Multiparametric MRI for Prostate Cancer Characterization: Combined Use of Radiomics Model with PI-RADS and Clinical Parameters
Abstract
1. Introduction
2. Results
2.1. Demographic Data
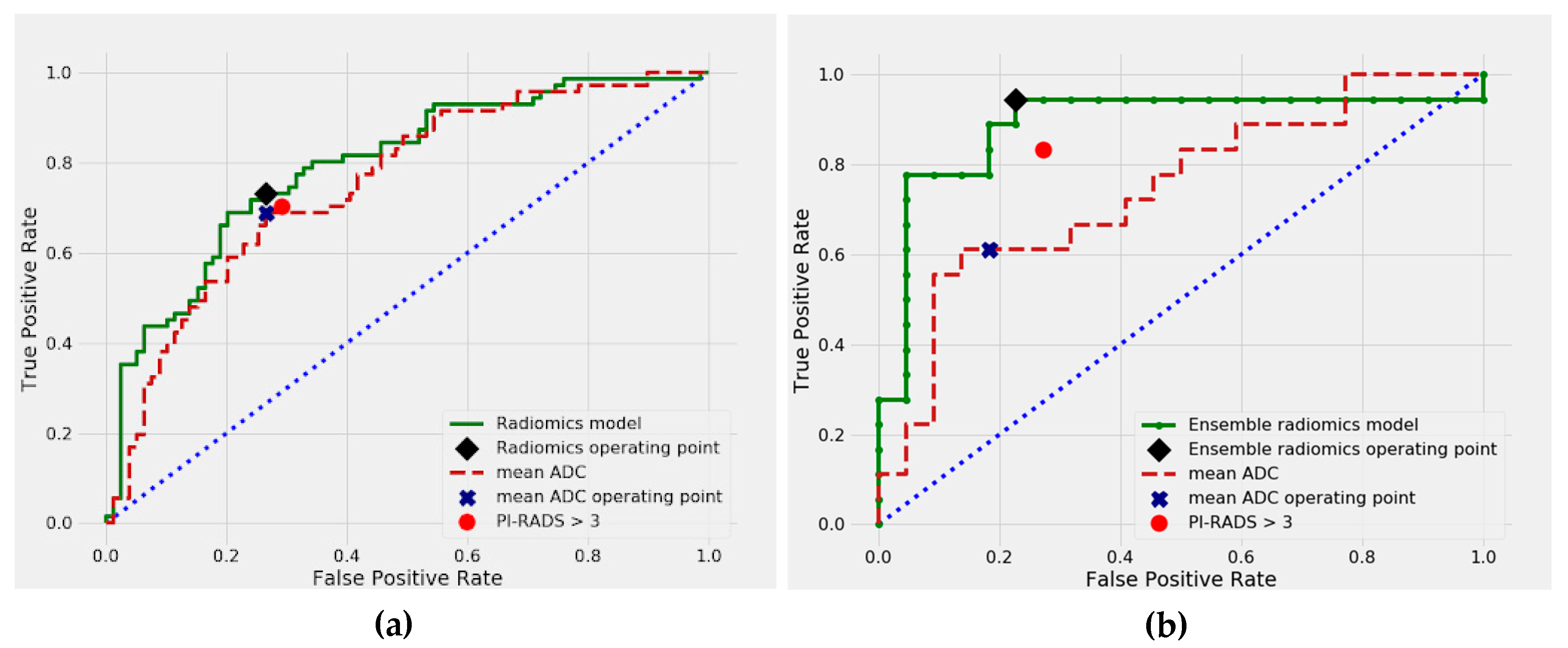
2.2. Differentiation between Malignant and Benign Prostate Lesions
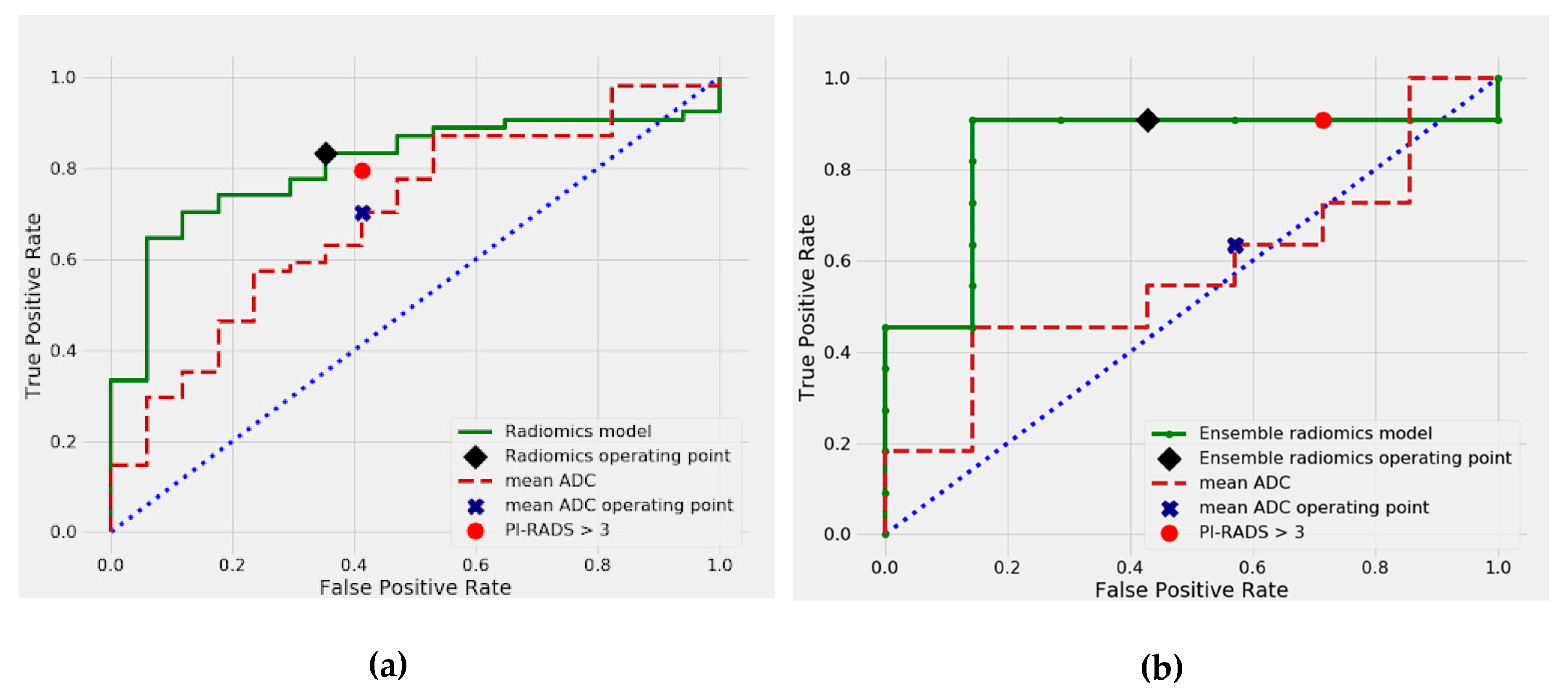
2.3. Differentiation between csPCa and cisPCa
3. Discussion
4. Materials and Methods
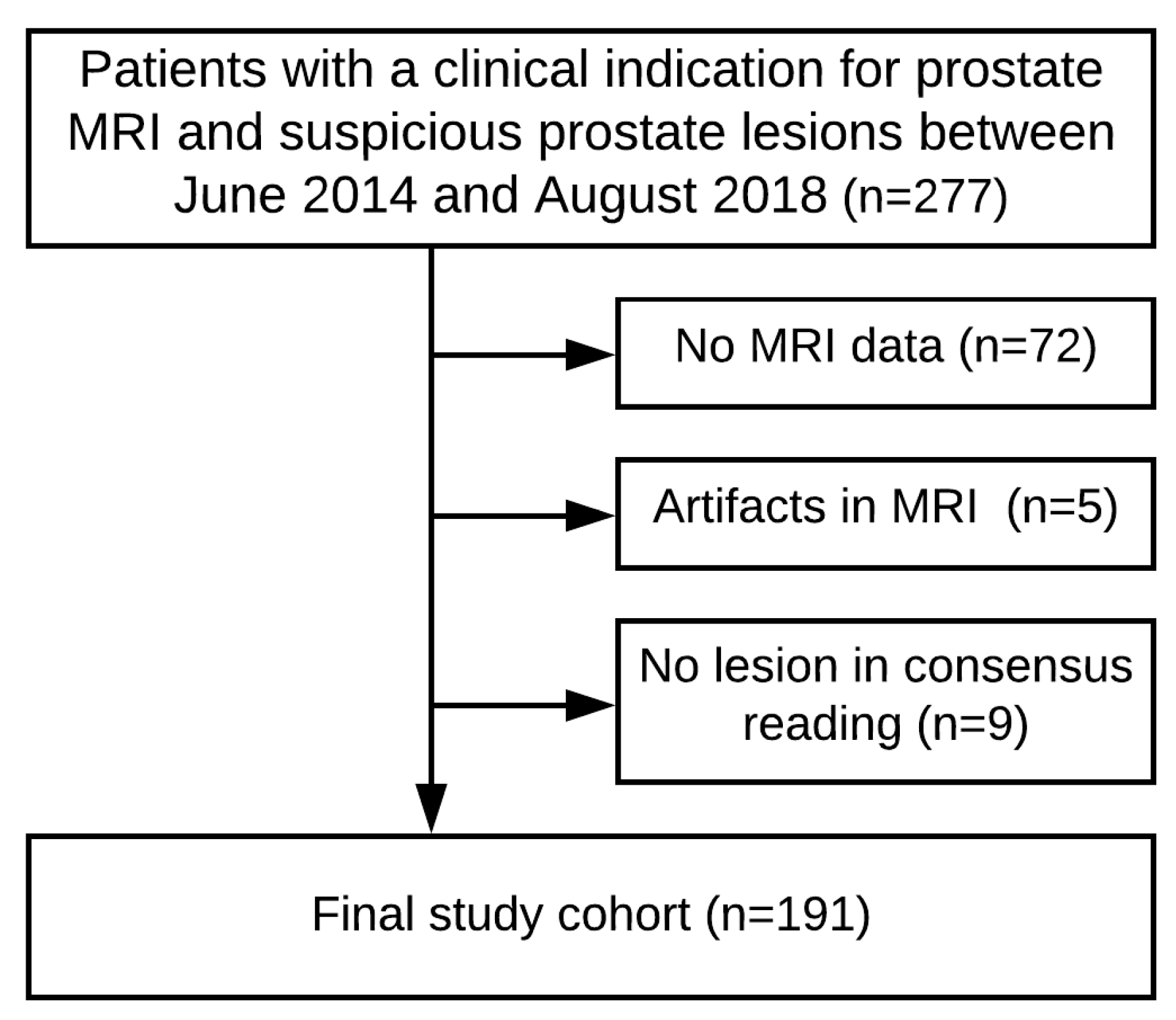
4.1. Patient Cohort
4.2. MRI Data Acquisition
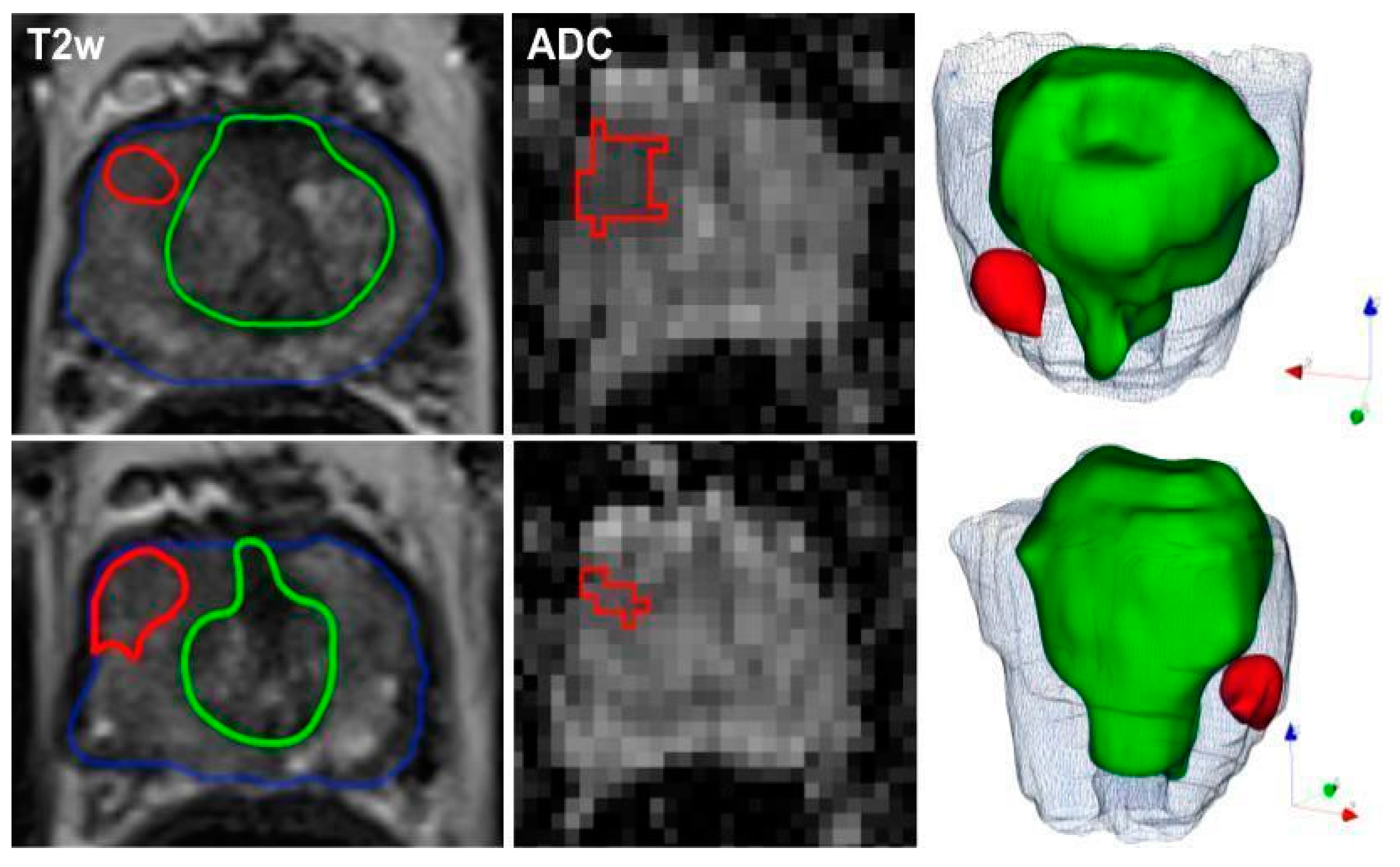
4.3. Image Segmentations
4.4. Radiomic Feature Extraction
4.5. Model Development
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
Appendix A.1. Details of Hyperparameter Tuning and Model Selection Process
References
- Rawla, P. Epidemiology of prostate cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef] [PubMed]
- Carter, H.B.; Partin, A.W.; Walsh, P.C.; Trock, B.J.; Veltri, R.W.; Nelson, W.G.; Coffey, D.S.; Singer, E.A.; Epstein, J.I. Gleason score 6 adenocarcinoma: Should it be labeled as cancer. J. Clin. Oncol. 2012, 30, 4294–4296. [Google Scholar] [CrossRef] [PubMed]
- Bergh, R.C.V.D.; Loeb, S.; Roobol, M.J. Impact of early diagnosis of prostate cancer on survival outcomes. Eur. Urol. Focus 2015, 1, 137–146. [Google Scholar] [CrossRef]
- Rooij, M.D.; Hamoen, E.H.J.; Fütterer, J.J.; Barentsz, J.O.; Rovers, M.M. Accuracy of multiparametric MRI for prostate cancer detection: A meta-analysis. Am. J. Roentgenol. 2014, 202, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.U.; Bosaily, A.E.-S.; Brown, L.C.; Gabe, R.; Kaplan, R.; Parmar, M.K.; Collaco-Moraes, Y.; Ward, K.; Hindley, R.G.; Freeman, A.; et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef]
- Rouvière, O.; Puech, P.; Renard-Penna, R.; Claudon, M.; Roy, C.; Mège-Lechevallier, F.; Decaussin-Petrucci, M.; Dubreuil-Chambardel, M.; Magaud, L.; Remontet, L.; et al. Use of prostate systematic and targeted biopsy on the basis of multiparametric MRI in biopsy-naive patients (MRI-FIRST): A prospective, multicentre, paired diagnostic study. Lancet Oncol. 2019, 20, 100–109. [Google Scholar] [CrossRef]
- Kasivisvanathan, V.; Rannikko, A.S.; Borghi, M. MRI-targeted biopsy for prostate-cancer diagnosis. N. Engl. J. Med. 2018, 379, 589–590. [Google Scholar]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef]
- Verma, A.; St Onge, J.; Dhillon, K.; Chorneyko, A. PSA density improves prediction of prostate cancer. Can. J. Urol. 2014, 21, 7312–7321. [Google Scholar]
- Distler, F.A.; Radtke, J.P.; Bonekamp, D.; Kesch, C.; Schlemmer, H.-P.; Wieczorek, K.; Kirchner, M.; Pahernik, S.; Hohenfellner, M.; Hadaschik, B.A. The value of PSA density in combination with PI-RADS™ for the accuracy of prostate cancer prediction. J. Urol. 2017, 198, 575–582. [Google Scholar] [CrossRef]
- Nordström, T.; Akre, O.; Aly, M.; Grönberg, H.; Eklund, M. Prostate-specific antigen (PSA) density in the diagnostic algorithm of prostate cancer. Prostate Cancer Prostatic Dis. 2017, 21, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Martorana, E.; Pirola, G.M.; Scialpi, M.; Micali, S.; Iseppi, A.; Bonetti, L.R.; Kaleci, S.; Torricelli, P.; Bianchi, G. Lesion volume predicts prostate cancer risk and aggressiveness: Validation of its value alone and matched with prostate imaging reporting and data system score. BJU Int. 2016, 120, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Naji, L.; Randhawa, H.; Sohani, Z.; Dennis, B.; Lautenbach, D.; Kavanagh, O.; Bawor, M.; Banfield, L.; Profetto, J. Digital rectal examination for prostate cancer screening in primary care: A systematic review and meta-analysis. Ann. Fam. Med. 2018, 16, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Cuocolo, R.; Cipullo, M.B.; Stanzione, A.; Ugga, L.; Romeo, V.; Radice, L.; Brunetti, A.; Imbriaco, M. Machine learning applications in prostate cancer magnetic resonance imaging. Eur. Radiol. Exp. 2019, 3, 35. [Google Scholar] [CrossRef]
- Sun, Y.; Reynolds, H.M.; Parameswaran, B.; Wraith, D.; Finnegan, M.E.; Williams, S.; Haworth, A. Multiparametric MRI and radiomics in prostate cancer: A review. Australas Phys. Eng. Sci. Med. 2019, 42, 3–25. [Google Scholar] [CrossRef]
- Schelb, P.; Kohl, S.; Radtke, J.P.; Wiesenfarth, M.; Kickingereder, P.; Bickelhaupt, S.; Kuder, T.A.; Stenzinger, A.; Hohenfellner, M.; Schlemmer, H.-P.; et al. Classification of cancer at prostate MRI: Deep learning versus clinical PI-RADS assessment. Radiology 2019, 293, 607–617. [Google Scholar] [CrossRef]
- Antonelli, M.; Johnston, E.W.; Dikaios, N.; Cheung, K.K.; Sidhu, H.S.; Appayya, M.B.; Giganti, F.; Simmons, L.A.M.; Freeman, A.; Allen, C.; et al. Machine learning classifiers can predict Gleason pattern 4 prostate cancer with greater accuracy than experienced radiologists. Eur. Radiol. 2019, 29, 4754–4764. [Google Scholar] [CrossRef]
- Bleker, J.; Kwee, T.C.; Dierckx, R.A.J.O.; Jong, I.J.D.; Huisman, H.; Yakar, D. Multiparametric MRI and auto-fixed volume of interest-based radiomics signature for clinically significant peripheral zone prostate cancer. Eur. Radiol. 2019, 30, 1313–1324. [Google Scholar] [CrossRef]
- Bonekamp, D.; Kohl, S.; Wiesenfarth, M.; Schelb, P.; Radtke, J.P.; Götz, M.; Kickingereder, P.; Yaqubi, K.; Hitthaler, B.; Gählert, N.; et al. Radiomic machine learning for characterization of prostate lesions with MRI: Comparison to ADC values. Radiology 2018, 289, 128–137. [Google Scholar] [CrossRef]
- Epstein, J.I. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. J. Am. Med. Assoc. 1994, 271, 368–374. [Google Scholar] [CrossRef]
- Stamey, T.A.; Freiha, F.S.; Mcneal, J.E.; Redwine, E.A.; Whittemore, A.S.; Schmid, H.-P. Localized prostate cancer. Relationship of tumor volume to clinical significance for treatment of prostate cancer. Cancer 1993, 71, 933–938. [Google Scholar] [CrossRef]
- Chaddad, A.; Kucharczyk, M.; Niazi, T. Multimodal radiomic features for the predicting gleason score of prostate cancer. Cancers 2018, 10, 249. [Google Scholar] [CrossRef] [PubMed]
- Monti, S.; Brancato, V.; Costanzo, G.D.; Basso, L.; Puglia, M.; Ragozzino, A.; Salvatore, M.; Cavaliere, C. Multiparametric MRI for prostate cancer detection: New insights into the combined use of a radiomic approach with advanced acquisition protocol. Cancers 2020, 12, 390. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, H.; Feng, Y.; Li, W. Prostate cancer diagnosis using deep learning with 3D multiparametric MRI. In Proceedings of the SPIE Medical Imaging, Orlando, FL, USA, 22 May 2017. [Google Scholar]
- Hötker, A.M.; Mazaheri, Y.; Aras, Ö.; Zheng, J.; Moskowitz, C.S.; Gondo, T.; Matsumoto, K.; Hricak, H.; Akin, O. Assessment of prostate cancer aggressiveness by use of the combination of quantitative DWI and dynamic contrast-enhanced MRI. Am. J. Roentgenol. 2016, 206, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Norris, J.M.; Echeverria, L.M.C.; Bott, S.R.; Brown, L.C.; Burns-Cox, N.; Dudderidge, T.; Bosaily, A.E.-S.; Frangou, E.; Freeman, A.; Ghei, M.; et al. What type of prostate cancer is systematically overlooked by multiparametric magnetic resonance imaging? An analysis from the PROMIS cohort. Eur. Urol. 2020. [CrossRef]
- Ito, K.; Furuta, A.; Kido, A.; Teramoto, Y.; Akamatsu, S.; Terada, N.; Yamasaki, T.; Inoue, T.; Ogawa, O.; Kobayashi, T. Detectability of prostate cancer in different parts of the gland with 3-Tesla multiparametric magnetic resonance imaging: Correlation with whole-mount histopathology. Int. J. Clin. Oncol. 2019, 25, 732–740. [Google Scholar] [CrossRef]
- Vignati, A.; Mazzetti, S.; Giannini, V.; Russo, F.; Bollito, E.; Porpiglia, F.; Stasi, M.; Regge, D. Texture features on T2-weighted magnetic resonance imaging: New potential biomarkers for prostate cancer aggressiveness. Phys. Med. Biol. 2015, 60, 2685–2701. [Google Scholar] [CrossRef]
- Fehr, D.; Veeraraghavan, H.; Wibmer, A.; Gondo, T.; Matsumoto, K.; Vargas, H.A.; Sala, E.; Hricak, H.; Deasy, J.O. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc. Natl. Acad. Sci. USA 2015, 112, E6265–E6273. [Google Scholar] [CrossRef]
- Wang, J.; Wu, C.-J.; Bao, M.-L.; Zhang, J.; Wang, X.-N.; Zhang, Y.-D. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur. Radiol. 2017, 27, 4082–4090. [Google Scholar] [CrossRef]
- Wibmer, A.; Hricak, H.; Gondo, T.; Matsumoto, K.; Veeraraghavan, H.; Fehr, D.; Zheng, J.; Goldman, D.; Moskowitz, C.; Fine, S.W.; et al. Haralick texture analysis of prostate MRI: Utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur. Radiol. 2015, 25, 2840–2850. [Google Scholar] [CrossRef] [PubMed]
- Schwier, M.; Griethuysen, J.V.; Vangel, M.G.; Pieper, S.; Peled, S.; Tempany, C.; Aerts, H.J.W.L.; Kikinis, R.; Fennessy, F.M.; Fedorov, A. Repeatability of multiparametric prostate MRI radiomics features. Sci. Rep. 2019, 9, 9441. [Google Scholar] [CrossRef] [PubMed]
- Tschudi, Y.; Pollack, A.; Punnen, S.; Ford, J.C.; Chang, Y.-C.; Soodana-Prakash, N.; Breto, A.L.; Kwon, D.; Munera, F.; Abramowitz, M.C.; et al. Automatic detection of prostate tumor habitats using diffusion MRI. Sci. Rep. 2018, 8, 16801. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.T.; Kauffman, C.R.; Garcia-Reyes, K.; Palmeri, M.L.; Madden, J.F.; Polascik, T.J.; Rosenkrantz, A.B. Apparent diffusion coefficient values of the benign central zone of the prostate: Comparison with low- and high-grade prostate cancer. Am. J. Roentgenol. 2015, 205, 331–336. [Google Scholar] [CrossRef]
- Mehralivand, S.; Shih, J.H.; Rais-Bahrami, S.; Oto, A.; Bednarova, S.; Nix, J.W.; Thomas, J.V.; Gordetsky, J.B.; Gaur, S.; Harmon, S.A.; et al. A magnetic resonance imaging–based prediction model for prostate biopsy risk stratification. JAMA Oncol. 2018, 4, 678. [Google Scholar] [CrossRef]
- Griethuysen, J.J.V.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Ding, C.; Peng, H. Minimum redundancy feature selection from microarray gene expression data. J. Bioinform. Comput. Biol. 2005, 3, 185–205. [Google Scholar] [CrossRef]
- Tsamardinos, I.; Greasidou, E.; Borboudakis, G. Bootstrapping the out-of-sample predictions for efficient and accurate cross-validation. Mach. Learn. 2018, 107, 1895–1922. [Google Scholar] [CrossRef]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008. [Google Scholar]
- Delong, E.R.; Delong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837. [Google Scholar] [CrossRef]
- Mcnemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef]
| Variable | Training Set | Test Set |
|---|---|---|
| Patients in total | 151 | 40 |
| PCa-negative | 80 | 22 |
| PCa-positive: | 71 | 18 |
| ISUP grade 1 (GS = 6) | 17 (24%) | 7 (39%) |
| ISUP grade ≥ 2 (GS ≥ 7) | 54 (76%) | 11 (61%) |
| ISUP grade 2 | 26 (37%) | 3 (17%) |
| ISUP grade 3 | 14 (20%) | 5 (28%) |
| ISUP grade 4 | 11 (15%) | 3 (17%) |
| ISUP grade 5 | 3 (4%) | − |
| Zonal distribution of index lesions: | ||
| Peripheral zone | 102 (67.55%) | 28 (70%) |
| Transition zone | 49 (32.45%) | 12 (30%) |
| Median age (years) | 68 (63–74) | 69 (63–72) |
| Median PSA value (ng/ml) | 7.60 (5.71–11.00) | 8.17 (6.82–11.85) |
| Median PSA density (ng/ml2) | 0.161 (0.108–0.241) | 0.194 (0.134–0.291) |
| Median prostate volume (ml) | 48.4 (33.4–67.9) | 42.6 (30.2–67.7) |
| Median lesion volume (ml) | 0.53 (0.34–0.91) | 0.53 (0.32–0.98) |
| MRI index lesion evaluation: | ||
| PI-RADS 2 | 8 (5%) | 5 (12%) |
| PI-RADS 3 | 70 (46%) | 14 (35%) |
| PI-RADS 4 | 43 (28%) | 14 (35%) |
| PI-RADS 5 | 30 (20%) | 7 (23%) |
| Prior biopsy status: | ||
| No biopsy | 73 | 13 |
| Prior biopsy negative | 59 | 22 |
| Prior biopsy positive | 19 | 5 |
| Prior transurethral resection of the prostate (TURP) | 17 | 5 |
| Predictor | Malignant vs. Benign Lesions | csPCa vs. cisPCa | ||||
|---|---|---|---|---|---|---|
| Cohort | mean AUC | 95% CI * | p-Value♱ | mean AUC | 95% CI * | p-Value♱ |
| Training | ||||||
| PI-RADS | 0.758 | (0.671–0.817) | 0.368 | 0.681 | (0.572–0.786) | 0.144 |
| mADC | 0.754 | (0.677–0.827) | 0.222 | 0.697 | (0.592–0.781) | 0.177 |
| PSAD | 0.780 | (0.704–0.859) | 0.714 | 0.644 | (0.545–0.740) | 0.039⤲ |
| DRE | 0.617 | (0.556–0.667) | <0.001⤲ | 0.666 | (0.605–0.721) | 0.039⤲ |
| Radiomics model | 0.783 | (0.682–0.875) | ref. | 0.807 | (0.691–0.906) | ref. |
| Test | ||||||
| PI-RADS | 0.779 | (0.603–0.922) | 0.054 | 0.688 | (0.431–0.889) | 0.209 |
| mADC | 0.745 | (0.583–0.887) | 0.067 | 0.571 | (0.277–0.691) | 0.022⤲ |
| Ensemble radiomics model | 0.889 | (0.751–0.990) | ref. | 0.844 | (0.6–1.0) | ref. |
| Predictor | Sensitivity (%) * | 95% CI (%) | p Value ♱ | Specificity (%) * | 95% CI (%) |
|---|---|---|---|---|---|
| Cohort | malignant vs. benign lesions | ||||
| Training | |||||
| PI-RADS | 70 (50/71) | (59–81) | 0.832 | 71 (56/79) | (60–81) |
| mADC | 69 (49/71) | (56–77) | 0.581 | 73 (58/79) | (65–84) |
| Radiomics model | 75 (53/71) | (57–87) | reference | 73 (58/79) | (56–83) |
| Test | |||||
| PI-RADS | 83 (15/18) | (74–91) | 0.500 | 73 (16/22) | (63–82) |
| mADC | 61 (11/18) | (50–73) | 0.031 | 82 (18/22) | (73–90) |
| Ensemble radiomics | 94 (17/18) | (88–99) | reference | 77 (17/22) | (68–86) |
| csPCa vs. cisPCa | |||||
| Training | |||||
| PI-RADS | 80 (43/54) | (69–90) | 0.803 | 59 (10/17) | (36–83) |
| mADC | 70 (38/54) | (58–81) | 0.092 | 59 (10/17) | (33–82) |
| Radiomics model | 83 (45/54) | (69–95) | reference | 65 (11/17) | (20–100) |
| Test | |||||
| PI-RADS | 91 (10/11) | (82–98) | 1.0 | 28 (2/7) | (13–46) |
| mADC | 64 (7/11) | (50–78) | 0.250 | 43 (3/7) | (24–60) |
| Ensemble radiomics | 91 (10/11) | (81–98) | reference | 57 (4/7) | (38–74) |
| Cohort * | Predictor | Mean AUC | 95% CI | p-Value ♱ |
|---|---|---|---|---|
| malignant vs. benign lesions | ||||
| Small lesions (71) | PI-RADS mADC Radiomics model | 0.694 0.662 0.678 | (0.582–0.803) (0.530–0.781) (0.560–0.814) | 0.964 0.574 reference |
| Large lesions (77) | PI-RADS mADC Radiomics model | 0.792 0.829 0.890 | (0.694–0.882) (0.736–0.915) (0.812–0.953) | 0.093 0.212 reference |
| csPCa vs. cisPCa | ||||
| Small lesions (32) | PI-RADS mADC Radiomics model | 0.707 0.731 0.708 | (0.506–0.875) (0.537–0.897)v(0.525–0.880) | 0.972 0.801 reference |
| Large lesions (39) | PI-RADS mADC Radiomics model | 0.619 0.633 0.873 | (0.385–0.853) (0.388–0.838) (0.701–0.993) | 0.030 0.086 reference |
| Feature | Sequence | VOI | ρ | p-Value |
|---|---|---|---|---|
| ‘original_shape_Maximum2DDiameterColumn’ | T2 | whole gland | −0.229 | 0.031 ⤲ |
| ‘original_firstorder_Maximum’ | ADC | lesion | −0.110 | 0.305 |
| ‘original_shape_Sphericity’ | T2 | whole gland | −0.154 | 0.149 |
| ‘original_glrlm_GrayLevelNonUniformityNormalized’ | T2 | lesion | −0.260 | 0.014 ⤲ |
| ‘original_shape_Elongation’ | ADC | lesion | −0.11 | 0.301 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woźnicki, P.; Westhoff, N.; Huber, T.; Riffel, P.; Froelich, M.F.; Gresser, E.; von Hardenberg, J.; Mühlberg, A.; Michel, M.S.; Schoenberg, S.O.; et al. Multiparametric MRI for Prostate Cancer Characterization: Combined Use of Radiomics Model with PI-RADS and Clinical Parameters. Cancers 2020, 12, 1767. https://doi.org/10.3390/cancers12071767
Woźnicki P, Westhoff N, Huber T, Riffel P, Froelich MF, Gresser E, von Hardenberg J, Mühlberg A, Michel MS, Schoenberg SO, et al. Multiparametric MRI for Prostate Cancer Characterization: Combined Use of Radiomics Model with PI-RADS and Clinical Parameters. Cancers. 2020; 12(7):1767. https://doi.org/10.3390/cancers12071767
Chicago/Turabian StyleWoźnicki, Piotr, Niklas Westhoff, Thomas Huber, Philipp Riffel, Matthias F. Froelich, Eva Gresser, Jost von Hardenberg, Alexander Mühlberg, Maurice Stephan Michel, Stefan O. Schoenberg, and et al. 2020. "Multiparametric MRI for Prostate Cancer Characterization: Combined Use of Radiomics Model with PI-RADS and Clinical Parameters" Cancers 12, no. 7: 1767. https://doi.org/10.3390/cancers12071767
APA StyleWoźnicki, P., Westhoff, N., Huber, T., Riffel, P., Froelich, M. F., Gresser, E., von Hardenberg, J., Mühlberg, A., Michel, M. S., Schoenberg, S. O., & Nörenberg, D. (2020). Multiparametric MRI for Prostate Cancer Characterization: Combined Use of Radiomics Model with PI-RADS and Clinical Parameters. Cancers, 12(7), 1767. https://doi.org/10.3390/cancers12071767





