. 2011, 2, e208. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.-L.; Xu, J.-F.; Hu, J. Regulation of Oral Squamous Cell Carcinoma Proliferation Through Crosstalk Between SMAD7 and CYLD. Cell. Physiol. Biochem. 2016, 38, 1209–1217. [Google Scholar] [CrossRef]
- Alameda, J.P.; Moreno-Maldonado, R.; Navarro, M.; Bravo, A.; Ramírez, Á.; Page, A.; Jorcano, J.L.; Fernández-Aceñero, M.J.; Casanova, M.L. An inactivating CYLD mutation promotes skin tumor progression by conferring enhanced proliferative, survival and angiogenic properties to epidermal cancer cells. Oncogene 2010, 29, 6522–6532. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; A Rivas, M.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Dai, W.; Ko, J.; Choi, S.S.A.; Yu, Z.; Ning, L.; Zheng, H.; Gopalan, V.; Chan, K.T.; Lee, N.P.; Chan, K.W.; et al. Whole-exome sequencing reveals critical genes underlying metastasis in oesophageal squamous cell carcinoma. J. Pathol. 2017, 242, 500–510. [Google Scholar] [CrossRef]
- Cheung, S.T.; Huang, D.P.; Hui, A.B.; Lo, K.W.; Ko, C.W.; Tsang, Y.S.; Wong, N.; Whitney, B.M.; Lee, J.C. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer 1999, 83, 121–126. [Google Scholar] [CrossRef]
- Lin, W.; Tsao, S.W.; Jia, L.; Deng, W.; Zheng, H.; Dai, W.; Ko, J.; Lo, K.W.; Chung, G.T.Y.; Yip, K.; et al. Establishment and characterization of new tumor xenografts and cancer cell lines from EBV-positive nasopharyngeal carcinoma. Nat. Commun. 2018, 9, 4663. [Google Scholar] [CrossRef]
- Yip, Y.L.; Lin, W.; Deng, W.; Jia, L.; Lo, K.W.; Busson, P.; Vérillaud, B.; Liu, X.; Tsang, C.M.; Lung, M.L.; et al. Establishment of a nasopharyngeal carcinoma cell line capable of undergoing lytic Epstein–Barr virus reactivation. Lab. Investig. 2018, 98, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.W.; Wang, X.; Liu, Y.; Cheung, Y.C.; Feng, H.; Zheng, Z.; Wong, N.; Yuen, P.W.; Lo, A.K.; Wong, Y.C.; et al. Establishment of two immortalized nasopharyngeal epithelial cell lines using SV40 large T and HPV16E6/E7 viral oncogenes. Biochim. Biophys. Acta 2002, 1590, 150–158. [Google Scholar] [CrossRef]
- Yu, V.Z.; Wong, V.C.-L.; Dai, W.; Ko, J.; Lam, A.K.; Chan, K.W.; Samant, R.S.; Lung, H.L.; Shuen, W.H.; Law, S.; et al. Nuclear Localization of DNAJB6 Is Associated With Survival of Patients with Esophageal Cancer and Reduces AKT Signaling and Proliferation of Cancer Cells. Gastroenterology 2015, 149, 1825.e5–1836.e5. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.K.L.; Lung, H.L.; Ko, J.M.Y.; Cheng, Y.; Stanbridge, E.J.; Zabarovsky, E.R.; Nicholls, J.M.; Chua, D.; Tsao, S.W.; Guan, X.-Y.; et al. Chromosome 14 transfer and functional studies identify a candidate tumor suppressor gene, Mirror image polydactyly 1, in nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA 2009, 106, 14478–14483. [Google Scholar] [CrossRef]
- Lung, H.L.; Cheung, A.K.L.; Xie, D.; Cheng, Y.; Kwong, F.M.; Murakami, Y.; Guan, X.-Y.; Sham, J.S.; Chua, D.; Protopopov, A.I.; et al. TSLC1Is a Tumor Suppressor Gene Associated with Metastasis in Nasopharyngeal Carcinoma. Cancer Res. 2006, 66, 9385–9392. [Google Scholar] [CrossRef]
- Cheung, A.K.L.; Ip, J.C.Y.; Chu, A.C.H.; Cheng, Y.; Leong, M.M.L.; Ko, J.; Shuen, W.H.; Lung, H.L.; Lung, M.L. PTPRG suppresses tumor growth and invasion via inhibition of Akt signaling in nasopharyngeal carcinoma. Oncotarget 2015, 6, 13434–13447. [Google Scholar] [CrossRef]
- Lung, H.L.; Cheung, A.K.L.; Cheng, Y.; Kwong, F.M.; Lo, P.H.Y.; Law, E.W.L.; Chua, D.; Zabarovsky, E.R.; Wang, N.; Tsao, S.W.; et al. Functional characterization ofTHY1as a tumor suppressor gene with antiinvasive activity in nasopharyngeal carcinoma. Int. J. Cancer 2010, 127, 304–312. [Google Scholar] [CrossRef]
- Wong, V.C.-L.; Chen, H.; Ko, J.; Chan, K.-W.; Chan, Y.P.; Law, S.; Chua, D.; Kwong, R.L.-W.; Lung, H.L.; Srivastava, G.; et al. Tumor suppressor dual-specificity phosphatase 6 (DUSP6) impairs cell invasion and epithelial-mesenchymal transition (EMT)-associated phenotype. Int. J. Cancer 2011, 130, 83–95. [Google Scholar] [CrossRef]
- Cheung, A.K.L.; Lung, H.L.; Hung, S.C.; Law, E.W.L.; Cheng, Y.; Yau, W.L.; Bangarusamy, D.K.; Miller, L.D.; Liu, E.T.-B.; Shao, J.-Y.; et al. Functional Analysis of a Cell Cycle-Associated, Tumor-Suppressive Gene, Protein Tyrosine Phosphatase Receptor Type G, in Nasopharyngeal Carcinoma. Cancer Res. 2008, 68, 8137–8145. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, I.; Becker, M.; Ociepka, K.; Friauf, E.; Nothwang, H.G. A subcellular prefractionation protocol for minute amounts of mammalian cell cultures and tissue. Proteomics 2005, 5, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Shuen, W.H.; Kan, R.; Yu, Z.; Lung, H.L.; Lung, M.L. Novel lentiviral-inducible transgene expression systems and versatile single-plasmid reporters for in vitro and in vivo cancer biology studies. Cancer Gene Ther. 2015, 22, 207–214. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, M.; Dai, W.; Yu, V.Z.; Tao, L.; Lung, M.L. Cylindromatosis Lysine 63 Deubiquitinase (CYLD) Regulates NF-kB Signaling Pathway and Modulates Fibroblast and Endothelial Cells Recruitment in Nasopharyngeal Carcinoma. Cancers 2020, 12, 1924. https://doi.org/10.3390/cancers12071924
Deng M, Dai W, Yu VZ, Tao L, Lung ML. Cylindromatosis Lysine 63 Deubiquitinase (CYLD) Regulates NF-kB Signaling Pathway and Modulates Fibroblast and Endothelial Cells Recruitment in Nasopharyngeal Carcinoma. Cancers. 2020; 12(7):1924. https://doi.org/10.3390/cancers12071924
Chicago/Turabian StyleDeng, Mingdan, Wei Dai, Valen Zhuoyou Yu, Lihua Tao, and Maria Li Lung. 2020. "Cylindromatosis Lysine 63 Deubiquitinase (CYLD) Regulates NF-kB Signaling Pathway and Modulates Fibroblast and Endothelial Cells Recruitment in Nasopharyngeal Carcinoma" Cancers 12, no. 7: 1924. https://doi.org/10.3390/cancers12071924
APA StyleDeng, M., Dai, W., Yu, V. Z., Tao, L., & Lung, M. L. (2020). Cylindromatosis Lysine 63 Deubiquitinase (CYLD) Regulates NF-kB Signaling Pathway and Modulates Fibroblast and Endothelial Cells Recruitment in Nasopharyngeal Carcinoma. Cancers, 12(7), 1924. https://doi.org/10.3390/cancers12071924




