Single Nucleotide Polymorphisms in MiRNA Binding Sites of Nucleotide Excision Repair-Related Genes Predict Clinical Benefit of Oxaliplatin in FOLFOXIRI Plus Bevacizumab: Analysis of the TRIBE Trial
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Patients
2.2. Selection of Candidate SNPs
2.3. DNA Extraction and Genotyping
2.4. Statistical Analysis
3. Results
3.1. Baseline Clinicopathological Characteristics of the Discovery and Control Cohorts
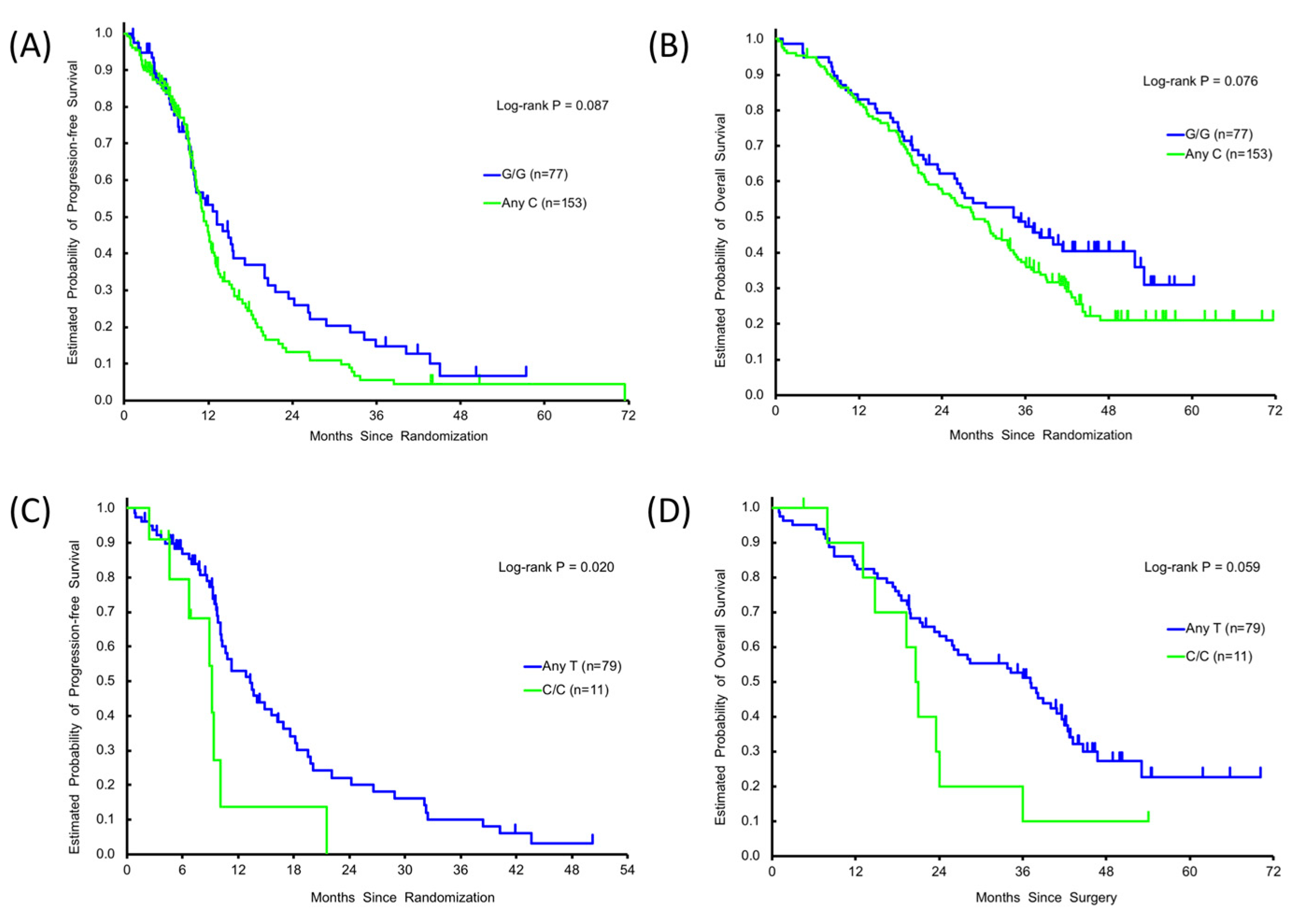
3.2. Association between Clinical Outcome and SNPs in the Discovery Cohort
3.3. Association between Clinical Outcome and SNPs in the Control Cohort
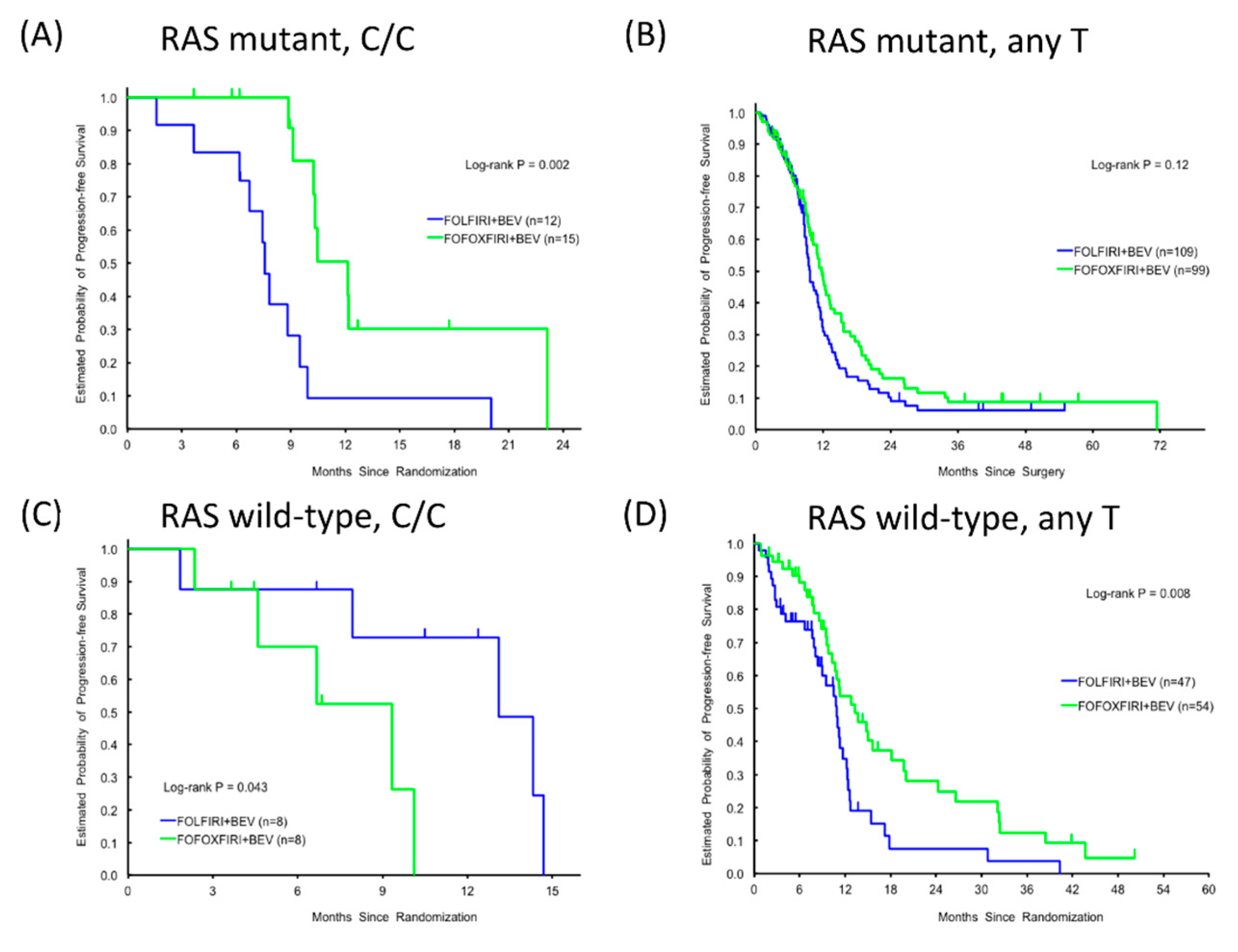
3.4. Clinical Significance of RPA2 SNP Alleles and RAS Mutational Status in the FOLFOXIRI + BEV and FOLFIRI + BEV Cohorts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheung-Ong, K.; Giaever, G.; Nislow, C. DNA-damaging agents in cancer chemotherapy: Serendipity and chemical biology. Chem. Biol. 2013, 20, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Maddukuri, L.; Dudzińska, D.; Tudek, B. Bacterial DNA repair genes and their eukaryotic homologues: 4. The role of nucleotide excision DNA repair (NER) system in mammalian cells. Acta Biochim. Pol. 2007, 54, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microRNA networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar] [CrossRef]
- Cipollini, M.; Landi, S.; Gemignani, F. MicroRNA binding site polymorphisms as biomarkers in cancer management and research. Pharmgenom. Pers. Med. 2014, 7, 173–191. [Google Scholar] [PubMed]
- Landi, D.; Gemignani, F.; Naccarati, A.; Pardini, B.; Vodicka, P.; Vodickova, L.; Novotny, J.; Försti, A.; Hemminki, K.; Canzian, F.; et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 2008, 29, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Naccarati, A.; Pardini, B.; Stefano, L.; Landi, D.; Slyskova, J.; Novotny, J.; Levy, M.; Polakova, V.; Lipska, L.; Vodicka, P. Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis 2012, 33, 1346–1351. [Google Scholar] [CrossRef] [PubMed]
- Loupakis, F.; Cremolini, C.; Masi, G.; Lonardi, S.; Zagonel, V.; Salvatore, L.; Cortesi, E.; Tomasello, G.; Ronzoni, M.; Spadi, R.; et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N. Engl. J. Med. 2014, 371, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Cremolini, C.; Loupakis, F.; Antoniotti, C.; Lupi, C.; Sensi, E.; Lonardi, S.; Mezi, S.; Tomasello, G.; Ronzoni, M.; Zaniboni, A.; et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: Updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015, 16, 1306–1315. [Google Scholar] [CrossRef]
- Lee, P.H.; Shatkay, H. F-SNP: Computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008, 36, D820–D824. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Balibrea, E.; Martínez-Cardús, A.; Ginés, A.; Ruiz de Porras, V.; Moutinho, C.; Layos, L.; Manzano, J.L.; Bugés, C.; Bystrup, S.; Esteller, M.; et al. Tumor-Related Molecular Mechanisms of Oxaliplatin Resistance. Mol. Cancer Ther. 2015, 14, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Shirota, Y.; Stoehlmacher, J.; Brabender, J.; Xiong, Y.P.; Uetake, H.; Danenberg, K.D.; Groshen, S.; Tsao-Wei, D.D.; Danenberg, P.V.; Lenz, H.J. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J. Clin. Oncol. 2001, 19, 4298–4304. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, A.; Di Salvatore, M.; Bagalà, C.; Basso, M.; Strippoli, A.; Plastino, F.; Calegari, M.A.; Cassano, A.; Astone, A.; Barone, C. ERCC1 Induction after Oxaliplatin Exposure May Depend on KRAS Mutational Status in Colorectal Cancer Cell Line: In Vitro Veritas. J. Cancer 2015, 6, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.; McLean, E.G.; Aroori, S.; Wilson, P.; McCulla, A.; Carey, P.D.; Longley, D.B.; Johnston, P.G. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin. Cancer Res. 2004, 10, 2158–2167. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 2001, 1, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.; Moggs, J.G.; Hwang, J.R.; Egly, J.M.; Wood, R.D. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997, 16, 6559–6573. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Choi, B.S. The protein shuffle. Sequential interactions among components of the human nucleotide excision repair pathway. FEBS J. 2006, 273, 1600–1608. [Google Scholar] [CrossRef] [PubMed]
- Jordheim, L.P.; Cros-Perrial, E.; Matera, E.L.; Bouledrak, K.; Dumontet, C. Expression of domains for protein-protein interaction of nucleotide excision repair proteins modifies cancer cell sensitivity to platinum derivatives and genomic stability. Clin. Exp. Pharmacol. Physiol. 2014, 4, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Saijo, M.; Takedachi, A.; Tanaka, K. Nucleotide excision repair by mutant xeroderma pigmentosum group A (XPA) proteins with deficiency in interaction with RPA. J. Biol. Chem. 2011, 286, 5476–5483. [Google Scholar] [CrossRef] [PubMed]
- Stigger, E.; Drissi, R.; Lee, S.H. Functional analysis of human replication protein A in nucleotide excision repair. J. Biol. Chem. 1998, 273, 9337–9343. [Google Scholar] [CrossRef] [PubMed]
- Araújo, S.J.; Nigg, E.A.; Wood, R.D. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell Biol. 2001, 21, 2281–2291. [Google Scholar] [CrossRef] [PubMed]
| SNPs | Patients | Tumor Response | Progression-Free Survival | Overall Survival | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variants | N | CR + PR | SD + PD | p Value * | Median, Months (95%CI) | HR (95%CI) † | p Value * | HR (95%CI) ‡ | p Value * | Median, Months (95%CI) | HR (95%CI) † | p Value * | HR (95%CI) ‡ | p Value * |
| Discovery cohort | ||||||||||||||
| GTF2H1 rs4596 | 0.21 | 0.21 | 0.18 | 0.20 | 0.15 | |||||||||
| G/G | 77 | 56(73.7%) | 20(26.3%) | 13.2(9.9,17.2) | Reference | Reference | 34.3(25.8,51.8) | Reference | Reference | |||||
| G/C | 117 | 74(66.7%) | 37(33.3%) | 1.3(10.1,12.2) | 1.30(0.92,1.83) | 1.32(0.90,1.93) | 30.2(24.0,33.4) | 1.38(0.97,1.97) | 1.37(0.93,2.04) | |||||
| C/C | 36 | 20(57.1%) | 15(42.9%) | 12.4(9.7,13.7) | 1.41(0.87,2.28) | 1.54(0.93,2.56) | 26.7(17.9,39.1) | 1.30(0.80,2.10) | 1.59(0.95,2.66) | |||||
| 0.16 | 0.087 | 0.085 | 0.076 | 0.066 | ||||||||||
| G/G | 77 | 56(73.7%) | 20(26.3%) | 13.2(9.9,17.2) | Reference | Reference | 34.3(25.8,51.8) | Reference | Reference | |||||
| AnyC | 153 | 94(64.4%) | 52(35.6%) | 11.3(10.3,12.5) | 1.32(0.95,1.84) | 1.37(0.96,1.96) | 28.6(23.4,33.4) | 1.36(0.97,1.92) | 1.42(0.98,2.07) | |||||
| RPA rs7356 | 0.25 | 0.47 | 0.31 | 0.60 | 0.92 | |||||||||
| T/T | 87 | 60(71.4%) | 24(28.6%) | 11.2(9.9,14.1) | Reference | Reference | 28.4(21.5,34.7) | Reference | Reference | |||||
| T/C | 111 | 66(62.3%) | 40(37.7%) | 12.5(10.9,13.7) | 1.22(0.87,1.72) | 1.29(0.90,1.84) | 33.4(25.2,37.1) | 1.18(0.84,1.66) | 1.08(0.76,1.53) | |||||
| C/C | 32 | 24(75.0%) | 8(25.0%) | 10.3(9.1,21.6) | 1.20(0.72,1.98) | 1.37(0.80,2.34) | 30.9(19.3,36.8) | 1.05(0.64,1.74) | 1.03(0.61,1.73) | |||||
| 0.33 | 0.76 | 0.48 | 0.83 | 0.96 | ||||||||||
| AnyT | 198 | 126(66.3%) | 64(33.7%) | 12.0(10.8,13.2) | Reference | Reference | 30.2(25.9,34.4) | Reference | Reference | |||||
| C/C | 32 | 24(75.0%) | 8(25.0%) | 10.3(9.1,21.6) | 1.07(0.68,1.70) | 1.20(0.73,1.96) | 30.9(19.3,36.8) | 0.95(0.60,1.51) | 0.99(0.61,1.60) | |||||
| Control cohort | ||||||||||||||
| GTF2H1 rs4596 | 0.90 | 0.59 | 0.27 | 0.60 | 0.077 | |||||||||
| G/G | 67 | 38(56.7%) | 29(43.3%) | 10.4(9.0,12.6) | Reference | Reference | 26.3(20.5,36.1) | Reference | Reference | |||||
| G/C | 110 | 64(59.8%) | 43(40.2%) | 9.7(8.6,10.8) | 1.17(0.83,1.66) | 1.34(0.93,1.94) | 25.6(20.8,30.8) | 1.07(0.76,1.52) | 1.39(0.97,2.01) | |||||
| C/C | 48 | 25(56.8%) | 19(43.2%) | 9.4(7.9,11.2) | 1.21(0.79,1.85) | 1.31(0.82,2.11) | 24.3(17.8,31.6) | 1.24(0.81,1.89) | 1.67(1.05,2.66) | |||||
| 0.76 | 0.31 | 0.11 | 0.50 | 0.038 | ||||||||||
| G/G | 67 | 38(56.7%) | 29(43.3%) | 10.4(9.0,12.6) | Reference | Reference | 26.3(20.5,36.1) | Reference | Reference | |||||
| AnyC | 158 | 89(58.9%) | 62(41.1%) | 9.5(8.7,10.8) | 1.18(0.85,1.64) | 1.33(0.94,1.89) | 25.1(21.1,29.1) | 1.12(0.81,1.55) | 1.45(1.02,2.07) | |||||
| RPA rs7356 | 0.45 | 0.38 | 0.61 | 0.55 | 0.77 | |||||||||
| T/T | 89 | 49(57.0%) | 37(43.0%) | 10.3(9.2,11.6) | Reference | Reference | 26.3(22.0,34.4) | Reference | Reference | |||||
| T/C | 113 | 67(61.5%) | 42(38.5%) | 9.5(8.7,11.1) | 1.18(0.86,1.64) | 1.18(0.84,1.66) | 26.2(21.1,32.5) | 0.97(0.71,1.33) | 1.01(0.72,1.40) | |||||
| C/C | 25 | 12(48.0%) | 13(52.0%) | 8.8(7.5,10.8) | 1.36(0.83,2.23) | 1.16(0.70,1.95) | 18.8(14.4,27.9) | 1.26(0.77,2.08) | 1.20(0.71,2.02) | |||||
| 0.27 | 0.35 | 0.80 | 0.28 | 0.47 | ||||||||||
| AnyT | 202 | 116(59.5%) | 79(40.5%) | 9.7(9.3,11.0) | Reference | Reference | 26.3(23.0,31.3) | Reference | Reference | |||||
| C/C | 25 | 12(48.0%) | 13(52.0%) | 8.8(7.5,10.8) | 1.24(0.78,1.96) | 1.06(0.66,1.71) | 18.8(14.4,27.9) | 1.29(0.81,2.06) | 1.20(0.73,1.95) | |||||
| SNPs | Patients | Tumor Response | Progression-Free Survival | Overall Survival | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variants | N | CR + PR | SD + PD | p Value * | Median, Months (95%CI) | HR (95%CI) † | p Value * | HR (95%CI) ‡ | p Value * | Median, Months (95%CI) | HR (95%CI) † | p Value * | HR (95%CI) ‡ | p Value * |
| KRAS wild-type | ||||||||||||||
| GTF2H1 rs4596 | 0.95 | 0.13 | 0.30 | 0.56 | 0.88 | |||||||||
| G/G | 32 | 21(67.7%) | 10(32.3%) | 13.3(9.3,24.2) | Reference | Reference | 37.1(19.7,NE) | Reference | Reference | |||||
| G/C | 47 | 30(65.2%) | 16(34.8%) | 10.7(9.5,14.1) | 1.60(0.91,2.82) | 1.69(0.85,3.36) | 33.8(21.0,40.9) | 1.34(0.76,2.34) | 1.15(0.62,2.17) | |||||
| C/C | 11 | 7(70.0%) | 3(30.0%) | 13.7(3.7,18.4) | 1.77(0.79,3.99) | 1.60(0.66,3.89) | 23.3(6.4,NE) | 1.36(0.60,3.11) | 1.20(0.49,2.94) | |||||
| 0.87 | 0.045 | 0.12 | 0.28 | 0.63 | ||||||||||
| G/G | 32 | 21(67.7%) | 10(32.3%) | 13.3(9.3,24.2) | Reference | Reference | 37.1(19.7,NE) | Reference | Reference | |||||
| AnyC | 58 | 37(66.1%) | 19(33.9%) | 10.8(9.7,14.1) | 1.64(0.95,2.81) | 1.66(0.87,3.17) | 28.5(21.0,40.9) | 1.34(0.78,2.30) | 1.16(0.63,2.13) | |||||
| RPA rs7356 | 0.48 | 0.061 | 0.042 | 0.13 | 0.056 | |||||||||
| T/T | 38 | 26(72.2%) | 10(27.8%) | 11.2(10.1,18.2) | Reference | Reference | 28.1(21.2,NE) | Reference | Reference | |||||
| T/C | 41 | 24(60.0%) | 16(40.0%) | 13.7(9.6,16.9) | 1.12(0.66,1.92) | 0.93(0.50,1.71) | 38.0(24.1,42.5) | 1.24(0.72,2.14) | 0.99(0.54,1.82) | |||||
| C/C | 11 | 8(72.7%) | 3(27.3%) | 9.1(4.6,10.1) | 2.47(1.08,5.65) | 2.85(1.16,7.03) | 20.8(7.9,24.0) | 2.19(1.00,4.80) | 2.56(1.13,5.82) | |||||
| 0.65 | 0.020 | 0.012 | 0.059 | 0.016 | ||||||||||
| AnyT | 79 | 50(65.8%) | 26(34.2%) | 13.3(10.2,16.9) | Reference | Reference | 37.1(25.9,42.0) | Reference | 37.1(25.9,42.0) | |||||
| C/C | 11 | 8(72.7%) | 3(27.3%) | 9.1(4.6,10.1) | 2.32(1.07,5.03) | 2.97(1.27,6.94) | 20.8(7.9,24.0) | 1.94(0.94,3.99) | 20.8(7.9,24.0) | |||||
| RAS wild-type | ||||||||||||||
| GTF2H1 rs4596 | 0.89 | 0.32 | 0.26 | 0.39 | 0.62 | |||||||||
| G/G | 22 | 14(66.7%) | 7(33.3%) | 14.8(8.9,24.2) | Reference | Reference | 37.1(19.4,NE) | Reference | Reference | |||||
| G/C | 34 | 20(60.6%) | 13(39.4%) | 10.8(8.6,18.2) | 1.50(0.77,2.95) | 1.87(0.77,4.57) | 34.3(21.0,42.5) | 1.39(0.71,2.73) | 1.12(0.50,2.48) | |||||
| C/C | 6 | 4(66.7%) | 2(33.3%) | 13.7(3.7,19.8) | 1.86(0.60,5.79) | 2.44(0.66,8.97) | 44.6(6.4,NE) | 0.71(0.20,2.47) | 0.58(0.15,2.25) | |||||
| 0.69 | 0.15 | 0.11 | 0.49 | 0.97 | ||||||||||
| G/G | 22 | 14(66.7%) | 7(33.3%) | 14.8(8.9,24.2) | Reference | 14.8(8.9,24.2) | 37.1(19.4,NE) | Reference | Reference | |||||
| AnyC | 40 | 24(61.5%) | 15(38.5%) | 10.8(9.3,15.0) | 1.55(0.81,2.98) | 10.8(9.3,15.0) | 35.9(21.0,42.6) | 1.26(0.65,2.44) | 0.98(0.46,2.11) | |||||
| RPA rs7356 | 0.59 | 0.015 | 0.008 | 0.18 | 0.22 | |||||||||
| T/T | 25 | 16(66.7%) | 8(33.3%) | 10.8(8.9,20.0) | Reference | Reference | 41.7(21.2,NE) | Reference | Reference | |||||
| T/C | 29 | 16(57.1%) | 12(42.9%) | 13.7(9.6,24.2) | 1.09(0.56,2.12) | 0.62(0.25,1.53) | 38.0(24.1,43.2) | 1.40(0.70,2.78) | 0.85(0.39,1.85) | |||||
| C/C | 8 | 6(75.0%) | 2(25.0%) | 9.3(2.4,10.1) | 3.70(1.18,11.56) | 4.63(1.33,16.14) | 20.6(7.9,23.6) | 2.44(0.91,6.54) | 2.16(0.73,6.35) | |||||
| 0.46 | 0.004 | 0.003 | 0.10 | 0.090 | ||||||||||
| AnyT | 54 | 32(61.5%) | 20(38.5%) | 13.3(10.3,18.2) | Reference | Reference | 38.0(26.1,43.2) | Reference | Reference | |||||
| C/C | 8 | 6(75.0%) | 2(25.0%) | 9.3(2.4,10.1) | 3.51(1.20,10.29) | 5.96(1.84,19.29) | 20.6(7.9,23.6) | 2.01(0.83,4.88) | 2.36(0.88,6.39) | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suenaga, M.; Schirripa, M.; Cao, S.; Zhang, W.; Yang, D.; Cremolini, C.; Murgioni, S.; Lonardi, S.; Ning, Y.; Okazaki, S.; et al. Single Nucleotide Polymorphisms in MiRNA Binding Sites of Nucleotide Excision Repair-Related Genes Predict Clinical Benefit of Oxaliplatin in FOLFOXIRI Plus Bevacizumab: Analysis of the TRIBE Trial. Cancers 2020, 12, 1742. https://doi.org/10.3390/cancers12071742
Suenaga M, Schirripa M, Cao S, Zhang W, Yang D, Cremolini C, Murgioni S, Lonardi S, Ning Y, Okazaki S, et al. Single Nucleotide Polymorphisms in MiRNA Binding Sites of Nucleotide Excision Repair-Related Genes Predict Clinical Benefit of Oxaliplatin in FOLFOXIRI Plus Bevacizumab: Analysis of the TRIBE Trial. Cancers. 2020; 12(7):1742. https://doi.org/10.3390/cancers12071742
Chicago/Turabian StyleSuenaga, Mitsukuni, Marta Schirripa, Shu Cao, Wu Zhang, Dongyun Yang, Chiara Cremolini, Sabina Murgioni, Sara Lonardi, Yan Ning, Satoshi Okazaki, and et al. 2020. "Single Nucleotide Polymorphisms in MiRNA Binding Sites of Nucleotide Excision Repair-Related Genes Predict Clinical Benefit of Oxaliplatin in FOLFOXIRI Plus Bevacizumab: Analysis of the TRIBE Trial" Cancers 12, no. 7: 1742. https://doi.org/10.3390/cancers12071742
APA StyleSuenaga, M., Schirripa, M., Cao, S., Zhang, W., Yang, D., Cremolini, C., Murgioni, S., Lonardi, S., Ning, Y., Okazaki, S., Berger, M. D., Miyamoto, Y., Barzi, A., Loupakis, F., Falcone, A., & Lenz, H.-J. (2020). Single Nucleotide Polymorphisms in MiRNA Binding Sites of Nucleotide Excision Repair-Related Genes Predict Clinical Benefit of Oxaliplatin in FOLFOXIRI Plus Bevacizumab: Analysis of the TRIBE Trial. Cancers, 12(7), 1742. https://doi.org/10.3390/cancers12071742






