NCL Inhibition Exerts Antineoplastic Effects against Prostate Cancer Cells by Modulating Oncogenic MicroRNAs
Abstract
1. Introduction
2. Results
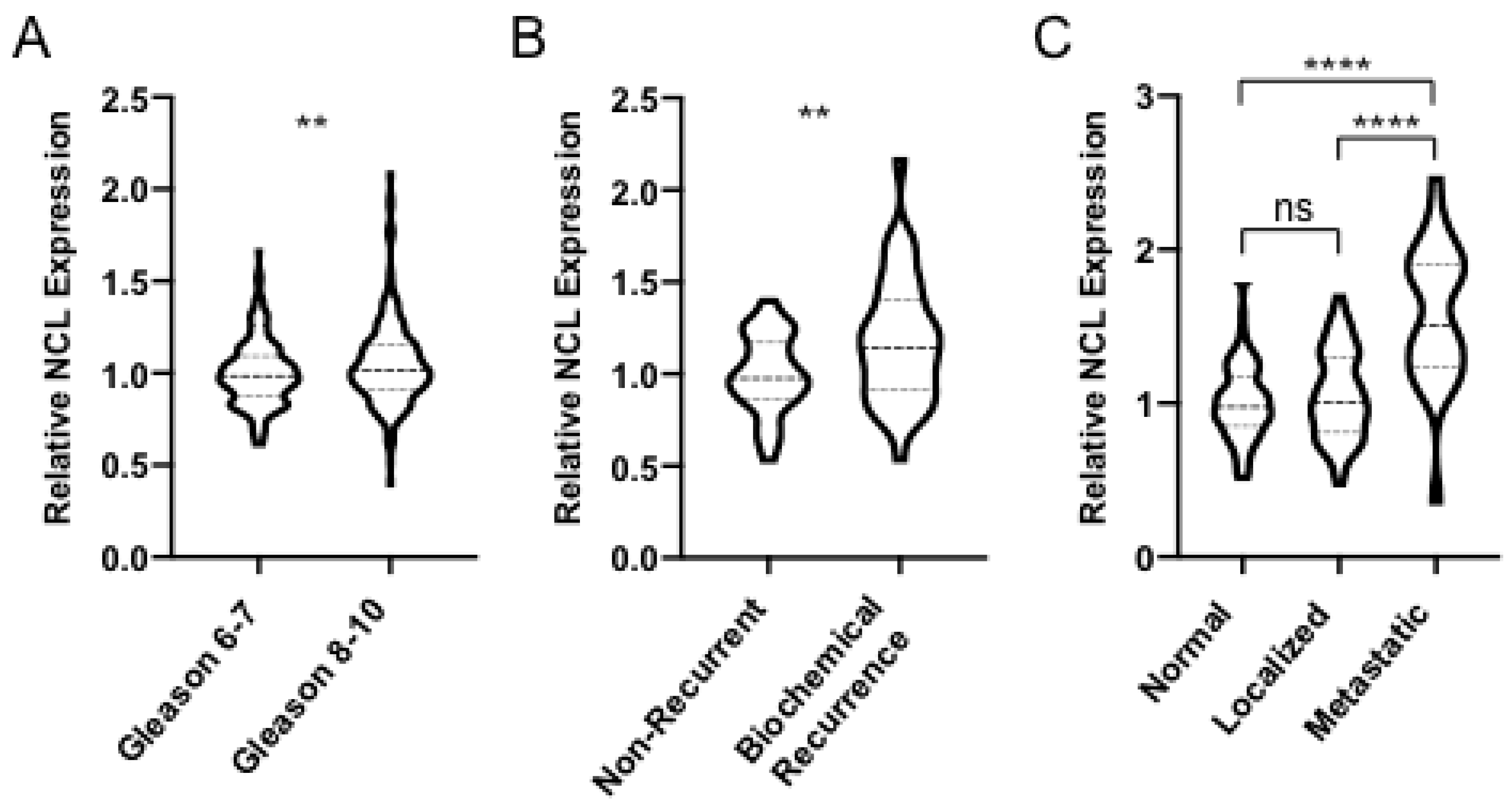
2.1. NCL is Upregulated in Aggressive Forms of PCa
2.2. 4LB5 Binds to PCa Cells and Inhibits Cell Proliferation
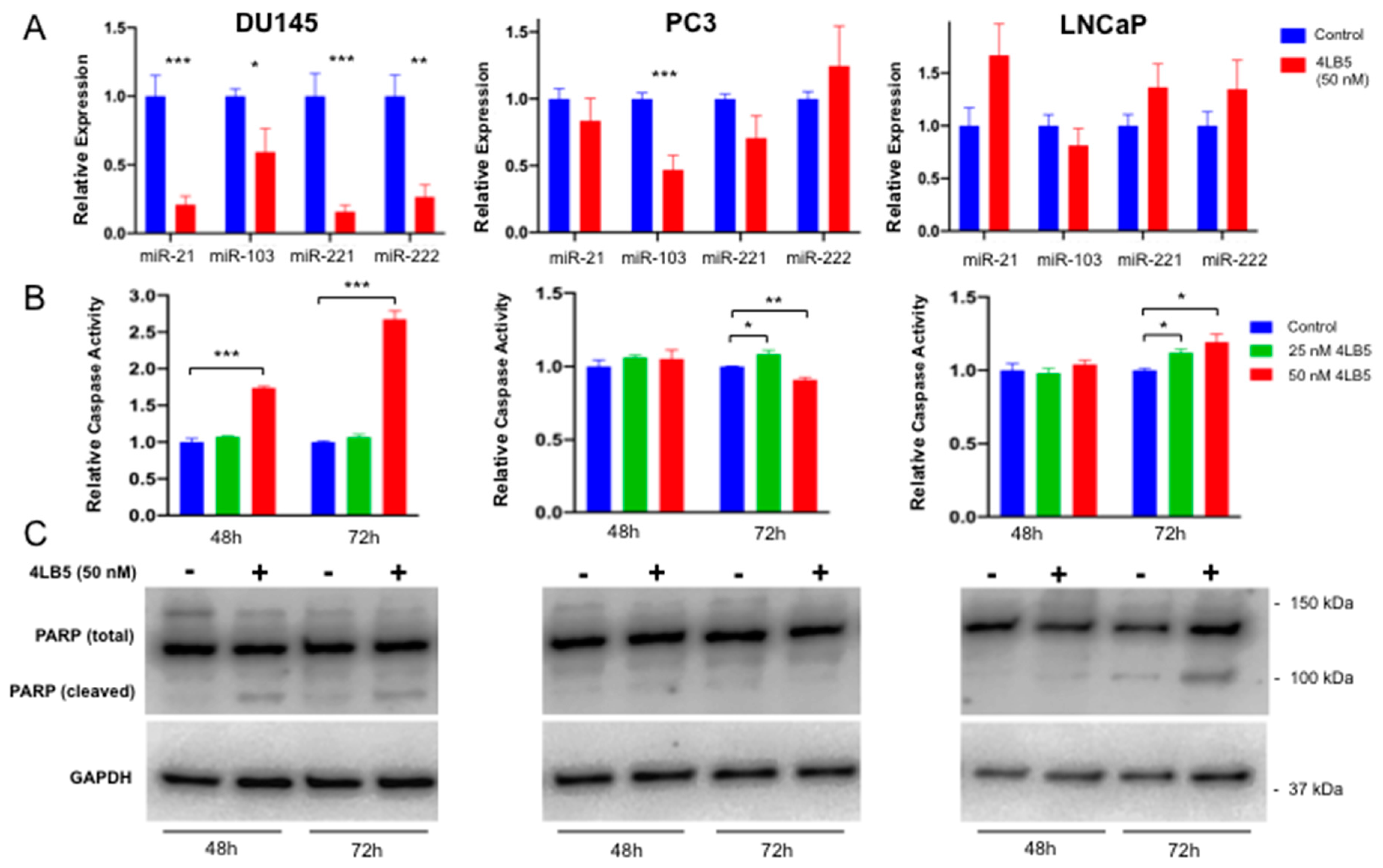
2.3. Treatment with 4LB5 Reduces Mature Forms of Oncogenic MicroRNAs and Initiates Apoptosis
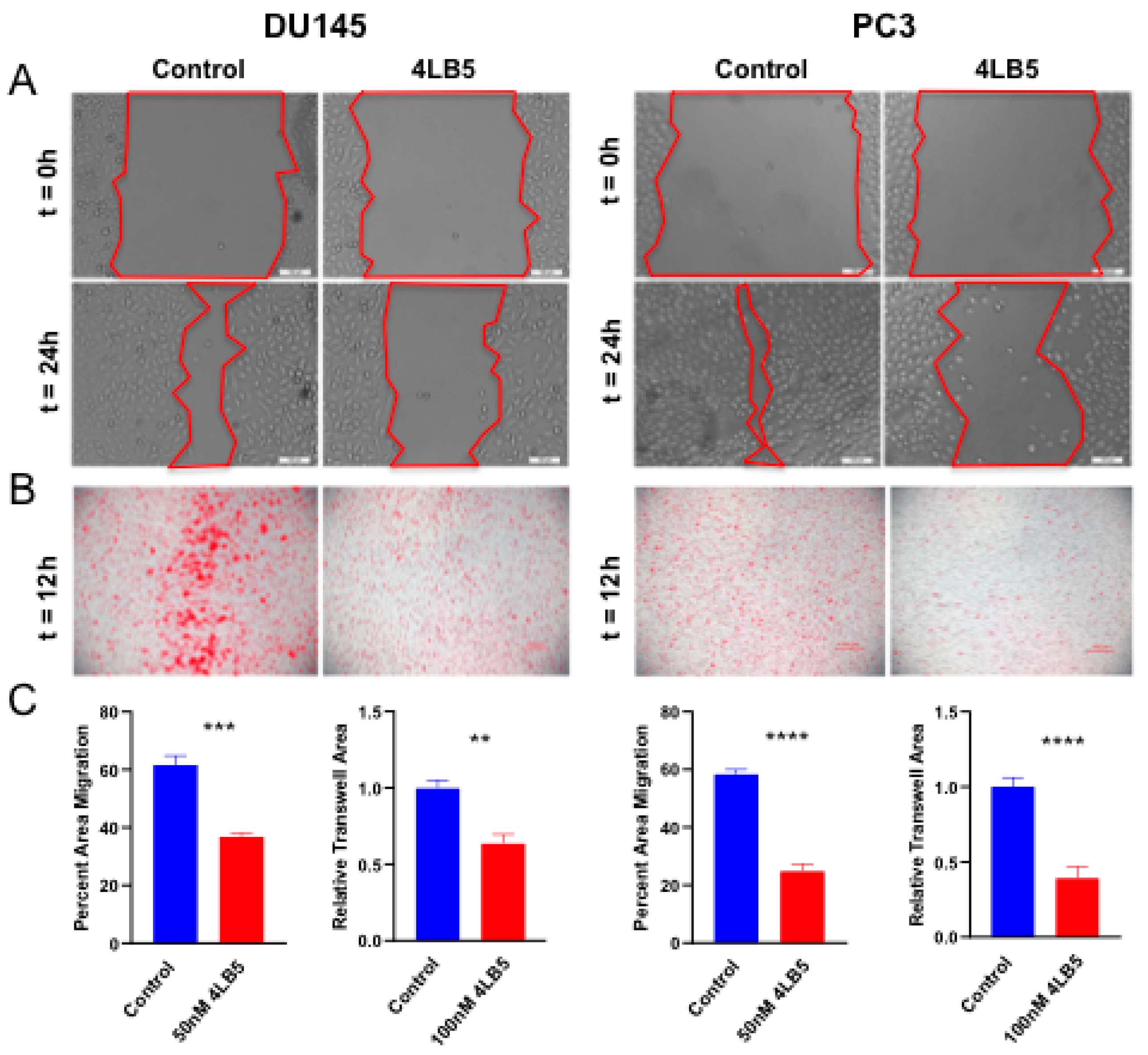
2.4. 4LB5 Impairs PCa Cell Migration
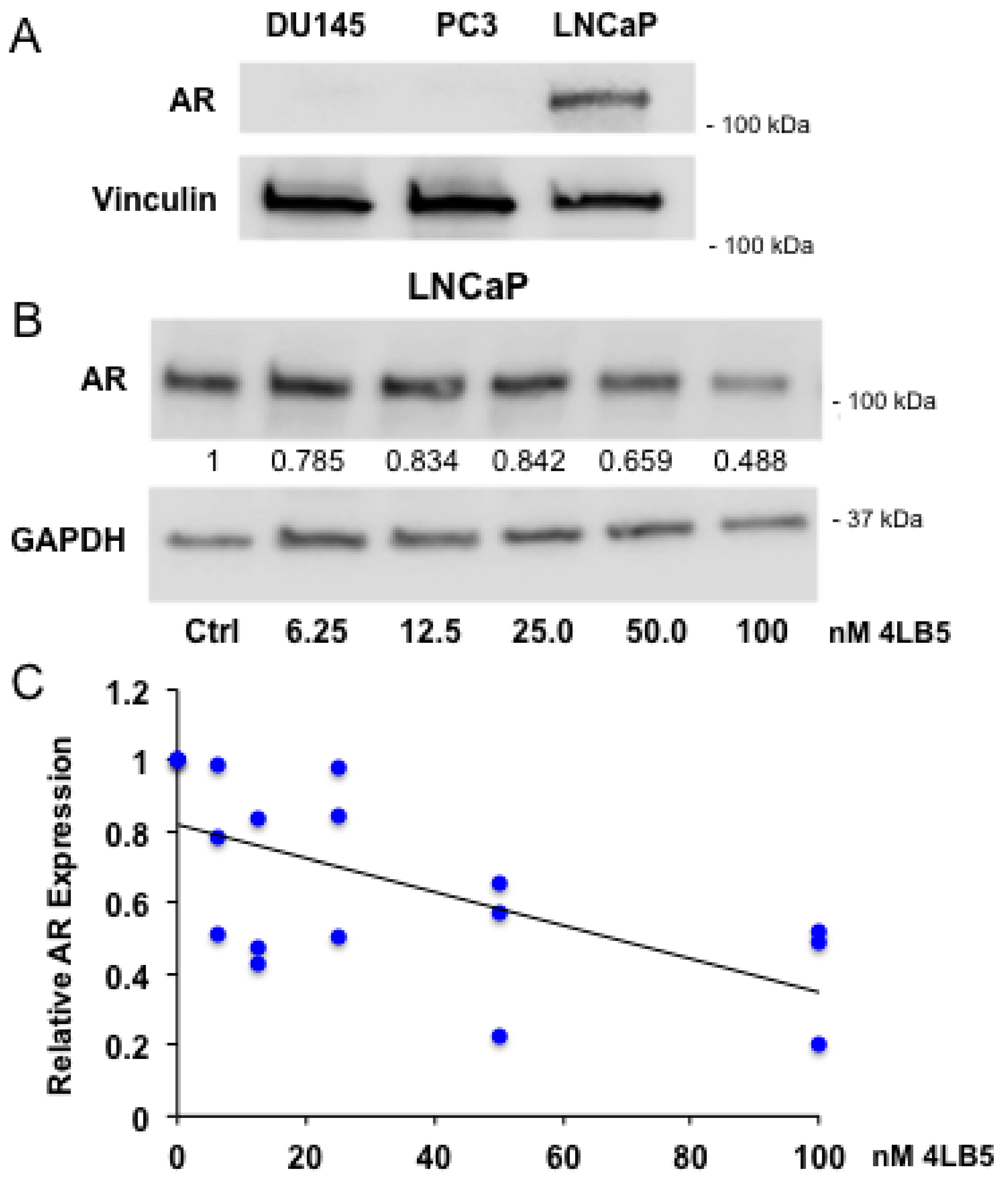
2.5. 4LB5 Reduces AR Expression in Hormone-Sensitive LNCaP Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Media
4.2. 4LB5 Production, Purification, and Treatments
4.3. Cell Surface ELISA
4.4. Cell Viability and Proliferation Assays
4.5. Western Blotting
4.6. Quantitative Real-Time PCR
4.7. Caspase Activity Assay
4.8. Trans-Well Migration and Wound Healing Assay
4.9. Bioinformatics Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- James, N.D.; Spears, M.R.; Clarke, N.W.; Dearnaley, D.P.; De Bono, J.S.; Gale, J.; Hetherington, J.; Hoskin, P.J.; Jones, R.J.; Laing, R.; et al. Survival with Newly Diagnosed Metastatic Prostate Cancer in the “Docetaxel Era”: Data from 917 Patients in the Control Arm of the STAMPEDE Trial ( MRC PR08, CRUK/06/019 ). Eur. Urol. 2015, 67, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Kirby, M.; Hirst, C.; Crawford, E.D. Characterising the castration-resistant prostate cancer population: A systematic review. Int. J. Clin. Pract. 2011, 65, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Shevrin, D. Genomic predictors for treatment of late stage prostate cancer. Asian J. Androl. 2016, 18, 586. [Google Scholar] [CrossRef] [PubMed]
- Shiota, M.; Yokomizo, A.; Naito, S. Increased androgen receptor transcription: A cause of castration-resistant prostate cancer and a possible therapeutic target. J. Mol. Endocrinol 2011, 47, R25–R41. [Google Scholar] [CrossRef] [PubMed]
- Dan, R.; Van Allen, E.M.; Wu, Y.-M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.-M.; Montgomery, B.; Taplin, M.-E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar]
- Grasso, C.S.; Wu, Y.; Robinson, D.R.; Cao, X.; Saravana, M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The Mutational Landscape of Lethal Castrate Resistant Prostate Cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Steentjes, L.; Siesling, S.; Drummond, F.J.; Van Manen, J.G.; Sharp, L.; Gavin, A. Factors associated with current and severe physical side-effects after prostate cancer treatment: What men report. Eur. J. Cancer Care 2018, 27, e12589. [Google Scholar] [CrossRef] [PubMed]
- Crawford, E.D.; Petrylak, D.; Sartor, O. Navigating the evolving therapeutic landscape in advanced prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2017, 35, S1–S13. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2015, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Vanacore, D.; Boccellino, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Di Franco, R.; Romano, F.J.; Montanari, M.; La Mantia, E.; Piscitelli, R.; et al. Micrornas in prostate cancer: An overview. Oncotarget 2017, 8, 50240–50251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Yang, L.F.; Zhu, Y.; Yao, X.D.; Zhang, S.L.; Dai, B.; Zhu, Y.-P.; Shen, Y.-J.; Shi, G.-H.; Ye, D.-W. Serum miRNA-21: Elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 2011, 71, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, R.S.; Li, Y.H.; Zhong, S.; Chen, Y.Y.; Zhang, C.M.; Hu, M.-M.; Shen, Z.-J. MiR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. J. Urol. 2012, 187, 1466–1472. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Danquah, M.; Chaudhary, A.K.; Mahato, R.I. Small Molecules Targeting MicroRNA for Cancer Therapy: Promises and Obstacles. J Controll. Release 2015, 219, 237–247. [Google Scholar] [CrossRef]
- Rothschild, S.I. microRNA therapies in cancer. Mol. Cell Ther. 2014, 2, 7. [Google Scholar] [CrossRef]
- Bugler, B.; Cauzergues-Ferrer, M.; Bouche, G.; Bourbon, H.; Amalric, F. Detection and Localization of a Class of Proteins Immunologically Related to a 100-kDa Nucleolar Protein. Eur. J. Biochem. 1982, 128, 475–480. [Google Scholar] [CrossRef]
- Borer, R.A.; Lehner, C.F.; Eppenberger, H.M.; Nigg, E.A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 1989, 56, 379–390. [Google Scholar] [CrossRef]
- Mongelard, F.; Bouvet, P. Nucleolin: A multiFACeTed protein. Trends Cell Biol. 2007, 17, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R. The nucleolus and ribosome formation. Curr. Opin. Cell Biol. 1990, 2, 521–527. [Google Scholar] [CrossRef]
- Srivastava, M.; Pollard, H.B. Molecular dissection of nucleolin’s role in growth and cell proliferation: New insights. FASEB J. 1999, 13, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xu, X.H. Roles of nucleolin: Focus on cancer and anti-cancer therapy. Saudi Med. J. 2016, 37, 1312–1318. [Google Scholar] [CrossRef]
- Hovanessian, A.G.; Puvion-Dutilleul, F.; Nisole, S.; Svab, J.; Perret, E.; Deng, J.-S.; Krust, B. The Cell-Surface-Expressed Nucleolin Is Associated with the Actin Cytoskeleton. Exp. Cell Res. 2000, 261, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Christian, S.; Pilch, J.; Akerman, M.E.; Porkka, K.; Laakkonen, P.; Ruoslahti, E. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J. Cell Biol. 2003, 163, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Tominaga, K.; Lee, E.K.; Srikantan, S.; Kang, M.J.; Kim, M.M.; Selimyan, R.; Martindale, J.L.; Yang, X.; Carrier, F.; et al. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 2011, 39, 8513–8530. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Gorospe, M. RNA-binding protein nucleolin in disease. RNA Biol. 2012, 9, 799–808. [Google Scholar] [CrossRef]
- Otake, Y.; Soundararajan, S.; Sengupta, T.K.; Kio, E.A.; Smith, J.C.; Roman, M.P.; Stuart, R.K.; Spicer, E.K.; Fernandes, D.J. Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood 2007, 109, 3069–3075. [Google Scholar] [CrossRef]
- Fernandes, E.; Freitas, R.; Ferreira, D.; Soares, J.; Azevedo, R.; Gaiteiro, C.; Peixoto, A.; Oliveira, S.; Cotton, S.; Relvas-Santos, M.; et al. Nucleolin-Sle A Glycoforms as E-Selectin Ligands and Potentially Targetable Biomarkers at the Cell Surface of Gastric Cancer Cells. Cancers 2020, 12, 861. [Google Scholar] [CrossRef]
- Chen, C.Y.; Gherzi, R.; Andersen, J.S.; Gaietta, G.; Jürchott, K.; Royer, H.-D.; Mann, M.; Karin, M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000, 14, 1236–1248. [Google Scholar]
- Reyes-Reyes, E.M.; Akiyama, S.K. Cell-surface nucleolin is a signal transducing P-selectin binding protein for human colon carcinoma cells. Exp. Cell Res. 2008, 314, 2212–2223. [Google Scholar] [CrossRef] [PubMed]
- Pichiorri, F.; Palmieri, D.; De Luca, L.; Consiglio, J.; You, J.; Rocci, A.; Talabere, T.; Piovan, C.; Lagana, A.; Cascione, L.; et al. In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. J. Exp. Med. 2013, 210, 951–968. [Google Scholar] [CrossRef] [PubMed]
- Drabik, A.; Ciołczyk-Wierzbicka, D.; Dulińska-Litewka, J.; Bodzoń-Kułakowska, A.; Suder, P.; Silberring, J.; Silberring, J.; Laidler, P. A comparative study of glycoproteomes in androgen-sensitive and -independent prostate cancer cell lines. Mol. Cell Biochem. 2014, 386, 189–198. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Pathol. Mech. Dis. 2014, 18, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Ye, D.; Yao, X.; Zhang, S.; Dai, B.; Zhang, H.; Shen, Y.-J.; Zhu, Y.; Zhu, Y.-P.; Xiao, W.-J.; et al. Involvement of microRNA-21 in mediating chemo-resistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacol. Sin. 2010, 31, 867–873. [Google Scholar] [CrossRef]
- Kopczyńska, E. Role of microRNAs in the resistance of prostate cancer to docetaxel and paclitaxel. Wspolczesna Onkol. 2015, 19, 423–427. [Google Scholar] [CrossRef]
- Ribas, J.; Ni, X.; Haffner, M.; Wentzel, E.A.; Salmasi, A.H.; Chowdhury, W.H.; Kudrolli, T.A.; Yegnasubramanian, S.; Luo, J.; Rodriguez, R.; et al. miR-21: An androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009, 69, 7165–7169. [Google Scholar] [CrossRef]
- Mishra, S.; Deng, J.J.; Gowda, P.S.; Rao, M.K.; Lin, C.-L.; Chen, C.L.; Huang, T.; Sun, L.-Z. Androgen Receptor and MicroRNA-21 axis down-regulates transforming growth factor beta receptor II (TGFBR2) expression in Prostate Cancer. Oncogene 2014, 33, 4097–4106. [Google Scholar] [CrossRef]
- Leite, K.R.M.; Reis, S.T.; Viana, N.; Morais, D.R.; Moura, C.M.; Silva, I.A.; Pontes, J., Jr.; Katz, B.; Srougi, M. Controlling RECK miR21 promotes tumor cell invasion and is related to biochemical recurrence in prostate cancer. J. Cancer 2015, 6, 292–301. [Google Scholar] [CrossRef]
- Guan, Y.; Wu, Y.; Liu, Y.; Ni, J.; Nong, S. Association of microRNA-21 expression with clinicopathological characteristics and the risk of progression in advanced prostate cancer patients receiving androgen deprivation therapy. Prostate 2016, 76, 986–993. [Google Scholar] [CrossRef]
- Galardi, S.; Mercatelli, N.; Giorda, E.; Massalini, S.; Frajese, G.V.; Ciafrè, S.A.; Farace, M.G. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007, 282, 23716–23724. [Google Scholar] [CrossRef] [PubMed]
- Le Sage, C.; Nagel., R.; Egan, D.A.; Schrier, M.; Mesman, E.; Mangiola, A.; Anile, C.; Maira, G.; Mercatelli, N.; Ciafrè, S.A.; et al. Regulation of the p27Kip1 tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007, 26, 3699–3708. [Google Scholar] [CrossRef]
- Wang, L.; Liu, C.; Li, C.; Xue, J.; Zhao, S.; Zhan, P.; Lin, Y.; Zhang, P.; Jiang, A.; Chen, W. Effects of microRNA-221/222 on cell proliferation and apoptosis in prostate cancer cells. Gene 2015, 572, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Thieu, W.; Tilki, D.; White, R.W.D.; Evans, C.P. The role of microRNA in castration-resistant prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, X.; He, H.H.; Sweeney, C.J.; Liu, S.X.; Brown, M.; Balk, S.; Lee, G.S.; Kantoff, P.W. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene 2014, 33, 2790–2800. [Google Scholar] [CrossRef]
- Sun, T.; Wang, Q.; Balk, S.; Brown, M.; Lee, G.-S.M.; Kantoff, P.W. The role of microRNA-221 and -222 in Androgen-independent Prostate Cancer Cell lines. Cancer. 2010, 69, 617–632. [Google Scholar] [CrossRef]
- Chen, Y.; Zaman, S.M.; Deng, G.; Majid, S.; Saini, S.; Liu, J.; Tanaka, Y.; Dahiya, R. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostate cancer. Cancer Prev. Res. 2011, 4, 76–86. [Google Scholar] [CrossRef]
- Sun, T.; Yang, M.; Chen, S.; Balk, S.; Pomerantz, M.; Hsieh, C.L.; Brown, M.; Lee, G.S.; Kantoff, P.W. The altered expression of MiR-221/-222 and MiR-23b/-27b is associated with the development of human castration resistant prostate cancer. Prostate 2012, 72, 1093–1103. [Google Scholar] [CrossRef]
- Palmieri, D.; Richmond, T.; Piovan, C.; Sheetz, T.; Zanesi, N.; Troise, F.; James, C.; Wernicke, D.; Nyei, F.; Gordon, T.J.; et al. Human anti-nucleolin recombinant immunoagent for cancer therapy. Proc. Natl. Acad. Sci. USA 2015, 112, 9418–9423. [Google Scholar] [CrossRef]
- Chalfin, H.J.; Verdone, J.E.; Van der Toom, E.E.; Glavaris, S.; Gorin, M.A.; Pienta, K.J. Nucleolin Staining May Aid in the Identification of Circulating Prostate Cancer Cells. Clin. Genitourin. Cancer 2017, 15, 477–481. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Teng, Y.; Bates, P.J. A new paradigm for aptamer therapeutic AS1411 action: Uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010, 70, 8617–8629. [Google Scholar] [CrossRef] [PubMed]
- Bonci, D.; Coppola, V.; Patrizii, M.; Addario, A.; Cannistraci, A.; Francescangeli, F.; Pecci, R.; Muto, G.; Collura, D.; Bedini, R.; et al. A microRNA code for prostate cancer metastasis. Oncogene 2016, 35, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, D.; Sha, J.; Sun, P.; Huang, Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem. Biophys. Res. Commun. 2009, 383, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Doldi, V.; Pennati, M.; Forte, B.; Gandellini, P.; Zaffaroni, N. Dissecting the role of microRNAs in prostate cancer metastasis: Implications for the design of novel therapeutic approaches. Cell. Mol. Life Sci. 2016, 73, 2531–2542. [Google Scholar] [CrossRef]
- Ayub, S.G.; Kaul, D.; Ayub, T. Microdissecting the role of microRNAs in the pathogenesis of prostate cancer. Cancer Genet. 2015, 208, 289–302. [Google Scholar] [CrossRef]
- Horoszewicz, J.S.; Leong, S.S.; Kawinski, E.; Karr, J.P.; Rosenthal, H.; Chu, T.M.; Mirand, E.A.; Murphy, G.P. LNCaP model of human prostatic carcinoma. Cancer Res. 1983, 43, 1809–1818. [Google Scholar]
- Pesarrodona, M.; Sánchez-García, L.; Seras-Franzoso, J.; Sánchez-Chardi, A.; Baltá-Foix, R.; Cámara-Sánchez, P.; Gener, P.; Jara, J.J.; Pulido, D.; Serna, N.; et al. Engineering a Nanostructured Nucleolin-Binding Peptide for Intracellular Drug Delivery in Triple-Negative Breast Cancer Stem Cells. ACS Appl. Mater. Interfaces 2020, 12, 5381–5388. [Google Scholar] [CrossRef]
- Xu, C.; Wang, Y.; Tu, Q.; Zhang, Z.; Chen, M.; Mwangi, J.; Li, Y.; Jin, Y.; Zhao, X.; Lai, R. Targeting surface nucleolin induces autophagy-dependent cell death in pancreatic cancer via AMPK activation. Oncogene 2019, 38, 1832–1844. [Google Scholar] [CrossRef]
- Ramos, K.S.; Moore, S.; Runge, I.; Tavera-Garcia, M.A.; Cascone, I.; Courty, J.; Reyes-Reyes, E.M. The nucleolin antagonist N6L inhibits LINE1 retrotransposon activity in non-small cell lung carcinoma cells. J. Cancer 2020, 11, 733–740. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2019, 155, 1420–1431. [Google Scholar] [CrossRef]
- Noaparast, Z.; Hosseinimehr, S.J.; Piramoon, M.; Abedi, S.M. Tumor targeting with a 99mTc-labeled AS1411 aptamer in prostate tumor cells. J. Drug Target 2015, 23, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Russell, P.J.; Kingsley, E.A. Human Prostate Cancer Cell Lines. Methods Mol. Med. Prostate Cancer Methods Protoc. 2003, 21–39. [Google Scholar]
- Wolfson, E.; Solomon, S.; Schmukler, E.; Goldshmit, Y.; Pinkas-Kramarski, R. Nucleolin and ErbB2 inhibition reduces tumorigenicity of ErbB2-positive breast cancer article. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Crivianu-Gaita, V.; Thompson, M. Aptamers, antibody scFv, and antibody Fab’ fragments: An overview and comparison of three of the most versatile biosensor biorecognition elements. Biosens. Bioelectron. 2016, 85, 32–45. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, C.; Palmieri, D.; Braddom, A.; Zanesi, N.; James, C.; Cole, S.; Salvatore, F.; Croce, C.M.; De Lorenzo, C. A novel fully human anti-NCL immunoRNase for triple-negative breast cancer therapy. Oncotarget 2016, 7, 87016–87030. [Google Scholar] [CrossRef]
- Mazzocco, C.; Fracasso, G.; Germain-Genevois, C.; Dugot-Senant, N.; Figini, M.; Colombatti, M.; Grenier, N.; Couillaud, F. In vivo imaging of prostate cancer using an anti-PSMA scFv fragment as a probe. Sci. Rep. 2016, 6, 23314. [Google Scholar] [CrossRef]
- Nawaz, S.; Mullen, G.E.D.; Blower, P.J.; Ballinger, J.R. A 99mTc-labelled scFv antibody fragment that binds to prostate-specific membrane antigen. Nucl. Med. Commun. 2017, 38, 666–671. [Google Scholar] [CrossRef]
- Sebaugh, J.L. Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 2011, 10, 128–134. [Google Scholar] [CrossRef]
- Limame, R.; Wouters, A.; Pauwels, B.; Fransen, E.; Peeters, M.; Lardon, F.; De Wever, O.; Pauwels, P. Comparative Analysis of Dynamic Cell Viability, Migration and Invasion Assessments by Novel Real-Time Technology and Classic Endpoint Assays. PLoS ONE 2012, 7, e46536. [Google Scholar] [CrossRef]
- Sun, Y.; Goodison, S. Optimizing Molecular Signatures for Predicting Prostate Cancer Recurrence. Prostate 2009, 69, 1119–1127. [Google Scholar] [CrossRef]
- Yu, Y.P.; Landsittel, D.; Jing, L.; Nelson, J.; Ren, B.; Liu, L.; McDonald, C.; Thomas, R.; Dhir, R.; Finkelstein, S.; et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 2004, 22, 2790–2799. [Google Scholar] [CrossRef] [PubMed]
- Chandran, U.R.; Ma, C.; Dhir, R.; Bisceglia, M.; Lyons-Weiler, M.; Liang, W.; Michalopoulos, G.; Becich, M.; Monzon, F.A. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007, 7, 64. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheetz, T.; Mills, J.; Tessari, A.; Pawlikowski, M.; Braddom, A.E.; Posid, T.; Zynger, D.L.; James, C.; Embrione, V.; Parbhoo, K.; et al. NCL Inhibition Exerts Antineoplastic Effects against Prostate Cancer Cells by Modulating Oncogenic MicroRNAs. Cancers 2020, 12, 1861. https://doi.org/10.3390/cancers12071861
Sheetz T, Mills J, Tessari A, Pawlikowski M, Braddom AE, Posid T, Zynger DL, James C, Embrione V, Parbhoo K, et al. NCL Inhibition Exerts Antineoplastic Effects against Prostate Cancer Cells by Modulating Oncogenic MicroRNAs. Cancers. 2020; 12(7):1861. https://doi.org/10.3390/cancers12071861
Chicago/Turabian StyleSheetz, Tyler, Joseph Mills, Anna Tessari, Megan Pawlikowski, Ashley E. Braddom, Tasha Posid, Debra L. Zynger, Cindy James, Valerio Embrione, Kareesma Parbhoo, and et al. 2020. "NCL Inhibition Exerts Antineoplastic Effects against Prostate Cancer Cells by Modulating Oncogenic MicroRNAs" Cancers 12, no. 7: 1861. https://doi.org/10.3390/cancers12071861
APA StyleSheetz, T., Mills, J., Tessari, A., Pawlikowski, M., Braddom, A. E., Posid, T., Zynger, D. L., James, C., Embrione, V., Parbhoo, K., Foray, C., Coppola, V., Croce, C. M., & Palmieri, D. (2020). NCL Inhibition Exerts Antineoplastic Effects against Prostate Cancer Cells by Modulating Oncogenic MicroRNAs. Cancers, 12(7), 1861. https://doi.org/10.3390/cancers12071861





