Hot Spot TERT Promoter Mutations Are Rare in Sporadic Pancreatic Neuroendocrine Neoplasms and Associated with Telomere Length and Epigenetic Expression Patterns
Abstract
1. Introduction
2. Results
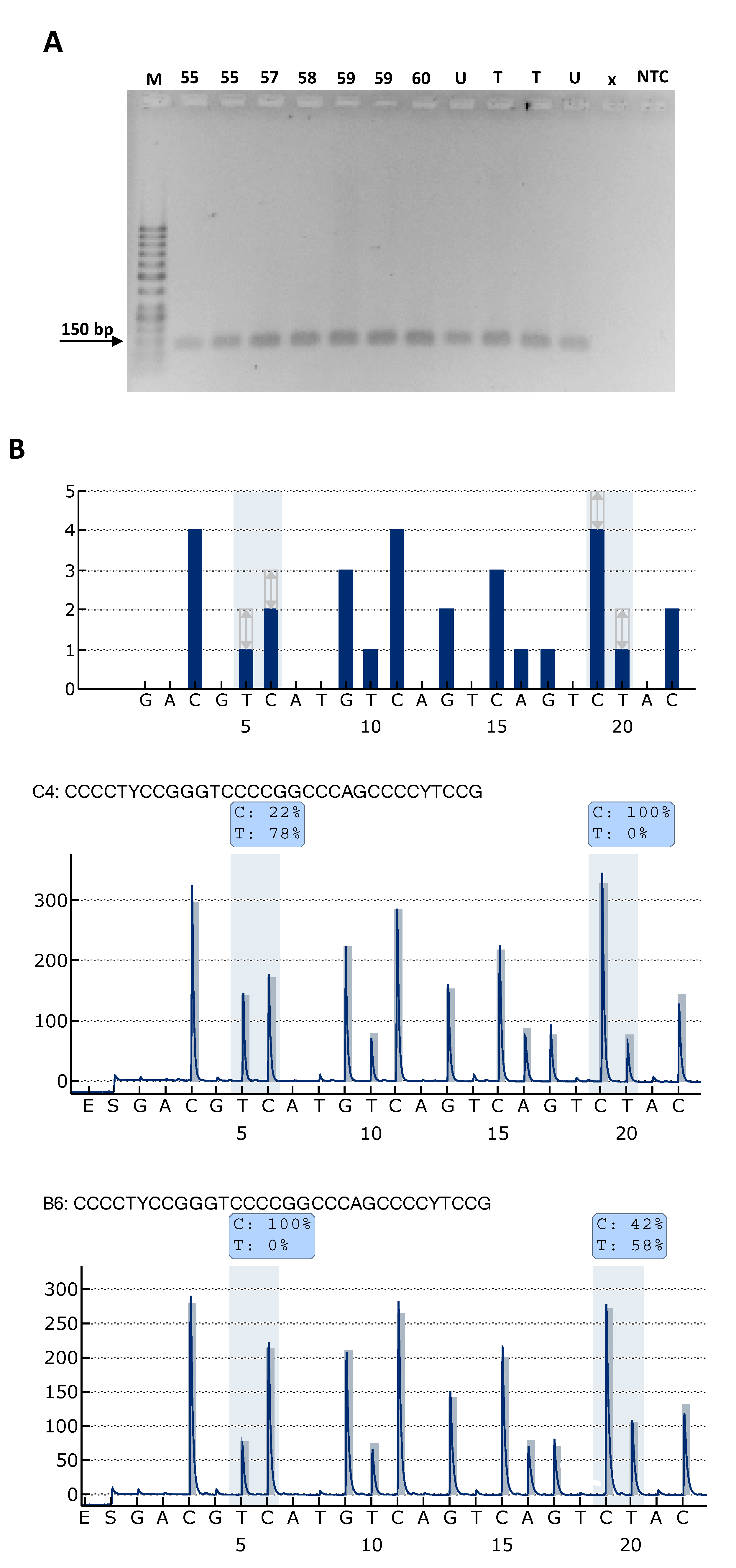
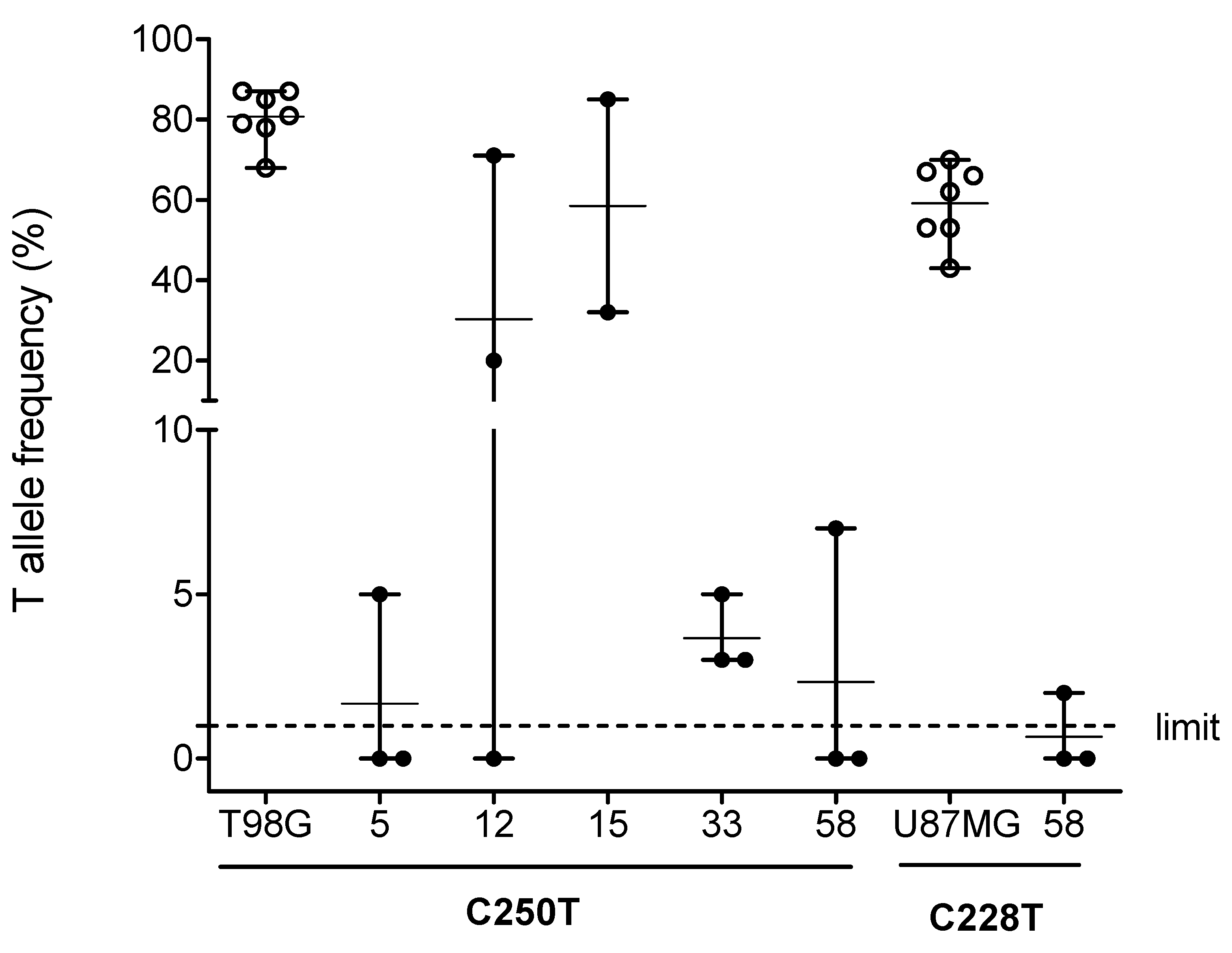
2.1. TPM Analysis and Clinicopathological Characteristics of Patients
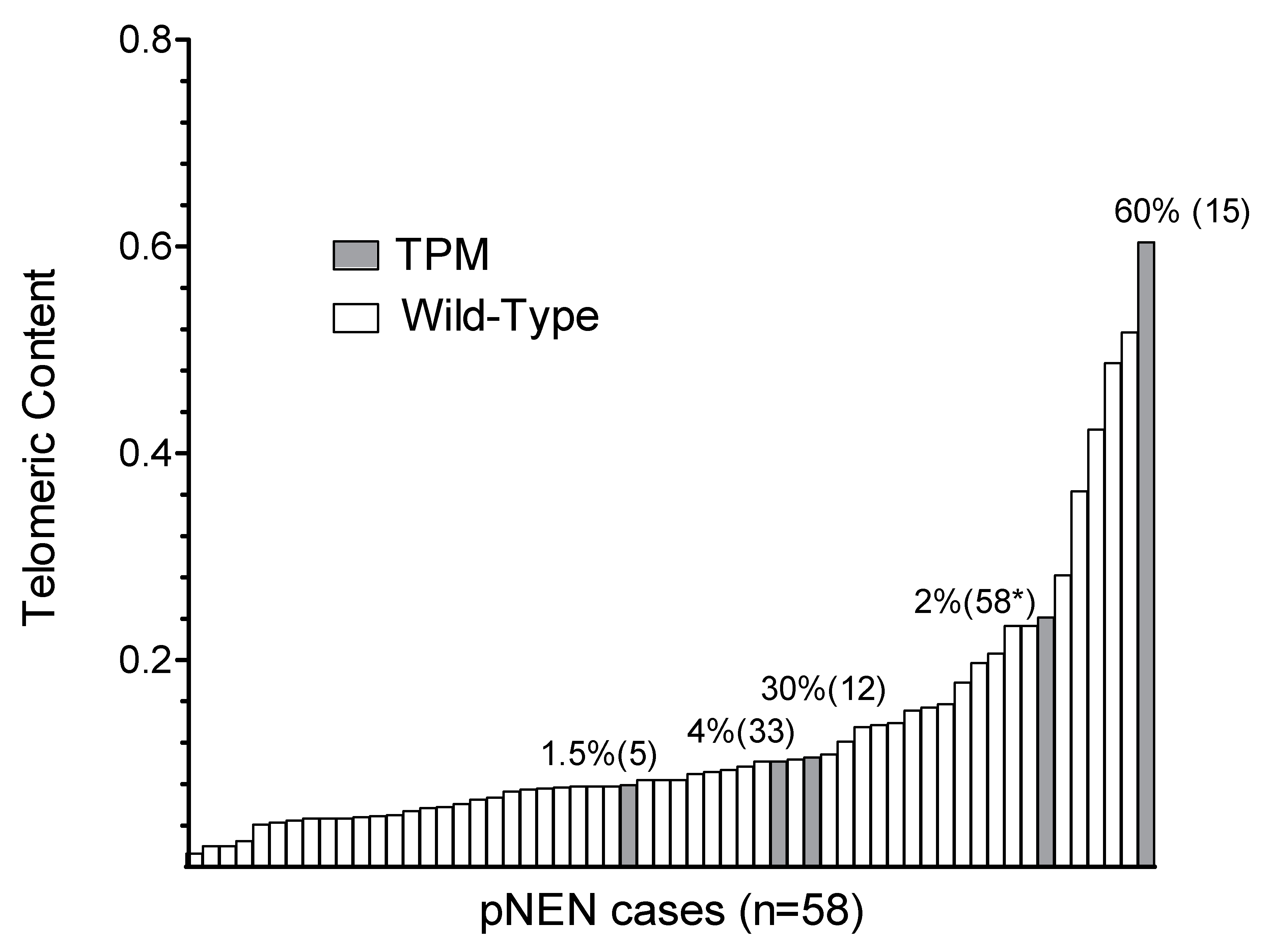
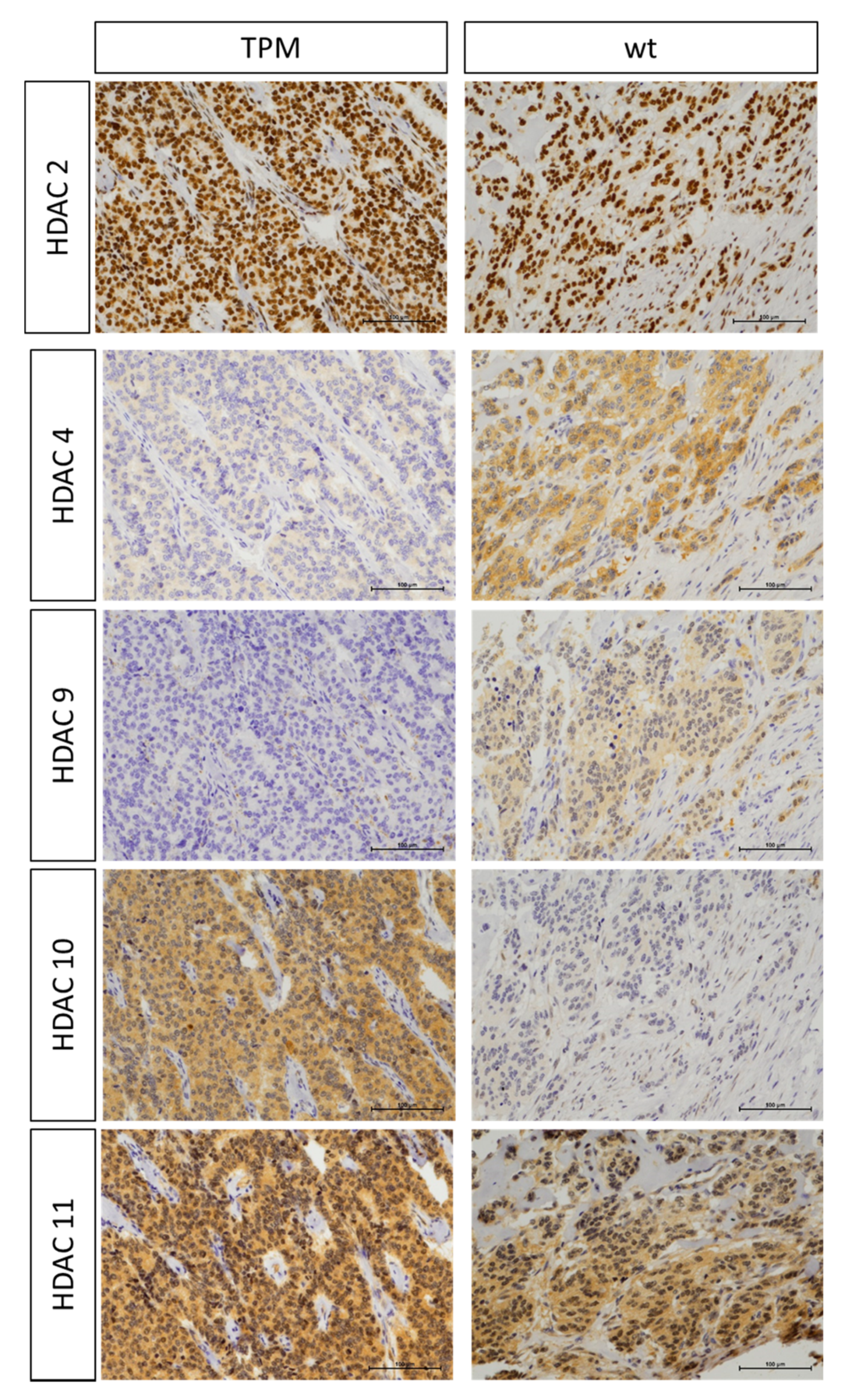
2.2. Association of TPM Status with Telomeric Content, miRNA and HDAC Expression
3. Discussion
4. Materials and Methods
4.1. Clinical and Pathological Characterization of Patients
4.2. Cell Line Controls
4.3. Isolation of DNA
4.4. TERT Promoter Mutation (TPM) Analysis by Pyrosequencing
4.5. Immunohistochemistry and Processing for Markers of HDACs (1-6, 8-11 and Sirt1)
4.6. Relative Telomere Length Analysis by qPCR
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; A Weinberg, R. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Gaspar, T.B.; Sá, A.; Lopes, J.M.; Sobrinho-Simões, M.; Soares, P.; Vinagre, J. Telomere Maintenance Mechanisms in Cancer. Genes 2018, 9, 241. [Google Scholar] [CrossRef]
- Roake, C.M.; Artandi, S.E. Regulation of human telomerase in homeostasis and disease. Nat. Rev. Mol. Cell Biol. 2020, 1–14. [Google Scholar] [CrossRef]
- Naderlinger, E.; Holzmann, K. Epigenetic Regulation of Telomere Maintenance for Therapeutic Interventions in Gliomas. Genes 2017, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- Lorbeer, F.K.; Hockemeyer, D. TERT promoter mutations and telomeres during tumorigenesis. Curr. Opin. Genet. Dev. 2020, 60, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Hackeng, W.M.; Hruban, R.H.; Offerhaus, G.J.A.; Brosens, L.A.A. Surgical and molecular pathology of pancreatic neoplasms. Diagn. Pathol. 2016, 11, 47. [Google Scholar] [CrossRef] [PubMed]
- Primavesi, F.; Klieser, E.; Cardini, B.; Marsoner, K.; Fröschl, U.; Thalhammer, S.; Fischer, I.; Hauer, A.; Urbas, R.; Kiesslich, T.; et al. Exploring the surgical landscape of pancreatic neuroendocrine neoplasia in Austria: Results from the ASSO pNEN study group. Eur. J. Surg. Oncol. 2019, 45, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Rindi, G.; Klersy, C.; Albarello, L.; Baudin, E.; Bianchi, A.; Büchler, M.W.; Caplin, M.E.; Couvelard, A.; Cros, J.; De Herder, W.; et al. Competitive Testing of the WHO 2010 versus the WHO 2017 Grading of Pancreatic Neuroendocrine Neoplasms: Data from a Large International Cohort Study. Neuroendocrinology 2018, 107, 375–386. [Google Scholar] [CrossRef]
- Mafficini, A.; Scarpa, A. Genetics and Epigenetics of Gastroenteropancreatic Neuroendocrine Neoplasms. Endocr. Rev. 2019, 40, 506–536. [Google Scholar] [CrossRef] [PubMed]
- Vinagre, J.; Nabais, J.; Pinheiro, J.; Batista, R.; Oliveira, R.C.; Gonçalves, A.P.; Pestana, A.; Reis, M.; Mesquita, B.; Pinto, V.; et al. TERT promoter mutations in pancreatic endocrine tumours are rare and mainly found in tumours from patients with hereditary syndromes. Sci. Rep. 2016, 6, 29714. [Google Scholar] [CrossRef] [PubMed]
- Klieser, E.; Urbas, R.; Swierczynski, S.; Stättner, S.; Primavesi, F.; Jäger, T.; Mayr, C.; Kiesslich, T.; Di Fazio, P.; Helm, K.; et al. HDAC-Linked “Proliferative” miRNA Expression Pattern in Pancreatic Neuroendocrine Tumors. Int. J. Mol. Sci. 2018, 19, 2781. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.J.A.; Rube, H.T.; Xavier-Magalhães, A.; Costa, B.M.; Mancini, A.; Song, J.S.; Costello, J.F. Understanding TERT Promoter Mutations: A Common Path to Immortality. Mol. Cancer Res. 2016, 14, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Arita, H.; Narita, Y.; Fukushima, S.; Tateishi, K.; Matsushita, Y.; Yoshida, A.; Miyakita, Y.; Ohno, M.; Collins, V.P.; Kawahara, N.; et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013, 126, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Lade-Keller, J.; Yuusufi, S.; Riber-Hansen, R.; Steiniche, T.; Stougaard, M. Telomerase reverse transcriptase promoter mutations and solar elastosis in cutaneous melanoma. Melanoma Res. 2018, 28, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Dang, S.; Wu, K.; Shao, Y.; Yang, Q.; Ji, M.; Shi, B.; Hou, P. TERT promoter mutations predict worse survival in laryngeal cancer patients. Int. J. Cancer 2014, 135, 1008–1010. [Google Scholar] [CrossRef]
- Qu, Y.; Shi, L.; Wang, D.; Zhang, B.; Yang, Q.; Ji, M.; Shi, B.; Hou, P. Low frequency of TERT promoter mutations in a large cohort of gallbladder and gastric cancers. Int. J. Cancer 2013, 134, 2993–2994. [Google Scholar] [CrossRef]
- Killela, P.J.; Reitman, Z.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef] [PubMed]
- Tsiatis, A.C.; Norris-Kirby, A.; Rich, R.G.; Hafez, M.J.; Gocke, C.D.; Eshleman, J.R.; Murphy, K.M. Comparison of Sanger Sequencing, Pyrosequencing, and Melting Curve Analysis for the Detection of KRAS Mutations. J. Mol. Diagn. 2010, 12, 425–432. [Google Scholar] [CrossRef]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [CrossRef]
- Sieverling, L.; PCAWG-Structural Variation Working Group; Hong, C.; Koser, S.D.; Ginsbach, P.; Kleinheinz, K.; Hutter, B.; Braun, D.M.; Cortes-Ciriano, I.; Xi, R.; et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; De Wilde, R.F.; Jiao, Y.; Klein, A.P.; Edil, B.H.; Shi, C.; Bettegowda, C.; Rodriguez, F.J.; Eberhart, C.G.; Hebbar, S.; et al. Altered Telomeres in Tumors with ATRX and DAXX Mutations. Science 2011, 333, 425. [Google Scholar] [CrossRef]
- Heaphy, C.M.; Subhawong, A.P.; Hong, S.-M.; Goggins, M.G.; Montgomery, E.A.; Gabrielson, E.; Netto, G.J.; Epstein, J.I.; Lotan, T.L.; Westra, W.H.; et al. Prevalence of the Alternative Lengthening of Telomeres Telomere Maintenance Mechanism in Human Cancer Subtypes. Am. J. Pathol. 2011, 179, 1608–1615. [Google Scholar] [CrossRef]
- Gerstung, M.; PCAWG Evolution & Heterogeneity Working Group; Jolly, C.; Leshchiner, I.; Dentro, S.C.; Gonzalez, S.; Rosebrock, D.; Mitchell, T.J.; Rubanova, Y.; Anur, P.; et al. The evolutionary history of 2,658 cancers. Nature 2020, 578, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Primavesi, F.; Andreasi, V.; Hoogwater, F.; Partelli, S.; Wiese, D.; Heidsma, C.; Cardini, B.; Klieser, E.; Marsoner, K.; Fröschl, U.; et al. A Preoperative Clinical Risk Score Including C-Reactive Protein Predicts Histological Tumor Characteristics and Patient Survival after Surgery for Sporadic Non-Functional Pancreatic Neuroendocrine Neoplasms: An International Multicenter Cohort Study. Cancers 2020, 12, 1235. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Kim, Y.-E.; Ahn, S.; Kim, J.-Y.; Ki, C.-S.; Oh, Y.L.; Kim, K.; Yun, J.W.; Park, W.-Y.; Choe, J.-H.; et al. TERT promoter mutations and long-term survival in patients with thyroid cancer. Endocr. Relat. Cancer 2016, 23, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.G.; Duong, U.N.; Altibi, A.M.; Ngo, H.T.; Pham, T.Q.; Tran, H.M.; Gandolfi, G.; Hassell, L. A meta-analysis of prognostic roles of molecular markers in papillary thyroid carcinoma. Endocr. Connect. 2017, 6, R8–R17. [Google Scholar] [CrossRef]
- Yin, D.-T.; Yu, K.; Lu, R.-Q.; Li, X.; Xu, J.; Lei, M.; Li, H.; Wang, Y.; Liu, Z.; De-Tao, Y.; et al. Clinicopathological significance of TERT promoter mutation in papillary thyroid carcinomas: a systematic review and meta-analysis. Clin. Endocrinol. 2016, 85, 299–305. [Google Scholar] [CrossRef]
- Gao, K.; Li, G.; Qu, Y.; Wang, M.; Cui, B.; Ji, M.; Shi, B.; Hou, P. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget 2015, 7, 8712–8725. [Google Scholar] [CrossRef] [PubMed]
- Batista, R.; Vinagre, N.; Meireles, S.; Vinagre, J.; Prazeres, H.; Leao, R.; Máximo, V.; Soares, P. Biomarkers for Bladder Cancer Diagnosis and Surveillance: A Comprehensive Review. Diagnostics 2020, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Chiba, K.; Lorbeer, F.K.; Shain, A.H.; McSwiggen, D.T.; Schruf, E.; Oh, A.; Ryu, J.; Darzacq, X.; Bastian, B.C.; Hockemeyer, D. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science 2017, 357, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Brosnan-Cashman, J.; An, S.; Kim, S.J.; Song, K.-B.; Kim, M.-S.; Kim, M.-J.; Hwang, D.W.; Meeker, A.K.; Yu, E.; et al. Alternative Lengthening of Telomeres in Primary Pancreatic Neuroendocrine Tumors Is Associated with Aggressive Clinical Behavior and Poor Survival. Clin. Cancer Res. 2016, 23, 1598–1606. [Google Scholar] [CrossRef] [PubMed]
- De Wilde, R.F.; Heaphy, C.M.; Maitra, A.; Meeker, A.K.; Edil, B.H.; Wolfgang, C.L.; A Ellison, T.; Schulick, R.D.; Molenaar, I.Q.; Valk, G.D.; et al. Loss of ATRX or DAXX expression and concomitant acquisition of the alternative lengthening of telomeres phenotype are late events in a small subset of MEN-1 syndrome pancreatic neuroendocrine tumors. Mod. Pathol. 2012, 25, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Marinoni, I.; Kurrer, A.S.; Vassella, E.; Dettmer, M.S.; Rudolph, T.; Banz, V.; Hunger, F.; Pasquinelli, S.; Speel, E.; Perren, A. Loss of DAXX and ATRX Are Associated With Chromosome Instability and Reduced Survival of Patients with Pancreatic Neuroendocrine Tumors. Gastroenterology 2014, 146, 453–460.e5. [Google Scholar] [CrossRef] [PubMed]
- Jamiruddin, M.R.; Kaitsuka, T.; Hakim, F.; Fujimura, A.; Wei, F.-Y.; Saitoh, H.; Tomizawa, K. HDAC9 regulates the alternative lengthening of telomere (ALT) pathway via the formation of ALT-associated PML bodies. Biochem. Biophys. Res. Commun. 2016, 481, 25–30. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA A Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.V.; Osamura, R.Y.; Klöppel, G.; Rosai, J. Who Classification of Tumours of Endocrine Organs, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017; pp. 209–240.
- Kreilmeier, T.; Mejri, D.; Hauck, M.; Kleiter, M.; Holzmann, K. Telomere Transcripts Target Telomerase in Human Cancer Cells. Genes 2016, 7, 46. [Google Scholar] [CrossRef]
- Kreilmeier, T.; Sampl, S.; Deloria, A.J.; Walter, I.; Reifinger, M.; Hauck, M.; Borst, L.; Holzmann, K.; Kleiter, M. Alternative lengthening of telomeres does exist in various canine sarcomas. Mol. Carcinog. 2016, 56, 923–935. [Google Scholar] [CrossRef]
- Nonoguchi, N.; Ohta, T.; Oh, J.-E.; Kim, Y.-H.; Kleihues, P.; Ohgaki, H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013, 126, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Johanns, T.M.; Fu, Y.; Kobayashi, D.K.; Mei, Y.; Dunn, I.F.; Mao, D.D.; Kim, A.H.; Dunn, G.P. High incidence of TERT mutation in brain tumor cell lines. Brain Tumor Pathol. 2016, 33, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Klieser, E.; Urbas, R.; Stättner, S.; Primavesi, F.; Jäger, T.; Dinnewitzer, A.; Mayr, C.; Kiesslich, T.; Holzmann, K.; Di Fazio, P.; et al. Comprehensive immunohistochemical analysis of histone deacetylases in pancreatic neuroendocrine tumors: HDAC5 as a predictor of poor clinical outcome. Hum. Pathol. 2017, 65, 41–52. [Google Scholar] [CrossRef]
- Henson, J.; Lau, L.; Koch, S.; La Rotta, N.M.; Dagg, R.A.; Reddel, R.R. The C-Circle Assay for alternative-lengthening-of-telomeres activity. Methods 2017, 114, 74–84. [Google Scholar] [CrossRef] [PubMed]
- The R Project for Statistical Computing. Available online: https://www.r-project.org (accessed on 18 June 2020).
| Parameters | Cases | p Value | ||
|---|---|---|---|---|
| with Data, n (%) | Wild-Type n = 53 (91%) | C250T/C228T n = 5 (9%) | ||
| Gender | 58 (100) | 0.536 | ||
| Male | 25 (43) | 24 (45) | 1 (20) | |
| Female | 33 (57) | 29 (55) | 4 (80) | |
| Age, years; mean ± SD | 58 (100) | 61.2 ± 14.4 | 68.4 ± 11.4 | 0.283 |
| Size, cm; mean ± SD | 57 (98) | 2.85 ± 2.38 | 1.85 ± 1.25 | 0.361 |
| Localization | 58 (100) | 0.613 | ||
| Caput | 23 (40) | 22 (42) | 1 (20) | |
| Corpus | 10 (17) | 9 (17) | 1 (20) | |
| Cauda | 22 (38) | 19 (36) | 3 (60) | |
| Unknown | 3 (5) | 3 (6) | 0 | |
| TNM-Staging (AJCC/UICC 2017) | 58 (100) | |||
| T-Category 1/2/3/4 | 19/13/16/5 | 3/0/1/1 | 0.433 | |
| N0/1 | 37/16 | 3/2 | 0.641 | |
| M0/1 | 44/9 | 3/2 | 0.237 | |
| M1a/1b/1c | 5/3/1 | 2/0/0 | 1.00 | |
| T-ENETS (2006) | 58 (100) | |||
| 1/2/3/4 | 19/13/16/5 | 3/0/1/1 | 0.434 | |
| Invasion of lymphatic vessels (L) | 58 (100) | 1.00 | ||
| L1 | 7 (12) | 7 (13) | 0 (0) | |
| L0 | 51(88) | 46 (87) | 5 (100) | |
| Vascular invasion (V) | 58 (100) | 0.433 | ||
| V1 | 6 (10) | 5 (9) | 1 (20) | |
| V0 | 52 (90) | 48 (91) | 4 (80) | |
| Grading (WHO 2017) | 58 (100) | 0.694 | ||
| NET G1 | 30 (52) | 27 (51) | 3 (60) | |
| NET G2 | 20 (34) | 19 (36) | 1 (20) | |
| NET G3 | 2 (4) | 2 (4) | 0 (0) | |
| NEC G3 | 6 (10) | 5 (9) | 1 (20) | |
| R status | 52 (90) | 1.00 | ||
| R0 | 47 (90) | 43 (90) | 4 (100) | |
| R1 | 5 (10) | 5 (10) | 0 (0) | |
| Overall survival (OS) | 51 (88) | 47 (89) | 4 (80) | |
| OS months, median (range) | 82 (6–232) | 77 (9–110) | 0.890 | |
| Disease-free survival (DFS) | 50 (86) | 46 (87) | 4 (80) | |
| DFS months, median (range) | 77 (0–194) | 56 (9–110) | 0.950 | |
| Ki67-index | 58 (100) | 53 | 5 | 0.657 |
| Mean ± SD | 10.7 ± 20.8 | 16.6 ± 20.9 | ||
| Hormonal activity | 44 (76) | 0.327 | ||
| Yes | 4 (9) | 3 (8) | 1 (25) | |
| No | 40 (91) | 37(93) | 3 (75) | |
| Parameters | Case Number | ||||
|---|---|---|---|---|---|
| 5 | 12 * | 15 | 33 | 58 | |
| TPMs | C250T | C250T | C250T | C250T | C250T and C228T |
| T allele frequency range (%) | 0–5 | 0–71 | 31–85 | 1–5 | 0–11 (C250T) 0–3 (C228T) |
| Tissue tumor cell content range (%) | 90–95 | 60–70 | 75–80 | 90–95 | 85–90 |
| Gender (male/female) | f | f | f | f | m |
| Age (years) | 55.7 | 84.7 | 59.7 | 62.8 | 79.3 |
| Tumor size (cm) | 4.00 | 2.50 | 0.95 | 1.20 | 0.60 |
| Localization | cauda | caput | cauda | corpus | cauda |
| TNM–Staging (AJCC/UICC 2017) | |||||
| T-category | 4 | 3 | 1 | 1 | 1 |
| N-status | 1 | 1 | 0 | 0 | 0 |
| M-status (a/b/c) | 1a | 1a | 0 | 0 | 0 |
| T-category (ENETS 2006) | 4 | 3 | 1 | 1 | 1 |
| Invasion of lymphatic vessels (L) | no | no | no | no | no |
| Vascular invasion (V) | no | yes | no | no | no |
| Grading (WHO 2017) | NEC G3 | NET G2 | NET G1 | NET G1 | NET G1 |
| R status | 0 | n.a. | 0 | 0 | n.a. |
| Ki67 (%) | 54.6 | 5.7 | 2.0 | 1.7 | 1.2 |
| Hormone syndrome association | no | n.a. | yes | no | no |
| Parameters * | Cases | p Value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPM Wild-Type | TPM Mutant C250T/C228T | ||||||||||||
| n | mean | median | SD | min | max | n | mean | median | SD | min | max | ||
| HDAC1 ncl | 43 | 153 | 150 | 75 | 5 | 300 | 5 | 159 | 128 | 93 | 40 | 285 | 0.852 |
| HDAC2 ncl | 44 | 193 | 170 | 66 | 90 | 300 | 5 | 181 | 190 | 50 | 113 | 238 | 0.778 |
| HDAC3 ncl | 43 | 102 | 88 | 84 | 0 | 285 | 4 | 73 | 35 | 96 | 10 | 213 | 0.434 |
| HDAC4 ncl | 41 | 32 | 30 | 26 | 0 | 83 | 5 | 17 | 10 | 24 | 0 | 60 | 0.146 |
| HDAC4 cyt | 41 | 72 | 70 | 48 | 0 | 213 | 5 | 47 | 15 | 52 | 10 | 128 | 0.297 |
| HDAC5 ncl | 42 | 166 | 170 | 62 | 3 | 300 | 5 | 175 | 190 | 48 | 98 | 213 | 0.862 |
| HDAC5 cyt | 42 | 96 | 98 | 44 | 10 | 190 | 5 | 117 | 113 | 59 | 50 | 190 | 0.521 |
| HDAC6 cyt | 44 | 105 | 113 | 48 | 20 | 213 | 5 | 112 | 120 | 81 | 10 | 190 | 0.728 |
| HDAC8 ncl | 46 | 10 | 5 | 19 | 0 | 113 | 5 | 4 | 0 | 6 | 0 | 10 | 0.347 |
| HDAC8 cyt | 43 | 15 | 10 | 23 | 0 | 130 | 3 | 17 | 0 | 29 | 0 | 50 | 0.665 |
| HDAC9 ncl | 46 | 4 | 5 | 6 | 0 | 30 | 5 | 1 | 0 | 2 | 0 | 5 | 0.149 |
| HDAC10 ncl | 40 | 100 | 120 | 71 | 0 | 250 | 5 | 64 | 60 | 62 | 10 | 160 | 0.384 |
| HDAC10 cyt | 40 | 138 | 165 | 74 | 5 | 300 | 5 | 119 | 135 | 70 | 10 | 190 | 0.549 |
| HDAC11 ncl | 42 | 132 | 128 | 56 | 3 | 238 | 5 | 124 | 113 | 42 | 75 | 190 | 0.704 |
| miR132-3p | 51 | 0.2038 | 0.0633 | 0.4358 | 0.0041 | 2.7602 | 4 | 0.0709 | 0.0290 | 0.0816 | 0.0138 | 0.2118 | 0.264 |
| miR145-5p | 51 | 1.0603 | 0.5649 | 1.1733 | 0.0453 | 4.7951 | 4 | 0.6500 | 0.5434 | 0.5141 | 0.1491 | 1.3640 | 0.427 |
| miR183-5p | 51 | 0.0741 | 0.0186 | 0.1900 | 0.0003 | 1.3351 | 4 | 0.0185 | 0.0188 | 0.0060 | 0.0098 | 0.0266 | 0.961 |
| miR34a-5p | 51 | 0.2545 | 0.1618 | 0.2536 | 0.0483 | 1.4541 | 4 | 0.1627 | 0.1386 | 0.0900 | 0.0681 | 0.3065 | 0.466 |
| miR449a | 51 | 0.0020 | 0.0008 | 0.0062 | 0.0001 | 0.0452 | 4 | 0.0040 | 0.0023 | 0.0040 | 0.0007 | 0.0106 | 0.157 |
| Telomeric content (TC) | 53 | 0.121 | 0.084 | 0.112 | 0.013 | 0.517 | 5 | 0.226 | 0.106 | 0.221 | 0.079 | 0.604 | 0.086 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Posch, A.; Hofer-Zeni, S.; Klieser, E.; Primavesi, F.; Naderlinger, E.; Brandstetter, A.; Filipits, M.; Urbas, R.; Swiercynski, S.; Jäger, T.; et al. Hot Spot TERT Promoter Mutations Are Rare in Sporadic Pancreatic Neuroendocrine Neoplasms and Associated with Telomere Length and Epigenetic Expression Patterns. Cancers 2020, 12, 1625. https://doi.org/10.3390/cancers12061625
Posch A, Hofer-Zeni S, Klieser E, Primavesi F, Naderlinger E, Brandstetter A, Filipits M, Urbas R, Swiercynski S, Jäger T, et al. Hot Spot TERT Promoter Mutations Are Rare in Sporadic Pancreatic Neuroendocrine Neoplasms and Associated with Telomere Length and Epigenetic Expression Patterns. Cancers. 2020; 12(6):1625. https://doi.org/10.3390/cancers12061625
Chicago/Turabian StylePosch, Alexandra, Sarah Hofer-Zeni, Eckhard Klieser, Florian Primavesi, Elisabeth Naderlinger, Anita Brandstetter, Martin Filipits, Romana Urbas, Stefan Swiercynski, Tarkan Jäger, and et al. 2020. "Hot Spot TERT Promoter Mutations Are Rare in Sporadic Pancreatic Neuroendocrine Neoplasms and Associated with Telomere Length and Epigenetic Expression Patterns" Cancers 12, no. 6: 1625. https://doi.org/10.3390/cancers12061625
APA StylePosch, A., Hofer-Zeni, S., Klieser, E., Primavesi, F., Naderlinger, E., Brandstetter, A., Filipits, M., Urbas, R., Swiercynski, S., Jäger, T., Winkelmann, P., Kiesslich, T., Lu, L., Neureiter, D., Stättner, S., & Holzmann, K. (2020). Hot Spot TERT Promoter Mutations Are Rare in Sporadic Pancreatic Neuroendocrine Neoplasms and Associated with Telomere Length and Epigenetic Expression Patterns. Cancers, 12(6), 1625. https://doi.org/10.3390/cancers12061625








