Tissue-Resident NK Cells: Development, Maturation, and Clinical Relevance
Abstract
1. Introduction
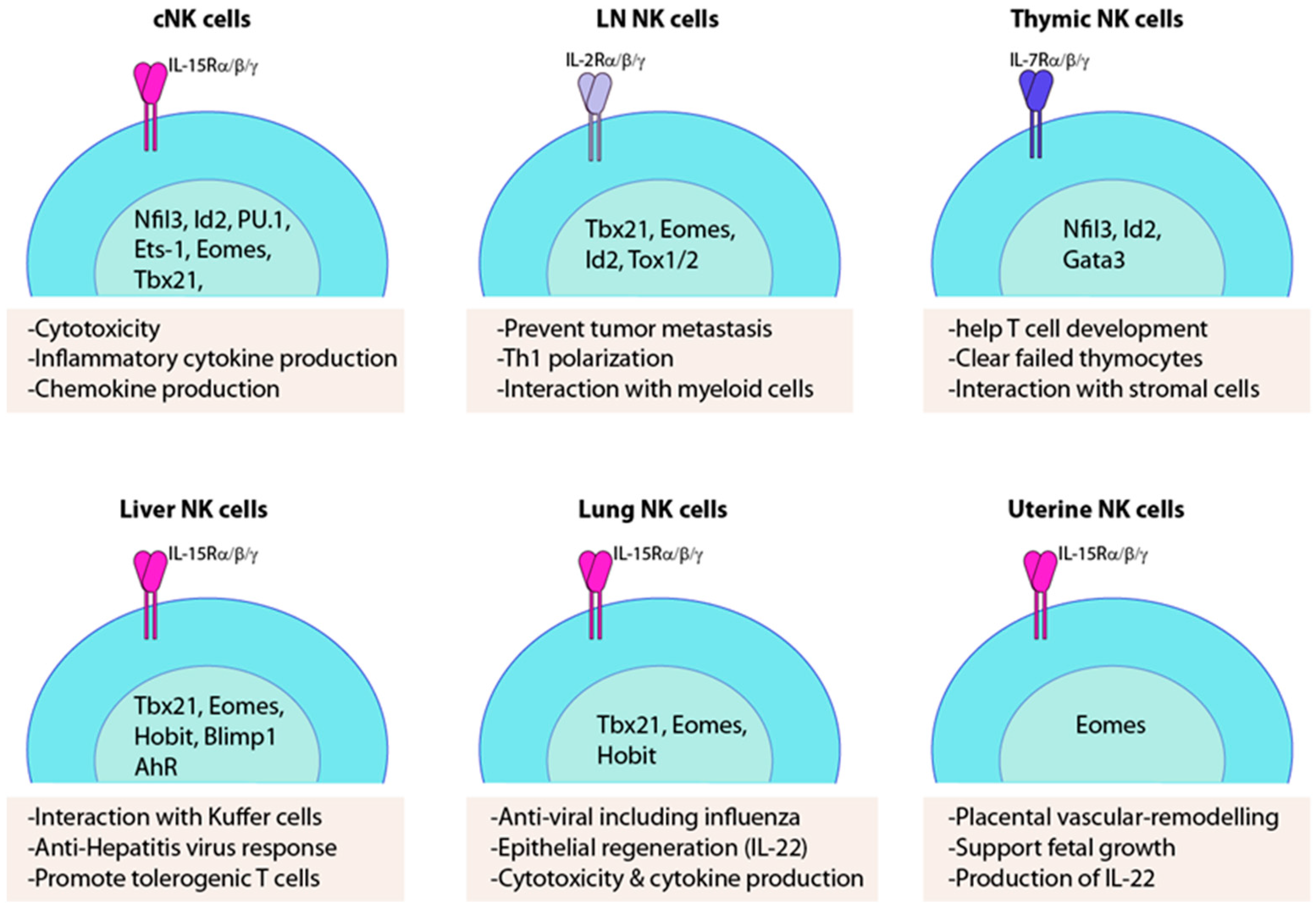
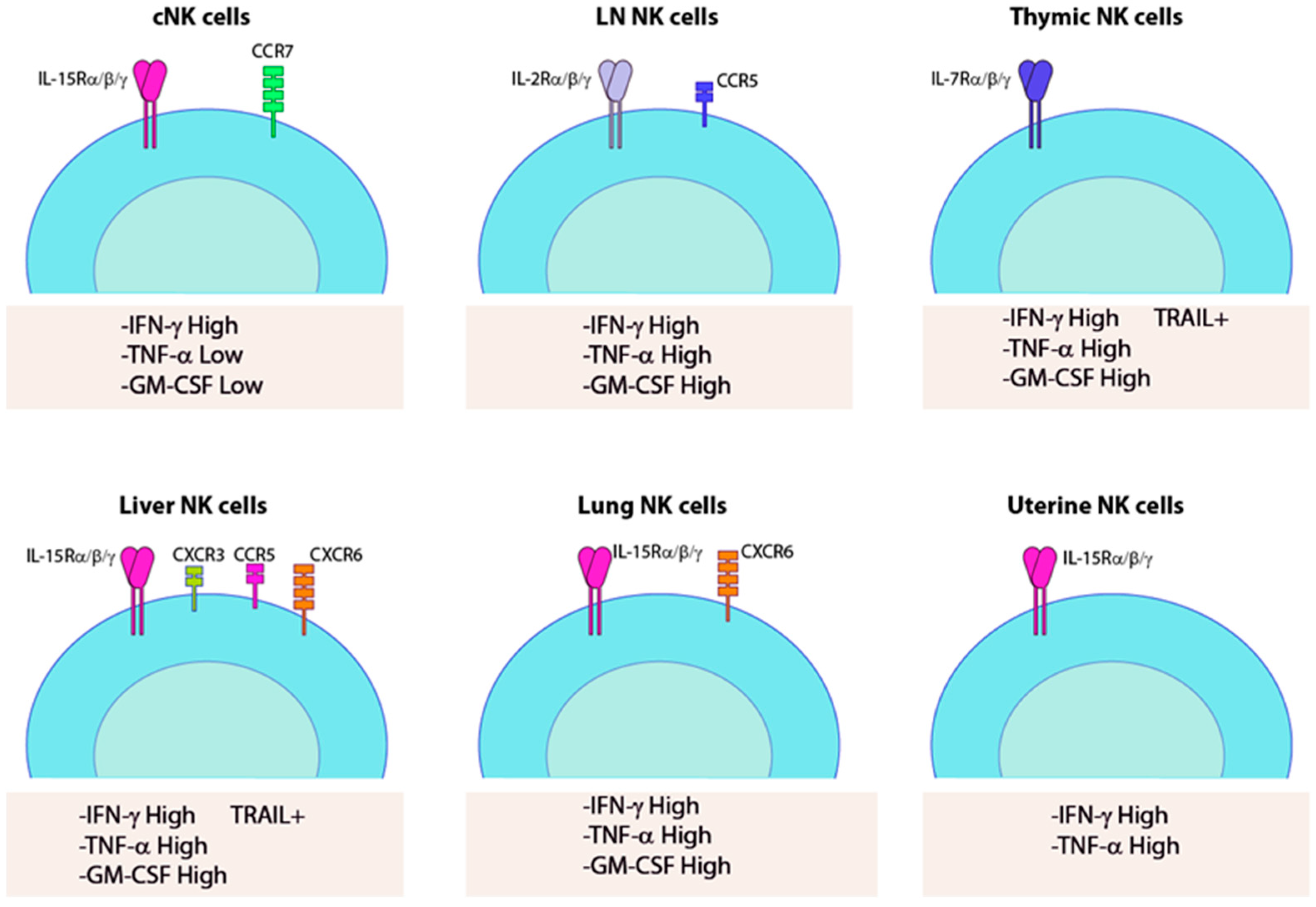
2. cNK Versus trNK Cells
3. Thymic NK Cells
4. Liver-Resident NK Cells
5. Lung-Resident NK Cells
6. Lymph Node-Resident NK Cells
7. Uterine NK Cells
8. Clinical Relevance of trNK Cells
9. Summary and Future Outlook
Funding
Conflicts of Interest
Abbreviation
| NK | Natural killer |
| ILC1 | Innate lymphoid cells |
| Tr | Tissue-resident |
| Prf1 | Perforin |
| IFN-γ | Interferon-γ |
| TNF-α | Tumor necrosis factor alpha |
| BM | Bone marrow |
| cNK cells | Convectional NK cells |
| PBMC | Peripheral blood mononuclear cells |
| HSCs | Hematopoietic stem cells |
| ELP | Early lymphoid progenitors |
| CLP | Common lymphoid progenitors |
| IL | Interleukin |
| KIR | Immunoglobulin-like inhibitory receptors |
| MHC-1 | MHC Class I |
| GM-CSF | Granulocyte–monocyte colony-stimulating factor |
| NFIL3 | Nuclear factor IL-3-regulated protein |
| DN | CD4−CD8− double negative |
| SCF | Stem cell factor |
| ETPs | Early thymic precursors |
| LNs | Lymph nodes |
| TRAIL | TNF-related apoptosis-inducing ligand |
| ihNK | Intrahepatic resident NK cells |
| TRM | Tissue-resident memory T cells |
| NSCLC | Non-small-cell lung cancer |
| IFZ | Interfollicular zone |
| NKT | Natural killer T cells |
| TRCs | T-zone reticular cells |
| HEV | High endothelial venules |
| Ltr | Lymphoid-tissue resident |
| DNAM1 | DNAX accessory molecule 1 |
| S1P | Sphingosine-1 phosphate |
| Endometrium | Non-pregnant uterus |
| Decidua | Pregnant uterus |
| gd | gestational day |
| MLAp | Mesometrial lymphoid aggregate of pregnancy |
References
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014, 32, 659–702. [Google Scholar] [CrossRef]
- Kim, C.H. Chemokine-chemokine receptor network in immune cell trafficking. Curr. Drug Targets Immune Endocr. Metabol. Disord. 2004, 4, 343–361. [Google Scholar] [CrossRef]
- Schulz, O.; Hammerschmidt, S.I.; Moschovakis, G.L.; Förster, R.J. Chemokines and chemokine receptors in lymphoid tissue dynamics. Annu. Rev. Immunol. 2016, 34, 203–242. [Google Scholar] [CrossRef]
- Sokol, C.L.; Luster, A.D. The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 2015, 7, a016303. [Google Scholar] [CrossRef]
- Fernandez-Ruiz, D.; Ng, W.Y.; Holz, L.E.; Ma, J.Z.; Zaid, A.; Wong, Y.C.; Lau, L.S.; Mollard, V.; Cozijnsen, A.; Collins, N.J. Liver-resident memory CD8+ T cells form a front-line defense against malaria liver-stage infection. Immunity 2016, 45, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Krueger, P.D.; Kim, T.S.; Sung, S.-S.J.; Braciale, T.J.; Hahn, Y.S. Liver-resident CD103+ dendritic cells prime antiviral CD8+ T cells in situ. J. Immunol. 2015, 194, 3213–3222. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, N.; Kekäläinen, E.; Chen, P.; Kvedaraite, E.; Wilson, J.N.; Ivarsson, M.A.; Mjösberg, J.; Berglin, L.; Säfholm, J.; Manson, M.L.J.; et al. Human lung natural killer cells are predominantly comprised of highly differentiated hypofunctional CD69− CD56dim cells. J. Allergy Clin. Immunol. 2017, 139, 1321–1330.e4. [Google Scholar] [CrossRef] [PubMed]
- Sojka, D.K.; Plougastel-Douglas, B.; Yang, L.; Pak-Wittel, M.A.; Artyomov, M.N.; Ivanova, Y.; Zhong, C.; Chase, J.M.; Rothman, P.B.; Yu, J.J. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 2014, 3, e01659. [Google Scholar] [CrossRef] [PubMed]
- Victorino, F.; Sojka, D.K.; Brodsky, K.S.; McNamee, E.N.; Masterson, J.C.; Homann, D.; Yokoyama, W.M.; Eltzschig, H.K.; Clambey, E.T. Tissue-resident NK cells mediate ischemic kidney injury and are not depleted by Anti–Asialo-GM1 antibody. J. Immunol. 2015, 195, 4973–4985. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Peng, H.; Li, K.; Qu, K.; Wang, B.; Wu, Y.; Ye, L.; Dong, Z.; Wei, H.; Sun, R.J.I. Liver-resident NK cells control antiviral activity of hepatic T cells via the PD-1-PD-L1 axis. Immunity 2019, 50, 403–417.e4. [Google Scholar] [CrossRef] [PubMed]
- Gwalani, L.A.; Orange, J.S. Single degranulations in NK cells can mediate target cell killing. J. Immunol. 2018, 200, 3231–3243. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Abad-Fernandez, M.; Tuyishime, M.; Pollara, J.J.; Ferrari, G.; Soriano-Sarabia, N.; Margolis, D.M. Interleukin-15-stimulated natural killer cells clear HIV-1-infected cells following latency reversal ex vivo. J. Virol. 2018, 92, e00235-18. [Google Scholar] [CrossRef] [PubMed]
- Mandelboim, O.; Lieberman, N.; Lev, M.; Paul, L.; Arnon, T.I.; Bushkin, Y.; Davis, D.M.; Strominger, J.L.; Yewdell, J.W.; Porgador, A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 2001, 409, 1055–1060. [Google Scholar] [CrossRef]
- Lodoen, M.B.; Lanier, L.L. Viral modulation of NK cell immunity. Nat. Rev. Microbiol. 2005, 3, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Fehniger, T.A.; Cai, S.F.; Cao, X.; Bredemeyer, A.J.; Presti, R.M.; French, A.R.; Ley, T.J. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity 2007, 26, 798–811. [Google Scholar] [CrossRef]
- Salcedo, T.; Azzoni, L.; Wolf, S.F.; Perussia, B. Modulation of perforin and granzyme messenger RNA expression in human natural killer cells. J. Immunol. 1993, 151, 2511–2520. [Google Scholar]
- Shresta, S.; MacIvor, D.M.; Heusel, J.W.; Russell, J.H.; Ley, T.J. Natural killer and lymphokine-activated killer cells require granzyme B for the rapid induction of apoptosis in susceptible target cells. Proc. Natl. Acad. Sci. USA 1995, 92, 5679–5683. [Google Scholar] [CrossRef]
- King, A.; Wooding, P.; Gardner, L.; Loke, Y.W. Immunology: Expression of perforin, granzyme A and TIA-1 by human uterine CD56+ NK cells implies they are activated and capable of effector functions. Hum. Reprod. 1993, 8, 2061–2067. [Google Scholar] [CrossRef]
- Vermijlen, D.; Luo, D.; Robaye, B.; Seynaeve, C.; Baekeland, M.; Wisse, E. Pit cells (Hepatic natural killer cells) of the rat induce apoptosis in colon carcinoma cells by the perforin/granzyme pathway. Hepatology 1999, 29, 51–56. [Google Scholar] [CrossRef]
- Arase, H.; Arase, N.; Saito, T. Interferon gamma production by natural killer (NK) cells and NK1. 1+ T cells upon NKR-P1 cross-linking. J. Exp. Med. 1996, 183, 2391–2396. [Google Scholar] [CrossRef] [PubMed]
- Luetke-Eversloh, M.; Hammer, Q.; Durek, P.; Nordström, K.; Gasparoni, G.; Pink, M.; Hamann, A.; Walter, J.; Chang, H.-D.; Dong, J. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. 2014, 10, e1004441. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, Å.; Pavlovic, V.; Flach, C.-F.; Sjöling, Å.; Lundin, S. Interferon-gamma secretion is induced in IL-12 stimulated human NK cells by recognition of Helicobacter pylori or TLR2 ligands. Innate Immun. 2011, 17, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Kokordelis, P.; Krämer, B.; Körner, C.; Boesecke, C.; Voigt, E.; Ingiliz, P.; Glässner, A.; Eisenhardt, M.; Wolter, F.; Kaczmarek, D. An effective interferon-gamma-mediated inhibition of hepatitis C virus replication by natural killer cells is associated with spontaneous clearance of acute hepatitis C in human immunodeficiency virus-positive patients. Hepatology 2014, 59, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Stein, N.; Berhani, O.; Schmiedel, D.; Duev-Cohen, A.; Seidel, E.; Kol, I.; Tsukerman, P.; Hecht, M.; Reches, A.; Gamliel, M. IFNG-AS1 Enhances Interferon Gamma Production in Human Natural Killer Cells. iScience 2019, 11, 466–473. [Google Scholar] [CrossRef]
- Makowska, A.; Franzen, S.; Braunschweig, T.; Denecke, B.; Shen, L.; Baloche, V.; Busson, P.; Kontny, U. Interferon beta increases NK cell cytotoxicity against tumor cells in patients with nasopharyngeal carcinoma via tumor necrosis factor apoptosis-inducing ligand. Cancer Immunol. Immunother. 2019, 68, 1317–1329. [Google Scholar] [CrossRef]
- Small, H.Y.; Nosalski, R.; Morgan, H.; Beattie, E.; Guzik, T.; Graham, D.; Delles, C. The Role of Tumor Necrosis Factor α and Natural Killer Cells in Uterine Artery Function During Pregnancy in the Stroke Prone Spontaneously Hypertensive Rat. Hypertension 2016, 68, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Parr, E.L.; Chen, H.-L.; Parr, M.B.; Hunt, J.S. Synthesis and granular localization of tumor necrosis factor-alpha in activated NK cells in the pregnant mouse uterus. J. Reprod. Immunol. 1995, 28, 31–40. [Google Scholar] [CrossRef]
- Takeda, K.; Hayakawa, Y.; Smyth, M.J.; Kayagaki, N.; Yamaguchi, N.; Kakuta, S.; Iwakura, Y.; Yagita, H.; Okumura, K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 2001, 7, 94–100. [Google Scholar] [CrossRef]
- Chan, A.; Hong, D.-L.; Atzberger, A.; Kollnberger, S.; Filer, A.D.; Buckley, C.D.; McMichael, A.; Enver, T.; Bowness, P. CD56bright human NK cells differentiate into CD56dim cells: Role of contact with peripheral fibroblasts. J. Immunol. 2007, 179, 89–94. [Google Scholar] [CrossRef]
- Romagnani, C.; Juelke, K.; Falco, M.; Morandi, B.; D’Agostino, A.; Costa, R.; Ratto, G.; Forte, G.; Carrega, P.; Lui, G. CD56brightCD16− killer Ig-like receptor—NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J. Immunol. 2007, 178, 4947–4955. [Google Scholar] [CrossRef]
- Melsen, J.E.; Lugthart, G.; Lankester, A.C.; Schilham, M.W. Human circulating and tissue-resident CD56bright natural killer cell populations. Front. Immunol. 2016, 7, 262. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, E.; Kuranaga, N.; Satoh, K.; Habu, Y.; Shinomiya, N.; Asano, T.; Seki, S.; Hayakawa, M. Induction of CD16+ CD56bright NK cells with antitumour cytotoxicity not only from CD16− CD56bright NK Cells but also from CD16− CD56dim NK cells. Scand. J. Immunol. 2007, 65, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Siebert, J.R.; Burns, R.; Gerbec, Z.J.; Bonacci, B.; Rymaszewski, A.; Rau, M.; Riese, M.J.; Rao, S.; Carlson, K.-S. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hudspeth, K.; Donadon, M.; Cimino, M.; Pontarini, E.; Tentorio, P.; Preti, M.; Hong, M.; Bertoletti, A.; Bicciato, S.; Invernizzi, P.; et al. Human liver-resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J. Autoimmun. 2016, 66, 40–50. [Google Scholar] [CrossRef]
- Lugthart, G.; Melsen, J.E.; Vervat, C.; van Ostaijen-ten Dam, M.M.; Corver, W.E.; Roelen, D.L.; van Bergen, J.; van Tol, M.J.; Lankester, A.C.; Schilham, M.W. Human lymphoid tissues harbor a distinct CD69+ CXCR6+ NK cell population. J. Immunol. 2016, 197, 78–84. [Google Scholar] [CrossRef]
- Rajasekaran, K.; Riese, M.J.; Rao, S.; Wang, L.; Thakar, M.S.; Sentman, C.L.; Malarkannan, S. Signaling in effector lymphocytes: Insights toward safer immunotherapy. Front. Immunol. 2016, 7, 176. [Google Scholar] [CrossRef]
- Vivier, E.; Nunès, J.A.; Vély, F. Natural killer cell signaling pathways. Science 2004, 306, 1517–1519. [Google Scholar] [CrossRef]
- Ho, E.L.; Heusel, J.W.; Brown, M.G.; Matsumoto, K.; Scalzo, A.A.; Yokoyama, W.M. Murine Nkg2d and Cd94 are clustered within the natural killer complex and are expressed independently in natural killer cells. Proc. Natl. Acad. Sci. USA 1998, 95, 6320–6325. [Google Scholar] [CrossRef]
- Bauer, S.; Groh, V.; Wu, J.; Steinle, A.; Phillips, J.H.; Lanier, L.L.; Spies, T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 1999, 285, 727–729. [Google Scholar] [CrossRef]
- Vitale, M.; Bottino, C.; Sivori, S.; Sanseverino, L.; Castriconi, R.; Marcenaro, E.; Augugliaro, R.; Moretta, L.; Moretta, A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J. Exp. Med. 1998, 187, 2065–2072. [Google Scholar] [CrossRef]
- Pende, D.; Parolini, S.; Pessino, A.; Sivori, S.; Augugliaro, R.; Morelli, L.; Marcenaro, E.; Accame, L.; Malaspina, A.; Biassoni, R.; et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 1999, 190, 1505–1516. [Google Scholar] [CrossRef] [PubMed]
- Sivori, S.; Parolini, S.; Marcenaro, E.; Castriconi, R.; Pende, D.; Millo, R.; Moretta, A. Involvement of natural cytotoxicity receptors in human natural killer cell-mediated lysis of neuroblastoma and glioblastoma cell lines. J. Neuroimmunol. 2000, 107, 220–225. [Google Scholar] [CrossRef]
- De Maria, A.; Biassoni, R.; Fogli, M.; Rizzi, M.; Cantoni, C.; Costa, P.; Conte, R.; Mavilio, D.; Ensoli, B.; Cafaro, A.; et al. Identification, molecular cloning and functional characterization of NKp46 and NKp30 natural cytotoxicity receptors in Macaca fascicularis NK cells. Eur. J. Immunol. 2001, 31, 3546–3556. [Google Scholar] [CrossRef]
- Arnon, T.I.; Lev, M.; Katz, G.; Chernobrov, Y.; Porgador, A.; Mandelboim, O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur. J. Immunol. 2001, 31, 2680–2689. [Google Scholar] [CrossRef]
- Diefenbach, A.; Jamieson, A.M.; Liu, S.D.; Shastri, N.; Raulet, D.H. Ligands for the murine NKG2D receptor: Expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol. 2000, 1, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Samarakoon, A.; Chu, H.; Malarkannan, S. Murine NKG2D ligands: “Double, double toil and trouble”. Mol. Immunol. 2009, 46, 1011–1019. [Google Scholar] [CrossRef]
- Steinle, A.; Li, P.; Morris, D.L.; Groh, V.; Lanier, L.L.; Strong, R.K.; Spies, T. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics 2001, 53, 279–287. [Google Scholar] [CrossRef]
- Li, P.; Willie, S.T.; Bauer, S.; Morris, D.L.; Spies, T.; Strong, R.K. Crystal structure of the MHC class I homolog MIC-A, a gammadelta T cell ligand. Immunity 1999, 10, 577–584. [Google Scholar] [CrossRef]
- Holmes, M.A.; Li, P.; Petersdorf, E.W.; Strong, R.K. Structural studies of allelic diversity of the MHC class I homolog MIC-B, a stress-inducible ligand for the activating immunoreceptor NKG2D. J. Immunol. 2002, 169, 1395–1400. [Google Scholar] [CrossRef]
- Groh, V.; Bahram, S.; Bauer, S.; Herman, A.; Beauchamp, M.; Spies, T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Natl. Acad. Sci. USA 1996, 93, 12445–12450. [Google Scholar] [CrossRef] [PubMed]
- Groh, V.; Steinle, A.; Bauer, S.; Spies, T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science 1998, 279, 1737–1740. [Google Scholar] [CrossRef] [PubMed]
- Malarkannan, S.; Shih, P.P.; Eden, P.A.; Horng, T.; Zuberi, A.R.; Christianson, G.; Roopenian, D.; Shastri, N. The molecular and functional characterization of a dominant minor H antigen, H60. J. Immunol. 1998, 161, 3501–3509. [Google Scholar] [PubMed]
- Malarkannan, S.; Horng, T.; Eden, P.; Gonzalez, F.; Shih, P.; Brouwenstijn, N.; Klinge, H.; Christianson, G.; Roopenian, D.; Shastri, N. Differences that matter: Major cytotoxic T cell-stimulating minor histocompatibility antigens. Immunity 2000, 13, 333–344. [Google Scholar] [CrossRef]
- Cerwenka, A.; Bakker, A.B.; McClanahan, T.; Wagner, J.; Wu, J.; Phillips, J.H.; Lanier, L.L. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity 2000, 12, 721–727. [Google Scholar] [CrossRef]
- Carayannopoulos, L.N.; Naidenko, O.V.; Fremont, D.H.; Yokoyama, W.M. Cutting edge: Murine UL16-binding protein-like transcript 1: A newly described transcript encoding a high-affinity ligand for murine NKG2D. J. Immunol. 2002, 169, 4079–4083. [Google Scholar] [CrossRef]
- Tokuyama, M.; Lorin, C.; Delebecque, F.; Jung, H.; Raulet, D.H.; Coscoy, L. Expression of the RAE-1 family of stimulatory NK-cell ligands requires activation of the PI3K pathway during viral infection and transformation. PLoS Pathog. 2011, 7, e1002265. [Google Scholar] [CrossRef]
- Nagler, A.; Lanier, L.L.; Cwirla, S.; Phillips, J.H. Comparative studies of human FcRIII-positive and negative natural killer cells. J. Immunol. 1989, 143, 3183–3191. [Google Scholar]
- Moretta, A.; Tambussi, G.; Bottino, C.; Tripodi, G.; Merli, A.; Ciccone, E.; Pantaleo, G.; Moretta, L. A novel surface antigen expressed by a subset of human CD3- CD16+ natural killer cells. Role in cell activation and regulation of cytolytic function. J. Exp. Med. 1990, 171, 695–714. [Google Scholar] [CrossRef]
- Carson, W.; Caligiuri, M. Natural Killer Cell Subsets and Development. Methods 1996, 9, 327–343. [Google Scholar] [CrossRef]
- Stewart, C.A.; Laugier-Anfossi, F.; Vély, F.; Saulquin, X.; Riedmuller, J.; Tisserant, A.; Gauthier, L.; Romagné, F.; Ferracci, G.; Arosa, F.A. Recognition of peptide–MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 13224–13229. [Google Scholar] [CrossRef]
- Parikh, B.A.; Bern, M.D.; Piersma, S.J.; Yang, L.; Beckman, D.L.; Poursine-Laurent, J.; Douglas, B.P.; Yokoyama, W.M. Major histocompatibility complex class I-restricted protection against murine cytomegalovirus requires missing-self recognition by the natural killer cell inhibitory Ly49 receptors. bioRxiv 2019, 753970. [Google Scholar]
- Herberman, R.B.; Nunn, M.E.; Lavrin, D.H. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. Int. J. Cancer 1975, 16, 216–229. [Google Scholar] [CrossRef]
- Orange, J.S. Human natural killer cell deficiencies and susceptibility to infection. Microb. Infect. 2002, 4, 1545–1558. [Google Scholar] [CrossRef]
- Seaman, W.; Sleisenger, M.; Eriksson, E.; Koo, G. Depletion of natural killer cells in mice by monoclonal antibody to NK-1.1. Reduction in host defense against malignancy without loss of cellular or humoral immunity. J. Immunol. 1987, 138, 4539–4544. [Google Scholar] [PubMed]
- Hobbs, J.A.; Cho, S.; Roberts, T.J.; Sriram, V.; Zhang, J.; Xu, M.; Brutkiewicz, R.R. Selective loss of natural killer T cells by apoptosis following infection with lymphocytic choriomeningitis virus. J. Virol. 2001, 75, 10746–10754. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.D.; Whitmire, J.K. The depletion of NK cells prevents T cell exhaustion to efficiently control disseminating virus infection. J. Immunol. 2013, 190, 641–649. [Google Scholar] [CrossRef]
- Bukowski, J.F.; Woda, B.A.; Habu, S.; Okumura, K.; Welsh, R.M. Natural killer cell depletion enhances virus synthesis and virus-induced hepatitis in vivo. J. Immunol. 1983, 131, 1531–1538. [Google Scholar]
- Cooper, M.A.; Fehniger, T.A.; Fuchs, A.; Colonna, M.; Caligiuri, M.A. NK cell and DC interactions. Trends Immunol. 2004, 25, 47–52. [Google Scholar] [CrossRef]
- Unanue, E.R. Inter-relationship among macrophages, natural killer cells and neutrophils in early stages of Listeria resistance. Curr. Opin. Immunol. 1997, 9, 35–43. [Google Scholar] [CrossRef]
- Lam, V.C.; Lanier, L.L. NK cells in host responses to viral infections. Curr. Opin. Immunol. 2017, 44, 43–51. [Google Scholar] [CrossRef]
- Degli-Esposti, M.A.; Smyth, M.J. Close encounters of different kinds: Dendritic cells and NK cells take centre stage. Nat. Rev. Immunol. 2005, 5, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Lucas, M.; Schachterle, W.; Oberle, K.; Aichele, P.; Diefenbach, A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity 2007, 26, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Kasaian, M.T.; Whitters, M.J.; Carter, L.L.; Lowe, L.D.; Jussif, J.M.; Deng, B.; Johnson, K.A.; Witek, J.S.; Senices, M.; Konz, R.F. IL-21 limits NK cell responses and promotes antigen-specific T cell activation: A mediator of the transition from innate to adaptive immunity. Immunity 2002, 16, 559–569. [Google Scholar] [CrossRef]
- Moretta, A.; Marcenaro, E.; Parolini, S.; Ferlazzo, G.; Moretta, L. NK cells at the interface between innate and adaptive immunity. Cell Death Differ. 2008, 15, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Koka, R.; Burkett, P.R.; Chien, M.; Chai, S.; Chan, F.; Lodolce, J.P.; Boone, D.L.; Ma, A. Interleukin (IL)-15R[alpha]-deficient natural killer cells survive in normal but not IL-15R[alpha]-deficient mice. J. Exp. Med. 2003, 197, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Koka, R.; Burkett, P.; Chien, M.; Chai, S.; Boone, D.L.; Ma, A. Cutting edge: Murine dendritic cells require IL-15R alpha to prime NK cells. J. Immunol. 2004, 173, 3594–3598. [Google Scholar] [CrossRef]
- Mailliard, R.B.; Son, Y.-I.; Redlinger, R.; Coates, P.T.; Giermasz, A.; Morel, P.A.; Storkus, W.J.; Kalinski, P. Dendritic cells mediate NK cell help for Th1 and CTL responses: Two-signal requirement for the induction of NK cell helper function. J. Immunol. 2003, 171, 2366–2373. [Google Scholar] [CrossRef]
- Biron, C.A.; Nguyen, K.B.; Pien, G.C.; Cousens, L.P.; Salazar-Mather, T.P. Natural killer cells in antiviral defense: Function and regulation by innate cytokines. Annu. Rev. Immunol. 1999, 17, 189–220. [Google Scholar] [CrossRef]
- Yu, J.; Freud, A.G.; Caligiuri, M.A. Location and cellular stages of natural killer cell development. Trends Immunol. 2013, 34, 573–582. [Google Scholar] [CrossRef]
- Kumar, R.; Fossati, V.; Israel, M.; Snoeck, H.-W. Lin− Sca1+ Kit− bone marrow cells contain early lymphoid-committed precursors that are distinct from common lymphoid progenitors. J. Immunol. 2008, 181, 7507–7513. [Google Scholar] [CrossRef]
- Adolfsson, J.; Borge, O.J.; Bryder, D.; Theilgaard-Mönch, K.; Åstrand-Grundström, I.; Sitnicka, E.; Sasaki, Y.; Jacobsen, S.E. Upregulation of Flt3 expression within the bone marrow Lin− Sca1+ c-kit+ stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 2001, 15, 659–669. [Google Scholar] [CrossRef]
- Karsunky, H.; Inlay, M.A.; Serwold, T.; Bhattacharya, D.; Weissman, I.L. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood J. Am. Soc. Hematol. 2008, 111, 5562–5570. [Google Scholar] [CrossRef] [PubMed]
- Nozad Charoudeh, H.; Tang, Y.; Cheng, M.; Cilio, C.M.; Jacobsen, S.E.W.; Sitnicka, E. Identification of an NK/T cell–restricted progenitor in adult bone marrow contributing to bone marrow–and thymic-dependent NK cells. Blood J. Am. Soc. Hematol. 2010, 116, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Abel, A.M.; Yang, C.; Thakar, M.S.; Malarkannan, S. Natural killer cells: Development, maturation, and clinical utilization. Front. Immunol. 2018, 9, 1869. [Google Scholar] [CrossRef]
- Liu, C.-C.; Perussia, B.; Young, J.D.-E. The emerging role of IL-15 in NK-cell development. Immunol. Today 2000, 21, 113–116. [Google Scholar] [CrossRef]
- Huntington, N.D.; Legrand, N.; Alves, N.L.; Jaron, B.; Weijer, K.; Plet, A.; Corcuff, E.; Mortier, E.; Jacques, Y.; Spits, H. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 2009, 206, 25–34. [Google Scholar] [CrossRef]
- Fathman, J.W.; Bhattacharya, D.; Inlay, M.A.; Seita, J.; Karsunky, H.; Weissman, I.L. Identification of the earliest natural killer cell–committed progenitor in murine bone marrow. Blood J. Am. Soc. Hematol. 2011, 118, 5439–5447. [Google Scholar] [CrossRef]
- Yokoyama, W.M.; Sojka, D.K.; Peng, H.; Tian, Z. Tissue-resident natural killer cells. Cold Spring Harb. Symp. Quant. Biol. 2013, 78, 149–156. [Google Scholar] [CrossRef]
- Di Santo, J.P.; Vosshenrich, C.A. Bone marrow versus thymic pathways of natural killer cell development. Immunol. Rev. 2006, 214, 35–46. [Google Scholar] [CrossRef]
- Salzberger, W.; Martrus, G.; Bachmann, K.; Goebels, H.; Heß, L.; Koch, M.; Langeneckert, A.; Lunemann, S.; Oldhafer, K.J.; Pfeifer, C. Tissue-resident NK cells differ in their expression profile of the nutrient transporters Glut1, CD98 and CD71. PLoS ONE 2018, 13, e0201170. [Google Scholar] [CrossRef]
- Wang, R.; Jaw, J.J.; Stutzman, N.C.; Zou, Z.; Sun, P.D.J. Natural killer cell-produced IFN-γ and TNF-α induce target cell cytolysis through up-regulation of ICAM-1. J. Leukoc. Biol. 2012, 91, 299–309. [Google Scholar] [CrossRef]
- Smyth, M.J.; Zachariae, C.O.; Norihisa, Y.; Ortaldo, J.R.; Hishinuma, A.; Matsushima, K. IL-8 gene expression and production in human peripheral blood lymphocyte subsets. J. Immunol. 1991, 146, 3815–3823. [Google Scholar] [PubMed]
- Cuturi, M.C.; Anegon, I.; Sherman, F.; Loudon, R.; Clark, S.C.; Perussia, B.; Trinchieri, G. Production of hematopoietic colony-stimulating factors by human natural killer cells. J. Exp. Med. 1989, 169, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Warren, H.S.; Kinnear, B.F.; Phillips, J.H.; Lanier, L.L. Production of IL-5 by human NK cells and regulation of IL-5 secretion by IL-4, IL-10, and IL-12. J. Immunol. 1995, 154, 5144–5152. [Google Scholar] [PubMed]
- Fehniger, T.A.; Herbein, G.; Yu, H.; Para, M.I.; Bernstein, Z.P.; O’Brien, W.A.; Caligiuri, M.A. Natural killer cells from HIV-1+ patients produce C-C chemokines and inhibit HIV-1 infection. J. Immunol. 1998, 161, 6433–6438. [Google Scholar] [PubMed]
- Fehniger, T.A.; Shah, M.H.; Turner, M.J.; VanDeusen, J.B.; Whitman, S.P.; Cooper, M.A.; Suzuki, K.; Wechser, M.; Goodsaid, F.; Caligiuri, M.A. Differential cytokine and chemokine gene expression by human NK cells following activation with IL-18 or IL-15 in combination with IL-12: Implications for the innate immune response. J. Immunol. 1999, 162, 4511–4520. [Google Scholar]
- Loza, M.J.; Perussia, B. Final steps of natural killer cell maturation: A model for type 1-type 2 differentiation? Nat. Immunol. 2001, 2, 917–924. [Google Scholar] [CrossRef]
- Wallace, K.L.; Marshall, M.A.; Ramos, S.I.; Lannigan, J.A.; Field, J.J.; Strieter, R.M.; Linden, J.J.B. NKT cells mediate pulmonary inflammation and dysfunction in murine sickle cell disease through production of IFN-γ and CXCR3 chemokines. Blood J. Am. Soc. Hematol. 2009, 114, 667–676. [Google Scholar]
- Bluman, E.M.; Bartynski, K.J.; Avalos, B.R.; Caligiuri, M.A. Human natural killer cells produce abundant macrophage inflammatory protein-1 alpha in response to monocyte-derived cytokines. J. Clin. Investig. 1996, 97, 2722–2727. [Google Scholar] [CrossRef]
- Gascoyne, D.M.; Long, E.; Veiga-Fernandes, H.; De Boer, J.; Williams, O.; Seddon, B.; Coles, M.; Kioussis, D.; Brady, H.J. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 2009, 10, 1118. [Google Scholar] [CrossRef]
- Cortez, V.S.; Fuchs, A.; Cella, M.; Gilfillan, S.; Colonna, M. Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. J. Immunol. 2014, 192, 4487–4491. [Google Scholar] [CrossRef] [PubMed]
- Vosshenrich, C.A.; García-Ojeda, M.E.; Samson-Villéger, S.I.; Pasqualetto, V.; Enault, L.; Richard-Le Goff, O.; Corcuff, E.; Guy-Grand, D.; Rocha, B.; Cumano, A. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat. Immunol. 2006, 7, 1217–1224. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.S.; Blatz, K.; d’Hargues, Y.; Hernandez, P.P.; Kofoed-Nielsen, M.; Ripka, J.F.; Ebert, K.; Arnold, S.J.; Diefenbach, A.; Palmer, E.; et al. The transcription factor T-bet is induced by IL-15 and thymic agonist selection and controls CD8alphaalpha(+) intraepithelial lymphocyte development. Immunity 2014, 41, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Tian, Z. Diversity of tissue-resident NK cells. Semin. Immunol. 2017, 31, 3–10. [Google Scholar] [CrossRef]
- Zhang, L.H.; Shin, J.H.; Haggadone, M.D.; Sunwoo, J.B. The aryl hydrocarbon receptor is required for the maintenance of liver-resident natural killer cells. J. Exp. Med. 2016, 213, 2249–2257. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fu, B.; Xu, X.; Zhang, J.; Tong, X.; Wang, Y.; Dong, Z.; Zhang, X.; Shen, N.; Zhai, Y.; et al. PBX1 expression in uterine natural killer cells drives fetal growth. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Cong, J.; Wei, H. Natural Killer Cells in the Lungs. Front. Immunol. 2019, 10, 1416. [Google Scholar] [CrossRef]
- Greenwood, J.D.; Minhas, K.; di Santo, J.P.; Makita, M.; Kiso, Y.; Croy, B.A. Ultrastructural studies of implantation sites from mice deficient in uterine natural killer cells. Placenta 2000, 21, 693–702. [Google Scholar] [CrossRef]
- Ashkar, A.A.; Di Santo, J.P.; Croy, B.A. Interferon gamma contributes to initiation of uterine vascular modification, decidual integrity, and uterine natural killer cell maturation during normal murine pregnancy. J. Exp. Med. 2000, 192, 259–270. [Google Scholar] [CrossRef]
- Ratsep, M.T.; Felker, A.M.; Kay, V.R.; Tolusso, L.; Hofmann, A.P.; Croy, B.A. Uterine natural killer cells: Supervisors of vasculature construction in early decidua basalis. Reproduction 2015, 149, R91–R102. [Google Scholar] [CrossRef]
- Carson, W.E.; Fehniger, T.A.; Caligiuri, M.A. CD56bright natural killer cell subsets: Characterization of distinct functional responses to interleukin-2 and the c-kit ligand. Eur. J. Immunol. 1997, 27, 354–360. [Google Scholar] [CrossRef]
- Jacobs, R.; Hintzen, G.; Kemper, A.; Beul, K.; Kempf, S.; Behrens, G.; Sykora, K.W.; Schmidt, R.E. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J. Immunol. 2001, 31, 3121–3127. [Google Scholar] [CrossRef]
- Carrega, P.; Bonaccorsi, I.; Di Carlo, E.; Morandi, B.; Paul, P.; Rizzello, V.; Cipollone, G.; Navarra, G.; Mingari, M.C.; Moretta, L. CD56brightperforinlow noncytotoxic human NK cells are abundant in both healthy and neoplastic solid tissues and recirculate to secondary lymphoid organs via afferent lymph. J. Immunol. 2014, 192, 3805–3815. [Google Scholar] [CrossRef] [PubMed]
- Fehniger, T.A.; Cooper, M.A.; Nuovo, G.J.; Cella, M.; Facchetti, F.; Colonna, M.; Caligiuri, M.A. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell–derived IL-2: A potential new link between adaptive and innate immunity. Blood J. Am. Soc. Hematol. 2003, 101, 3052–3057. [Google Scholar] [CrossRef] [PubMed]
- Manaster, I.; Mizrahi, S.; Goldman-Wohl, D.; Sela, H.Y.; Stern-Ginossar, N.; Lankry, D.; Gruda, R.; Hurwitz, A.; Bdolah, Y.; Haimov-Kochman, R. Endometrial NK cells are special immature cells that await pregnancy. J. Immunol. 2008, 181, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Ferlazzo, G.; Thomas, D.; Lin, S.-L.; Goodman, K.; Morandi, B.; Muller, W.A.; Moretta, A.; Münz, C. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. J. Immunol. 2004, 172, 1455–1462. [Google Scholar] [CrossRef]
- Dogra, P.; Rancan, C.; Ma, W.; Toth, M.; Senda, T.; Carpenter, D.J.; Kubota, M.; Matsumoto, R.; Thapa, P.; Szabo, P.A. Tissue Determinants of Human NK Cell Development, Function, and Residence. Cell 2020, 180, 749–763.e13. [Google Scholar] [CrossRef] [PubMed]
- Boulenouar, S.; Doisne, J.-M.; Sferruzzi-Perri, A.; Gaynor, L.M.; Kieckbusch, J.; Balmas, E.; Yung, H.W.; Javadzadeh, S.; Volmer, L.; Hawkes, D.A. The residual innate lymphoid cells in NFIL3-deficient mice support suboptimal maternal adaptations to pregnancy. Front. Immunol. 2016, 7, 43. [Google Scholar] [CrossRef]
- Matson, B.C.; Caron, K.M. Uterine natural killer cells as modulators of the maternal-fetal vasculature. Int. J. Dev. Biol. 2014, 58, 199–204. [Google Scholar] [CrossRef]
- Lash, G.E.; Schiessl, B.; Kirkley, M.; Innes, B.A.; Cooper, A.; Searle, R.F.; Robson, S.C.; Bulmer, J.N. Expression of angiogenic growth factors by uterine natural killer cells during early pregnancy. J. Leukoc. Biol. 2006, 80, 572–580. [Google Scholar] [CrossRef]
- Gamliel, M.; Goldman-Wohl, D.; Isaacson, B.; Gur, C.; Stein, N.; Yamin, R.; Berger, M.; Grunewald, M.; Keshet, E.; Rais, Y. Trained memory of human uterine NK cells enhances their function in subsequent pregnancies. Immunity 2018, 48, 951–962.e5. [Google Scholar] [CrossRef] [PubMed]
- Rodewald, H.R.; Moingeon, P.; Lucich, J.L.; Dosiou, C.; Lopez, P.; Reinherz, E.L. A population of early fetal thymocytes expressing Fc gamma RII/III contains precursors of T lymphocytes and natural killer cells. Cell 1992, 69, 139–150. [Google Scholar] [CrossRef]
- Douagi, I.; Colucci, F.; Di Santo, J.P.; Cumano, A.J.B. Identification of the earliest prethymic bipotent T/NK progenitor in murine fetal liver. J. Am. Soc. Hematol. 2002, 99, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, T.; Kawamoto, H.; Fujimoto, S.; Katsura, Y.J.T. Commitment of common T/natural killer (NK) progenitors to unipotent T and NK progenitors in the murine fetal thymus revealed by a single progenitor assay. J. Exp. Med. 1999, 190, 1617–1626. [Google Scholar] [CrossRef]
- Michie, A.M.; Carlyle, J.R.; Schmitt, T.M.; Ljutic, B.; Cho, S.K.; Fong, Q.; Zúñiga-Pflücker, J.C.J.T. Clonal characterization of a bipotent T cell and NK cell progenitor in the mouse fetal thymus. J. Immunol. 2000, 164, 1730–1733. [Google Scholar] [CrossRef]
- Hosoya, T.; Kuroha, T.; Moriguchi, T.; Cummings, D.; Maillard, I.; Lim, K.-C.; Engel, J.D. GATA-3 is required for early T lineage progenitor development. J. Exp. Med. 2009, 206, 2987–3000. [Google Scholar] [CrossRef]
- Barik, S.; Miller, M.M.; Cattin-Roy, A.N.; Ukah, T.K.; Chen, W.; Zaghouani, H. IL-4/IL-13 Signaling Inhibits the Potential of Early Thymic Progenitors to Commit to the T Cell Lineage. J. Immunol. 2017, 199, 2767–2776. [Google Scholar] [CrossRef]
- Vargas, C.L.; Poursine-Laurent, J.; Yang, L.; Yokoyama, W.M.J.B. Development of thymic NK cells from double negative 1 thymocyte precursors. J. Am. Soc. Hematol. 2011, 118, 3570–3578. [Google Scholar] [CrossRef]
- Yui, M.A.; Feng, N.; Rothenberg, E.V. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J. Immunol. 2010, 185, 284–293. [Google Scholar] [CrossRef]
- Gabrielli, S.; Sun, M.; Bell, A.; Zook, E.C.; de Pooter, R.F.; Zamai, L.; Kee, B.L. Murine thymic NK cells are distinct from ILC1s and have unique transcription factor requirements. Eur J. Immunol. 2017, 47, 800–805. [Google Scholar] [CrossRef]
- Crotta, S.; Gkioka, A.; Male, V.; Duarte, J.H.; Davidson, S.; Nisoli, I.; Brady, H.J.; Wack, A. The transcription factor E4BP4 is not required for extramedullary pathways of NK cell development. J. Immunol. 2014, 192, 2677–2688. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, K.; Chandler, K.J.; Spaulding, C.; Zandi, S.; Sigvardsson, M.; Graves, B.J.; Kee, B.L. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity 2012, 36, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Schotte, R.; Dontje, W.; Nagasawa, M.; Yasuda, Y.; Bakker, A.Q.; Spits, H.; Blom, B. Synergy between IL-15 and Id2 promotes the expansion of human NK progenitor cells, which can be counteracted by the E protein HEB required to drive T cell development. J. Immunol. 2010, 184, 6670–6679. [Google Scholar] [CrossRef]
- Smyth, M.J.; Nutt, S.L. IL-7 and the thymus dictate the NK cell’labor market’. Nat. Immunol. 2006, 7, 1134–1136. [Google Scholar] [CrossRef] [PubMed]
- García-Peydró, M.; de Yébenes, V.G.; Toribio, M.L. Notch1 and IL-7 receptor interplay maintains proliferation of human thymic progenitors while suppressing non-T cell fates. J. Immunol. 2006, 177, 3711–3720. [Google Scholar] [CrossRef]
- Wisse, E.v.; Van’t Noordende, J.; Van der Meulen, J.; Daems, W.T. The pit cell: Description of a new type of cell occurring in rat liver sinusoids and peripheral blood. Cell Tissue Res. 1976, 173, 423–435. [Google Scholar] [CrossRef]
- Racanelli, V.; Rehermann, B.J. The liver as an immunological organ. Hepatology 2006, 43, S54–S62. [Google Scholar] [CrossRef]
- Mikulak, J.; Bruni, E.; Oriolo, F.; Di Vito, C.; Mavilio, D. Hepatic Natural Killer Cells: Organ-Specific Sentinels of Liver Immune Homeostasis and Physiopathology. Front. Immunol. 2019, 10, 946. [Google Scholar] [CrossRef]
- Phillips, J.H.; Hori, T.; Nagler, A.; Bhat, N.; Spits, H.; Lanier, L.L.J.T. Ontogeny of human natural killer (NK) cells: Fetal NK cells mediate cytolytic function and express cytoplasmic CD3 epsilon, delta proteins. J. Exp. Med. 1992, 175, 1055–1066. [Google Scholar] [CrossRef]
- Serafini, N.; Vosshenrich, C.A.; Di Santo, J.P. Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol. 2015, 15, 415–428. [Google Scholar] [CrossRef]
- Huntington, N.D.; Vosshenrich, C.A.; Di Santo, J.P. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat. Rev. Immunol. 2007, 7, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Crispe, I.N. The liver as a lymphoid organ. Annu. Rev. Immunol. 2009, 27, 147–163. [Google Scholar] [CrossRef]
- Bouwens, L.; Remels, L.; Baekeland, M.; Van Bossuyt, H.; Wisse, E.J. Large granular lymphocytes or “pit cells” from rat liver: Isolation, ultrastructural characterization and natural killer activity. Eur. J. Immunol. 1987, 17, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, F.; Cameron, T.O.; Sidobre, S.; Manlongat, N.; Kronenberg, M.; Briskin, M.J.; Dustin, M.L.; Littman, D.R. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005, 3, e113. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Jiang, X.; Chen, Y.; Sojka, D.K.; Wei, H.; Gao, X.; Sun, R.; Yokoyama, W.M.; Tian, Z. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Investig. 2013, 123, 1444–1456. [Google Scholar] [CrossRef]
- Takeda, K.; Cretney, E.; Hayakawa, Y.; Ota, T.; Akiba, H.; Ogasawara, K.; Yagita, H.; Kinoshita, K.; Okumura, K.; Smyth, M.J. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood 2005, 105, 2082–2089. [Google Scholar] [CrossRef] [PubMed]
- Harmon, C.; Robinson, M.W.; Fahey, R.; Whelan, S.; Houlihan, D.D.; Geoghegan, J.; O’Farrelly, C. Tissue-resident Eomeshi T-betlo CD56bright NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur. J. Immunol. 2016, 46, 2111–2120. [Google Scholar] [CrossRef]
- Stegmann, K.A.; Robertson, F.; Hansi, N.; Gill, U.; Pallant, C.; Christophides, T.; Pallett, L.J.; Peppa, D.; Dunn, C.; Fusai, G. CXCR6 marks a novel subset of T-bet lo Eomes hi natural killer cells residing in human liver. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Lunemann, S.; Martrus, G.; Goebels, H.; Kautz, T.; Langeneckert, A.; Salzberger, W.; Koch, M.; Bunders, M.J.; Nashan, B.; van Gisbergen, K.P. Hobit expression by a subset of human liver-resident CD56 bright natural killer cells. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Mackay, L.K.; Minnich, M.; Kragten, N.A.; Liao, Y.; Nota, B.; Seillet, C.; Zaid, A.; Man, K.; Preston, S.; Freestone, D. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 2016, 352, 459–463. [Google Scholar] [CrossRef]
- Martrus, G.; Kautz, T.; Lunemann, S.; Richert, L.; Glau, L.; Salzberger, W.; Goebels, H.; Langeneckert, A.; Hess, L.; Poch, T.; et al. Proliferative capacity exhibited by human liver-resident CD49a+CD25+ NK cells. PLoS ONE 2017, 12, e0182532. [Google Scholar] [CrossRef] [PubMed]
- Culley, F.J. Natural killer cells in infection and inflammation of the lung. Immunology 2009, 128, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Carrega, P.; Morandi, B.; Costa, R.; Frumento, G.; Forte, G.; Altavilla, G.; Ratto, G.B.; Mingari, M.C.; Moretta, L.; Ferlazzo, G. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56brightCD16− cells and display an impaired capability to kill tumor cells. Cancer 2008, 112, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, D.R.; Lanier, L.L.; Greenland, J.R. Natural killer cells in lung transplantation. Thorax 2019, 74, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Jaffar, Z.; Ferrini, M.; Roberts, K. Natural killer cells regulate allergic lung inflammation by acting on group 2 innate lymphoid cells. J. Am. Assoc. Immnol. 2018, 200, 44.5. [Google Scholar]
- Vanderven, H.A.; Jegaskanda, S.; Wheatley, A.K.; Kent, S.J. Antibody-dependent cellular cytotoxicity and influenza virus. Curr. Opin. Virol. 2017, 22, 89–96. [Google Scholar] [CrossRef]
- Wang, J.; Li, F.; Zheng, M.; Sun, R.; Wei, H.; Tian, Z. Lung natural killer cells in mice: Phenotype and response to respiratory infection. Immunology 2012, 137, 37–47. [Google Scholar] [CrossRef]
- Bankovich, A.J.; Shiow, L.R.; Cyster, J.G. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J. Biol. Chem. 2010, 285, 22328–22337. [Google Scholar] [CrossRef]
- Shiow, L.R.; Rosen, D.B.; Brdičková, N.; Xu, Y.; An, J.; Lanier, L.L.; Cyster, J.G.; Matloubian, M. CD69 acts downstream of interferon-α/β to inhibit S1P 1 and lymphocyte egress from lymphoid organs. Nature 2006, 440, 540–544. [Google Scholar] [CrossRef]
- Mackay, L.K.; Braun, A.; Macleod, B.L.; Collins, N.; Tebartz, C.; Bedoui, S.; Carbone, F.R.; Gebhardt, T. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J. Immunol. 2015, 194, 2059–2063. [Google Scholar] [CrossRef]
- Marquardt, N.; Kekäläinen, E.; Chen, P.; Lourda, M.; Wilson, J.N.; Scharenberg, M.; Bergman, P.; Al-Ameri, M.; Hård, J.; Mold, J.E. Unique transcriptional and protein-expression signature in human lung tissue-resident NK cells. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.V.; Ma, W.; Miron, M.; Granot, T.; Guyer, R.S.; Carpenter, D.J.; Senda, T.; Sun, X.; Ho, S.-H.; Lerner, H. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017, 20, 2921–2934. [Google Scholar] [CrossRef] [PubMed]
- Hombrink, P.; Helbig, C.; Backer, R.A.; Piet, B.; Oja, A.E.; Stark, R.; Brasser, G.; Jongejan, A.; Jonkers, R.E.; Nota, B. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 2016, 17, 1467. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Miyazato, K.; Takahashi, K.; Yoshimura, N.; Tahara, H.; Hayakawa, Y. Lung-resident natural killer cells control pulmonary tumor growth in mice. Cancer Sci. 2018, 109, 2670–2676. [Google Scholar] [CrossRef] [PubMed]
- Junqueira-Kipnis, A.P.; Kipnis, A.; Jamieson, A.; Juarrero, M.G.; Diefenbach, A.; Raulet, D.H.; Turner, J.; Orme, I.M. NK cells respond to pulmonary infection with Mycobacterium tuberculosis, but play a minimal role in protection. J. Immunol. 2003, 171, 6039–6045. [Google Scholar] [CrossRef] [PubMed]
- Katchar, K.; Söderström, K.; Wahlstrom, J.; Eklund, A.; Grunewald, J. Characterisation of natural killer cells and CD56+ T-cells in sarcoidosis patients. Eur. Respir. J. 2005, 26, 77–85. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Smyth, M.J. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J. Immunol. 2006, 176, 1517–1524. [Google Scholar] [CrossRef]
- Girard, J.-P.; Moussion, C.; Förster, R. HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat. Rev. Immunol. 2012, 12, 762–773. [Google Scholar] [CrossRef]
- Kohrt, H.E.; Nouri, N.; Nowels, K.; Johnson, D.; Holmes, S.; Lee, P.P. Profile of immune cells in axillary lymph nodes predicts disease-free survival in breast cancer. PLoS Med. 2005, 2, e284. [Google Scholar] [CrossRef]
- Förster, R.; Braun, A.; Worbs, T. Lymph node homing of T cells and dendritic cells via afferent lymphatics. Trends Immunol. 2012, 33, 271–280. [Google Scholar] [CrossRef]
- Louie, D.A.P.; Liao, S. Lymph node subcapsular sinus macrophages as the frontline of lymphatic immune defense. Front. Immunol. 2019, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Moore Jr, J.E.; Bertram, C.D. Lymphatic system flows. Annu. Rev. Fluid Mech. 2018, 50, 459–482. [Google Scholar] [CrossRef] [PubMed]
- Rezende, R.M.; Lopes, M.E.; Menezes, G.B.; Weiner, H.L. Visualizing Lymph Node Structure and Cellular Localization using Ex-Vivo Confocal Microscopy. J. Vis. Exp. Jove 2019, 150, e59335. [Google Scholar] [CrossRef] [PubMed]
- Suo, Y.; Yang, W.; Lu, F.; Xie, X.S. Label-free imaging of lymph nodes with stimulated Raman scattering microscopy. Proceedings of Optics in Health Care and Biomedical Optics IX, Hangzhou, China, 29 November 2019; p. 111900J. [Google Scholar]
- Knoblich, K.; Migoni, S.C.; Siew, S.M.; Jinks, E.; Kaul, B.; Jeffery, H.C.; Baker, A.T.; Suliman, M.; Vrzalikova, K.; Mehenna, H. The human lymph node microenvironment unilaterally regulates T-cell activation and differentiation. PLoS Biol. 2018, 16, e2005046. [Google Scholar] [CrossRef]
- Westermann, J.; Pabst, R. Distribution of lymphocyte subsets and natural killer cells in the human body. Clin. Investig. 1992, 70, 539–544. [Google Scholar] [CrossRef]
- Grant, S.M.; Lou, M.; Yao, L.; Germain, R.N.; Radtke, A.J. The lymph node at a glance–how spatial organization optimizes the immune response. J. Cell Sci. 2020, 133, jcs241828. [Google Scholar] [CrossRef]
- Kastenmüller, W.; Torabi-Parizi, P.; Subramanian, N.; Lämmermann, T.; Germain, R.N. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell 2012, 150, 1235–1248. [Google Scholar] [CrossRef]
- Rodda, L.B.; Lu, E.; Bennett, M.L.; Sokol, C.L.; Wang, X.; Luther, S.A.; Barres, B.A.; Luster, A.D.; Ye, C.J.; Cyster, J.G. Single-cell RNA sequencing of lymph node stromal cells reveals niche-associated heterogeneity. Immunity 2018, 48, 1014–1028.e6. [Google Scholar] [CrossRef]
- Eissens, D.N.; Spanholtz, J.; Van Der Meer, A.; Van Cranenbroek, B.; Dolstra, H.; Kwekkeboom, J.; Preijers, F.W.; Joosten, I. Defining early human NK cell developmental stages in primary and secondary lymphoid tissues. PLoS ONE 2012, 7, e30930. [Google Scholar] [CrossRef]
- Ferlazzo, G.; Carrega, P. Natural killer cell distribution and trafficking in human tissues. Front. Immunol. 2012, 3, 347. [Google Scholar]
- Campbell, J.J.; Qin, S.; Unutmaz, D.; Soler, D.; Murphy, K.E.; Hodge, M.R.; Wu, L.; Butcher, E.C. Unique subpopulations of CD56+ NK and NK-T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. J. Immunol. 2001, 166, 6477–6482. [Google Scholar] [CrossRef] [PubMed]
- Frey, M.; Packianathan, N.B.; Fehniger, T.A.; Ross, M.E.; Wang, W.-C.; Stewart, C.C.; Caligiuri, M.A.; Evans, S.S. Differential expression and function of L-selectin on CD56bright and CD56dim natural killer cell subsets. J. Immunol. 1998, 161, 400–408. [Google Scholar] [PubMed]
- Freud, A.G.; Becknell, B.; Roychowdhury, S.; Mao, H.C.; Ferketich, A.K.; Nuovo, G.J.; Hughes, T.L.; Marburger, T.B.; Sung, J.; Baiocchi, R.A. A human CD34 (+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity 2005, 22, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.M.; Chaix, J.; Rupp, L.J.; Wu, J.; Madera, S.; Sun, J.C.; Lindsten, T.; Reiner, S.L. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 2012, 36, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Luevano, M.E.; Madrigal, A.; Saudemont, A. Transcription factors involved in the regulation of natural killer cell development and function: An update. Front. Immunol. 2012, 3, 319. [Google Scholar] [CrossRef]
- Aliahmad, P.; De La Torre, B.; Kaye, J. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue–inducer cell and NK cell lineages. Nat. Immunol. 2010, 11, 945–952. [Google Scholar] [CrossRef]
- Sojka, D.K.; Yang, L.; Plougastel-Douglas, B.; Higuchi, D.A.; Croy, B.A.; Yokoyama, W.M. Cutting edge: Local proliferation of uterine tissue-resident NK cells during decidualization in mice. J. Immunol. 2018, 201, 2551–2556. [Google Scholar] [CrossRef] [PubMed]
- Sojka, D.K.; Yang, L.; Yokoyama, W.M. Uterine natural killer cells: To protect and to nurture. Birth Defects Res. 2018, 110, 1531–1538. [Google Scholar] [CrossRef] [PubMed]
- Kalkunte, S.; Chichester, C.O.; Gotsch, F.; Sentman, C.L.; Romero, R.; Sharma, S. Evolution of non-cytotoxic uterine natural killer cells. Am. J. Reprod. Immunol. 2008, 59, 425–432. [Google Scholar] [CrossRef]
- Keskin, D.B.; Allan, D.S.; Rybalov, B.; Andzelm, M.M.; Stern, J.N.; Kopcow, H.D.; Koopman, L.A.; Strominger, J.L. TGFβ promotes conversion of CD16+ peripheral blood NK cells into CD16− NK cells with similarities to decidual NK cells. Proc. Natl. Acad. Sci. USA 2007, 104, 3378–3383. [Google Scholar] [CrossRef]
- Sharkey, A.M.; Xiong, S.; Kennedy, P.R.; Gardner, L.; Farrell, L.E.; Chazara, O.; Ivarsson, M.A.; Hiby, S.E.; Colucci, F.; Moffett, A. Tissue-specific education of decidual NK cells. J. Immunol. 2015, 195, 3026–3032. [Google Scholar] [CrossRef] [PubMed]
- Björkström, N.K.; Ljunggren, H.-G.; Michaelsson, J. Emerging insights into natural killer cells in human peripheral tissues. Nat. Rev. Immunol. 2016, 16, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.; Guo, C.; Sun, R.; Fu, B.; Yang, Y.; Wu, L.; Ren, S.; Tian, Z.; Wei, H. MicroRNA transcriptomes of distinct human NK cell populations identify miR-362-5p as an essential regulator of NK cell function. Sci. Rep. 2015, 5, 9993. [Google Scholar] [CrossRef] [PubMed]
- Muzumdar, M.D.; Tasic, B.; Miyamichi, K.; Li, L.; Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 2007, 45, 593–605. [Google Scholar] [CrossRef]
- Sojka, D.K.; Yang, L.; Yokoyama, W.M. Uterine Natural Killer Cells. Front. Immunol. 2019, 10, 960. [Google Scholar] [CrossRef]
- Sojka, D.K. Uterine Natural Killer Cell Heterogeneity: Lessons From Mouse Models. Front. Immunol. 2020, 11, 290. [Google Scholar] [CrossRef]
- Kather, A.; Chantakru, S.; He, H.; Minhas, K.; Foster, R.; Markert, U.R.; Pfeffer, K.; Croy, B.A. Neither lymphotoxin α nor lymphotoxin β receptor expression is required for biogenesis of lymphoid aggregates or differentiation of natural killer cells in the pregnant mouse uterus. Immunology 2003, 108, 338–345. [Google Scholar] [CrossRef]
- Redhead, M.L.; Portilho, N.A.; Felker, A.M.; Mohammad, S.; Mara, D.L.; Croy, B.A. The transcription factor NFIL3 is essential for normal placental and embryonic development but not for uterine natural killer (UNK) cell differentiation in mice. Biol. Reprod. 2016, 101, 101–116. [Google Scholar] [CrossRef]
- Tayade, C.; Fang, Y.; Black, G.P.; Paffaro, V.A., Jr.; Erlebacher, A.; Croy, B.A. Differential transcription of Eomes and T-bet during maturation of mouse uterine natural killer cells. J. Leukoc. Biol. 2005, 78, 1347–1355. [Google Scholar] [CrossRef]
- Hanna, J.; Goldman-Wohl, D.; Hamani, Y.; Avraham, I.; Greenfield, C.; Natanson-Yaron, S.; Prus, D.; Cohen-Daniel, L.; Arnon, T.I.; Manaster, I. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat. Med. 2006, 12, 1065–1074. [Google Scholar] [CrossRef]
- Moffett, A.; Colucci, F. Uterine NK cells: Active regulators at the maternal-fetal interface. J. Clin. Investig. 2014, 124, 1872–1879. [Google Scholar] [CrossRef]
- Fu, B.; Li, X.; Sun, R.; Tong, X.; Ling, B.; Tian, Z.; Wei, H. Natural killer cells promote immune tolerance by regulating inflammatory TH17 cells at the human maternal-fetal interface. Proc. Natl. Acad. Sci. USA 2013, 110, E231–E240. [Google Scholar] [CrossRef] [PubMed]
- Blois, S.M.; Freitag, N.; Tirado-González, I.; Cheng, S.-B.; Heimesaat, M.M.; Bereswill, S.; Rose, M.; Conrad, M.L.; Barrientos, G.; Sharma, S. NK cell-derived IL-10 is critical for DC-NK cell dialogue at the maternal-fetal interface. Sci. Rep. 2017, 7, 2189. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.W.; Sabatini, B.L. Single-Cell Analysis of Neuroinflammatory Responses Following Intracranial Injection of G-Deleted Rabies Viruses. Front. Cell. Neurosci. 2020, 14, 65. [Google Scholar] [CrossRef] [PubMed]
- Langers, I.; Renoux, V.M.; Thiry, M.; Delvenne, P.; Jacobs, N. Natural killer cells: Role in local tumor growth and metastasis. Biologics 2012, 6, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, A.; Jensen, E.R.; Jamieson, A.M.; Raulet, D.H. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature 2001, 413, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Ljunggren, H.G.; Karre, K. Host resistance directed selectively against H-2-deficient lymphoma variants. Analysis of the mechanism. J. Exp. Med. 1985, 162, 1745–1759. [Google Scholar] [CrossRef]
- Imai, K.; Matsuyama, S.; Miyake, S.; Suga, K.; Nakachi, K. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: An 11-year follow-up study of a general population. Lancet 2000, 356, 1795–1799. [Google Scholar] [CrossRef]
- Bruno, A.; Mortara, L.; Baci, D.; Noonan, D.M.; Albini, A. Myeloid Derived Suppressor Cells Interactions with Natural Killer Cells and Pro-angiogenic Activities: Roles in Tumor Progression. Front. Immunol. 2019, 10, 771. [Google Scholar] [CrossRef]
- Sun, C.; Sun, H.; Zhang, C.; Tian, Z. NK cell receptor imbalance and NK cell dysfunction in HBV infection and hepatocellular carcinoma. Cell Mol. Immunol. 2015, 12, 292–302. [Google Scholar] [CrossRef]
- Sun, C.; Sun, H.Y.; Xiao, W.H.; Zhang, C.; Tian, Z.G. Natural killer cell dysfunction in hepatocellular carcinoma and NK cell-based immunotherapy. Acta Pharm. Sin. 2015, 36, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Coca, S.; Perez-Piqueras, J.; Martinez, D.; Colmenarejo, A.; Saez, M.A.; Vallejo, C.; Martos, J.A.; Moreno, M. The prognostic significance of intratumoral natural killer cells in patients with colorectal carcinoma. Cancer 1997, 79, 2320–2328. [Google Scholar] [CrossRef]
- Ishigami, S.; Natsugoe, S.; Tokuda, K.; Nakajo, A.; Che, X.; Iwashige, H.; Aridome, K.; Hokita, S.; Aikou, T. Prognostic value of intratumoral natural killer cells in gastric carcinoma. Cancer 2000, 88, 577–583. [Google Scholar] [CrossRef]
- Villegas, F.R.; Coca, S.; Villarrubia, V.G.; Jimenez, R.; Chillon, M.J.; Jareno, J.; Zuil, M.; Callol, L. Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer 2002, 35, 23–28. [Google Scholar] [CrossRef]
- Torelli, G.F.; Peragine, N.; Raponi, S.; Pagliara, D.; De Propris, M.S.; Vitale, A.; Bertaina, A.; Barberi, W.; Moretta, L.; Basso, G.; et al. Recognition of adult and pediatric acute lymphoblastic leukemia blasts by natural killer cells. Haematologica 2014, 99, 1248–1254. [Google Scholar] [CrossRef][Green Version]
- Handgretinger, R.; Lang, P.; Andre, M.C. Exploitation of natural killer cells for the treatment of acute leukemia. Blood 2016, 127, 3341–3349. [Google Scholar] [CrossRef]
- Mehta, R.S.; Randolph, B.; Daher, M.; Rezvani, K. NK cell therapy for hematologic malignancies. Int J. Hematol 2018, 107, 262–270. [Google Scholar] [CrossRef]
- Lim, O.; Jung, M.Y.; Hwang, Y.K.; Shin, E.C. Present and Future of Allogeneic Natural Killer Cell Therapy. Front. Immunol. 2015, 6, 286. [Google Scholar] [CrossRef]
- Li, Y.; Yin, J.; Li, T.; Huang, S.; Yan, H.; Leavenworth, J.; Wang, X. NK cell-based cancer immunotherapy: From basic biology to clinical application. Sci China Life Sci 2015, 58, 1233–1245. [Google Scholar] [CrossRef]
- Lee, D.A.; Denman, C.J.; Rondon, G.; Woodworth, G.; Chen, J.; Fisher, T.; Kaur, I.; Fernandez-Vina, M.; Cao, K.; Ciurea, S.; et al. Haploidentical Natural Killer Cells Infused before Allogeneic Stem Cell Transplantation for Myeloid Malignancies: A Phase I Trial. Biol. Blood Marrow Transpl. 2016, 22, 1290–1298. [Google Scholar] [CrossRef]
- Fang, F.; Xiao, W.; Tian, Z. NK cell-based immunotherapy for cancer. Semin. Immunol. 2017, 31, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Li, L.; McCarty, J.; Kaur, I.; Yvon, E.; Shaim, H.; Muftuoglu, M.; Liu, E.; Orlowski, R.Z.; Cooper, L.; et al. Phase I study of cord blood-derived natural killer cells combined with autologous stem cell transplantation in multiple myeloma. Br. J. Haematol. 2017, 177, 457–466. [Google Scholar] [CrossRef]
- Liu, E.; Marin, D.; Banerjee, P.; Macapinlac, H.A.; Thompson, P.; Basar, R.; Nassif Kerbauy, L.; Overman, B.; Thall, P.; Kaplan, M.; et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N. Engl. J. Med. 2020, 382, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Amadei, B.; Urbani, S.; Cazaly, A.; Fisicaro, P.; Zerbini, A.; Ahmed, P.; Missale, G.; Ferrari, C.; Khakoo, S.I. Activation of natural killer cells during acute infection with hepatitis C virus. Gastroenterology 2010, 138, 1536–1545. [Google Scholar] [CrossRef] [PubMed]
- Hudspeth, K.; Pontarini, E.; Tentorio, P.; Cimino, M.; Donadon, M.; Torzilli, G.; Lugli, E.; Della Bella, S.; Gershwin, M.E.; Mavilio, D. The role of natural killer cells in autoimmune liver disease: A comprehensive review. J. Autoimmun 2013, 46, 55–65. [Google Scholar] [CrossRef]
- Cooper, G.E.; Ostridge, K.; Khakoo, S.I.; Wilkinson, T.M.A.; Staples, K.J. Human CD49a(+) Lung Natural Killer Cell Cytotoxicity in Response to Influenza A Virus. Front. Immunol. 2018, 9, 1671. [Google Scholar] [CrossRef]
- Kumar, P.; Thakar, M.S.; Ouyang, W.; Malarkannan, S. IL-22 from conventional NK cells is epithelial regenerative and inflammation protective during influenza infection. Mucosal. Immunol. 2013, 6, 69–82. [Google Scholar] [CrossRef]
- Kumar, P.; Rajasekaran, K.; Palmer, J.M.; Thakar, M.S.; Malarkannan, S. IL-22: An Evolutionary Missing-Link Authenticating the Role of the Immune System in Tissue Regeneration. J. Cancer 2013, 4, 57–65. [Google Scholar] [CrossRef]
- Grundy, M.A.; Zhang, T.; Sentman, C.L. NK cells rapidly remove B16F10 tumor cells in a perforin and interferon-gamma independent manner in vivo. Cancer Immunol. Immunother. 2007, 56, 1153–1161. [Google Scholar] [CrossRef]
- Shen, H.; Kanoh, M.; Maruyama, S.; Matsumoto, A.; Zhang, W.; Asano, Y. Attenuated Listeria infection activates natural killer cell cytotoxicity to regress melanoma growth in vivo. MicroBiol. Immunol. 2008, 52, 107–117. [Google Scholar] [CrossRef]
- Nanbakhsh, A.; Srinivasamani, A.; Holzhauer, S.; Riese, M.J.; Zheng, Y.; Wang, D.; Burns, R.; Reimer, M.H.; Rao, S.; Lemke, A.; et al. Mirc11 Disrupts Inflammatory but Not Cytotoxic Responses of NK Cells. Cancer Immunol. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Scharton, T.M.; Scott, P. Natural killer cells are a source of interferon gamma that drives differentiation of CD4+ T cell subsets and induces early resistance to Leishmania major in mice. J. Exp. Med. 1993, 178, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Scharton-Kersten, T.; Caspar, P.; Sher, A.; Denkers, E.Y. Toxoplasma gondii: Evidence for interleukin-12-dependent and-independent pathways of interferon-gamma production induced by an attenuated parasite strain. Exp. Parasitol. 1996, 84, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Martín-Fontecha, A.; Thomsen, L.L.; Brett, S.; Gerard, C.; Lipp, M.; Lanzavecchia, A.; Sallusto, F. Induced recruitment of NK cells to lymph nodes provides IFN-γ for TH 1 priming. Nat. Immunol. 2004, 5, 1260–1265. [Google Scholar] [CrossRef] [PubMed]
- Fang, V.; Chaluvadi, V.S.; Ramos-Perez, W.D.; Mendoza, A.; Baeyens, A.; Rivera, R.; Chun, J.; Cammer, M.; Schwab, S.R. Gradients of the signaling lipid S1P in lymph nodes position natural killer cells and regulate their interferon-γ response. Nat. Immunol. 2017, 18, 15. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, S.; Chan, M.F.; Li, J.; King, M.R. Super natural killer cells that target metastases in the tumor draining lymph nodes. Biomaterials 2016, 77, 66–76. [Google Scholar] [CrossRef]
- Frazao, A.; Messaoudene, M.; Nunez, N.; Dulphy, N.; Roussin, F.; Sedlik, C.; Zitvogel, L.; Piaggio, E.; Toubert, A.; Caignard, A. CD16+ NKG2Ahigh Natural Killer Cells Infiltrate Breast Cancer–Draining Lymph Nodes. Cancer Immunol. Res. 2019, 7, 208–218. [Google Scholar] [CrossRef]
- Cross, J.C.; Hemberger, M.; Lu, Y.; Nozaki, T.; Whiteley, K.; Masutani, M.; Adamson, S.L. Trophoblast functions, angiogenesis and remodeling of the maternal vasculature in the placenta. Mol. Cell. Endocrinol. 2002, 187, 207–212. [Google Scholar] [CrossRef]
- Fu, B.; Zhou, Y.; Ni, X.; Tong, X.; Xu, X.; Dong, Z.; Sun, R.; Tian, Z.; Wei, H. Natural killer cells promote fetal development through the secretion of growth-promoting factors. Immunity 2017, 47, 1100–1113.e6. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashemi, E.; Malarkannan, S. Tissue-Resident NK Cells: Development, Maturation, and Clinical Relevance. Cancers 2020, 12, 1553. https://doi.org/10.3390/cancers12061553
Hashemi E, Malarkannan S. Tissue-Resident NK Cells: Development, Maturation, and Clinical Relevance. Cancers. 2020; 12(6):1553. https://doi.org/10.3390/cancers12061553
Chicago/Turabian StyleHashemi, Elaheh, and Subramaniam Malarkannan. 2020. "Tissue-Resident NK Cells: Development, Maturation, and Clinical Relevance" Cancers 12, no. 6: 1553. https://doi.org/10.3390/cancers12061553
APA StyleHashemi, E., & Malarkannan, S. (2020). Tissue-Resident NK Cells: Development, Maturation, and Clinical Relevance. Cancers, 12(6), 1553. https://doi.org/10.3390/cancers12061553




