1. Introduction
The leading cause of cancer-related deaths is lung cancer [
1], which can be divided into small-cell lung cancers and non-small-cell lung cancers. Accounting for 85% of lung cancers [
2], non-small-cell lung cancers can be further subdivided into lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large-cell lung cancer [
3]. LUAD accounts for about 40% of lung cancer cases [
4], whereas LUSC accounts for around 25% of non-small-cell lung cancer occurrences [
3]. Overall, the 5-year survival rate of lung cancer is only 17% [
4]. The broad impact and low survival rates of lung cancer highlight the critical importance of studying the etiologies and risk factors for lung cancer.
The human lungs have a diverse microbiota that is composed of many types of organisms. One study found over nine genera of bacteria in the lungs [
5,
6]. It has been found that the microbiome plays a significant role in disease and immunity in the lungs [
6]. Furthermore, studies have supported an association between the microbiome and cancer, including lung cancer [
7]. Factors including age have been implicated in the microbiome where it has been discovered that community diversity lowers in older age groups when specifically studying the lung microbiome [
8,
9]. Gender also plays a significant role in regards to the microbiome, as the microbiota associated with each gender is unique because of the hormones specific to each gender [
10]. Although there are many studies conducted on LUAD and LUSC, relatively few focus on the association of these cancers with their respective microbiome [
11].
Clinical variables, including age and gender, play significant roles in the development of cancers. Approximately 90% of lung cancer cases occur in individuals over the age of 55, whereas only around 10% of lung cancer cases occur in individuals under the age of 55 [
12]. Younger lung cancer patients also tend to have higher survival rates. This difference in cancer incidence and patient survival rates indicates that age could determine the microbiome, which, in turn, regulates the pathogenesis and progression of LUAD and LUSC. Gender is also an important factor to consider in cancer pathogenesis. Men tend to have higher mortality rates and a higher incidence of lung cancer than women. This could be explained partially by the fact that men tend to smoke more tobacco. Curiously, incidence of lung cancer is higher in non-smoking women than in non-smoking men [
13]. Younger lung cancer patients are also more likely to be female [
12]. Thus, it is imperative to investigate and characterize the association between the lung microbiome and lung cancers such as LUAD and LUSC in the context of both age and gender. In this study, we investigated the lung microbiome in LUAD and LUSC patients by dividing patients into eight cohorts based on age and gender and comparing the presence of similar or unique bacteria across cohorts. From these comparisons, we correlated the abundance of these microbes to patient survival, immune cell populations, immune signatures, immune and cancer pathways, and genomic alterations.
3. Discussion
Ongoing research and recent studies have indicated that the microbiome plays an important role in regulating and contributing to the progression of cancer [
16]. Although most studies have focused primarily on the gut microbiome in relation to cancer, several studies have begun attempting to identify key microbial species that lead to cancer in other tissues, including the skin, colon, liver and lungs [
17,
18,
19]. In particular, studies on the lung microbiome have correlated the lung microbiota in lung cancer with patient survival and established potential mechanisms for its relation to lung cancer progression via specific immune pathways [
20,
21]. A study by Greathouse et al. analyzed TCGA LUSC and LUAD data to discern the combinatorial effects of somatic mutations and smoking while adjusting for age and gender as co-variates, but the study did not identify microbes associated with age and gender [
22]. In this study, we focus on identifying and comparing unique and common microbial species in a total of 16 LUAD and LUSC patient cohorts that are stratified according to age and gender using TCGA data and correlating the abundance of the associated microbes to patient survival, immune cell populations, immune signatures, immune and cancer pathways, and genomic alterations. To the best of our knowledge, we have not found any other study that comparatively analyzes the LUAD and LUSC-associated microbiota when taking age and gender into account.
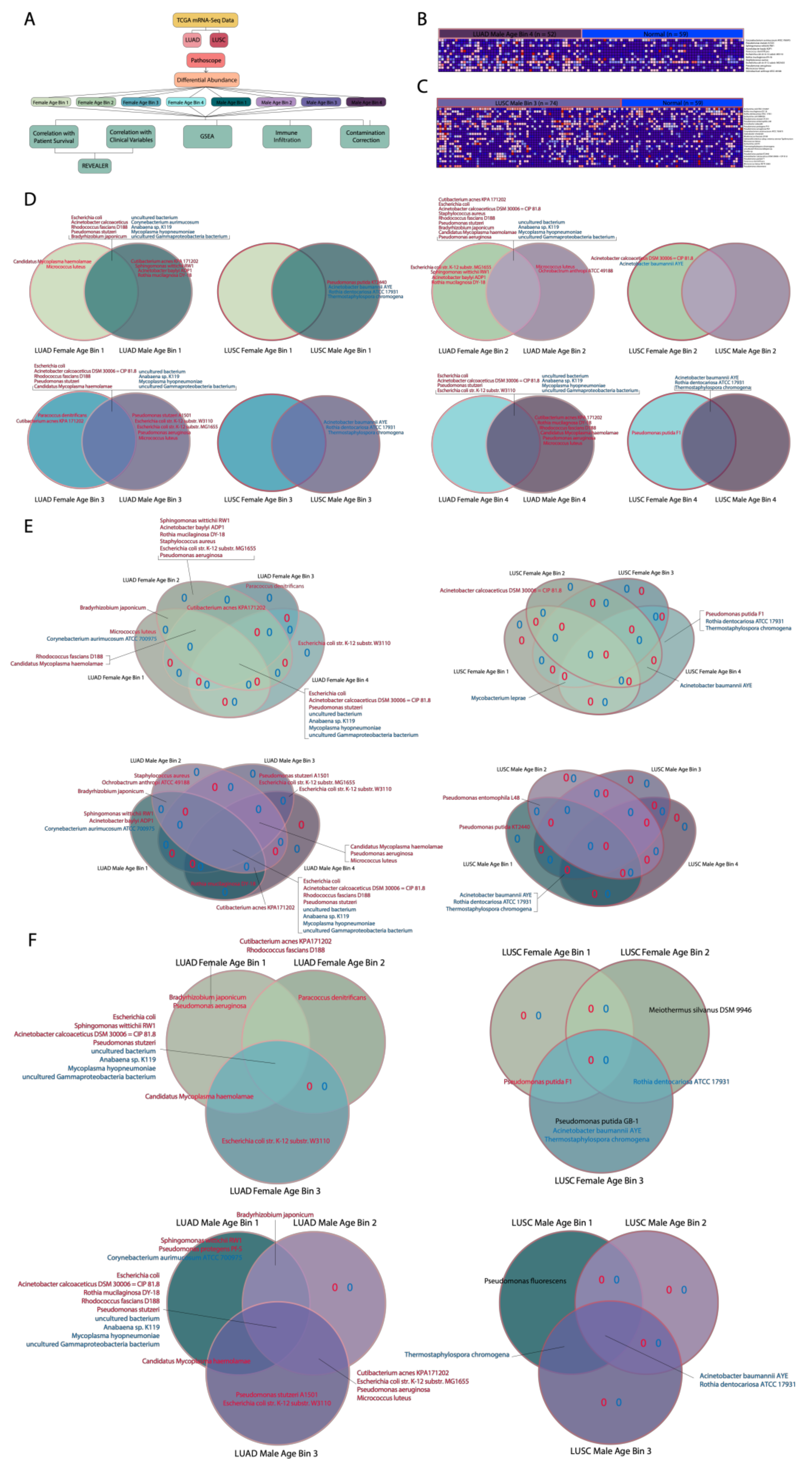
After rigorously identifying and removing potential microbial contaminants, we separated LUAD and LUSC patients into eight cohorts each based on four different age groups and two genders and compared each cohort’s associated microbial abundance to the microbial abundance of corresponding adjacent normal samples. From this comparison, we took differentially abundant microbes in each cohort and performed gender comparisons and age bin comparisons, which involve contrasting genders with the same age bin and age bins within one gender, respectively, within individual cancers. We also compared between cancers for microbes uniquely associated with a certain cohort or in common with two or four cohorts. For all comparisons, LUSC cohorts generally had fewer differentially abundant microbes compared to LUAD cohorts. For gender comparisons, in particular, we noted that it was more common for LUAD samples to share differentially abundant microbes between genders in each age bin than LUSC samples. LUAD cohorts were more likely than LUSC cohorts to contain significant unique microbes with high abundance whereas only LUSC cohorts contained significant unique microbes with low abundance. We found that, for age bin comparisons, only LUAD contained a significant microbe with high and low abundance. LUAD also had a greater number of cohorts uniquely associated with microbes with high abundance than LUSC, which only had three cohorts. No unique microbes or microbes in common across all four age bins were shared between both cancers.
Overall, when comparing between genders, all LUAD female age bins contained uniquely implicated microbes except for LUAD Female Age Bin 4 (73–88 years). All LUAD male age bins contained uniquely implicated microbes where most contained more uniquely associated microbes than their female counterparts except for LUAD Male Age Bin 2 (33–58 years). In the LUSC gender comparisons, only two LUSC female age bins and two LUSC male age bins contained uniquely implicated microbes, which were LUSC Female Age Bin 2 (62–67 years) and 4 (73–85 years) and LUSC Male Age Bin 1 (45–61 years) and 3 (68–72 years). The uniquely implicated microbes in most LUAD and LUSC cohorts could explain why certain genders are more prone to being diagnosed with either LUAD or LUSC.
When comparing between age bins, all LUAD female cohorts and LUAD male cohorts had at least one uniquely associated microbe except for LUAD Male Age Bin 4 (73–88 years). On the other hand, for LUSC age bin comparisons, only LUSC Female Age Bin 2 (A. calcoaceticus str. DSM 20,006 = CIP 81.8), LUSC Female Age Bin 4 (P. putida str. F1, R. dentocariosa str. ATCC 17931, T. chromogena) and LUSC Male Age Bin 1 contained uniquely implicated microbes (P. putida str. KT2440).
The majority of cohorts in LUAD and LUSC, which contained unique significantly implicated microbes in gender and age bins comparisons, demonstrates that there are significant microbial differences between age groups and gender in both cancers. Thus, a panel of predictive biomarkers based on specific age groups and gender is needed to perform early diagnoses of lung cancers. In addition to the microbes we identified using bulk RNA sequencing, other microbial species could potentially be identified by using 16S sequencing as it can provide a more specific identification of bacteria species that were unable to be cultured.
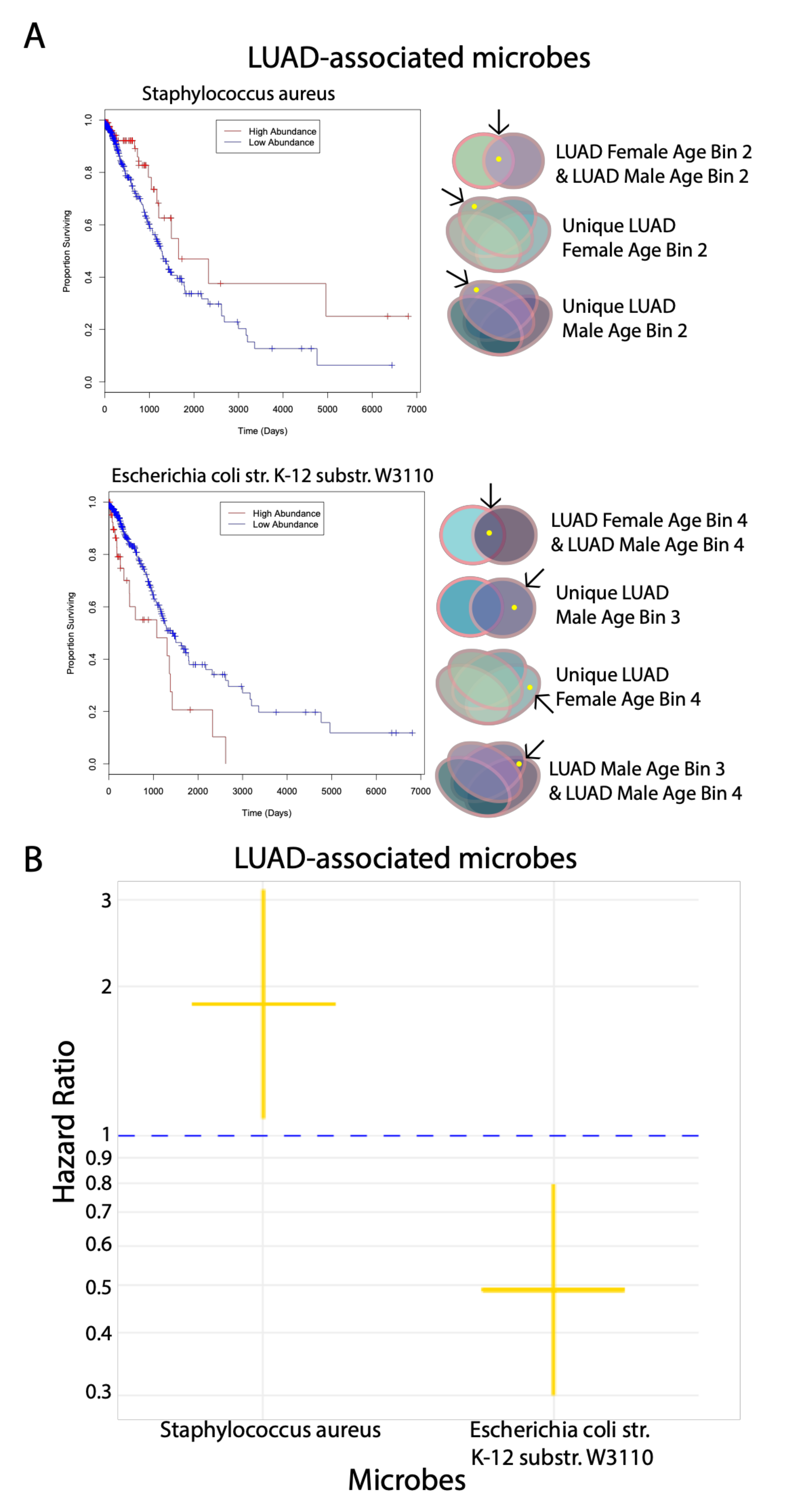
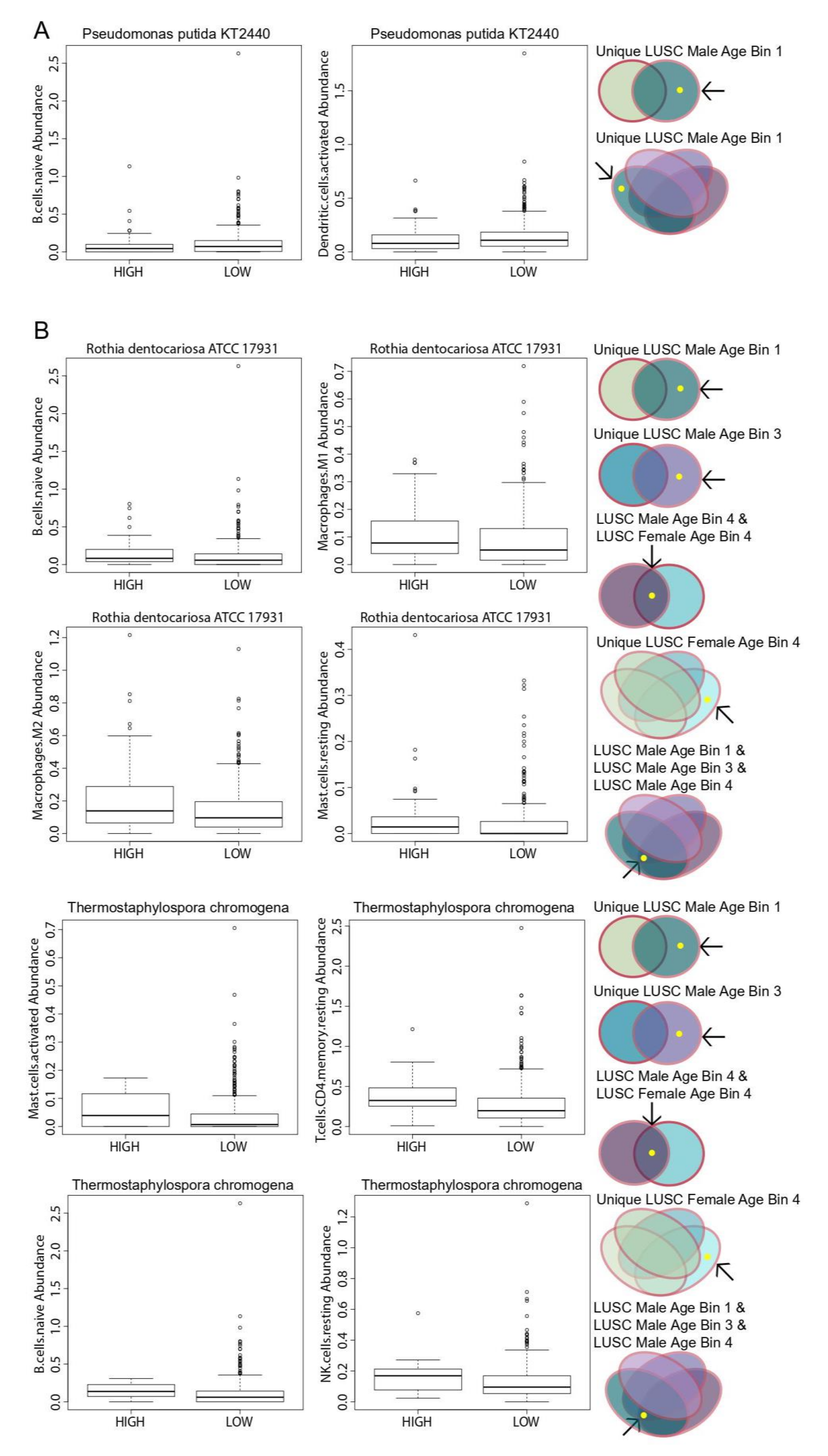
To assess the potential prognostic significance of age- and gender-associated microbes and generate hypotheses related to their function, we correlated the significant microbes that we compared in the gender and age bin comparisons to patient survival, immune cell populations, immune signatures, immune and cancer pathways and genomic alterations. In most analyses, only a select few microbes exhibited significant correlations where they belonged to either LUAD or LUSC. Based on the Cox proportional hazards regression model, we determined that the LUAD-associated microbe S. aureus displayed tumor-suppressive properties as it significantly increased patient survival rates when its abundance levels were high. On the other hand, E. coli str. K-12 substr. W3110, which is also an LUAD-associated microbe, may function like an oncogene in that patient survival rates were significantly decreased when its abundance levels were high. For correlations with immune cell populations, lower abundance levels of both R. dentocariosa str. ATCC 17931 and T. chromogena and higher abundance levels of P. putida str. KT2440, which are both LUSC-associated microbes, were correlated with lower expression of various immune cell populations.
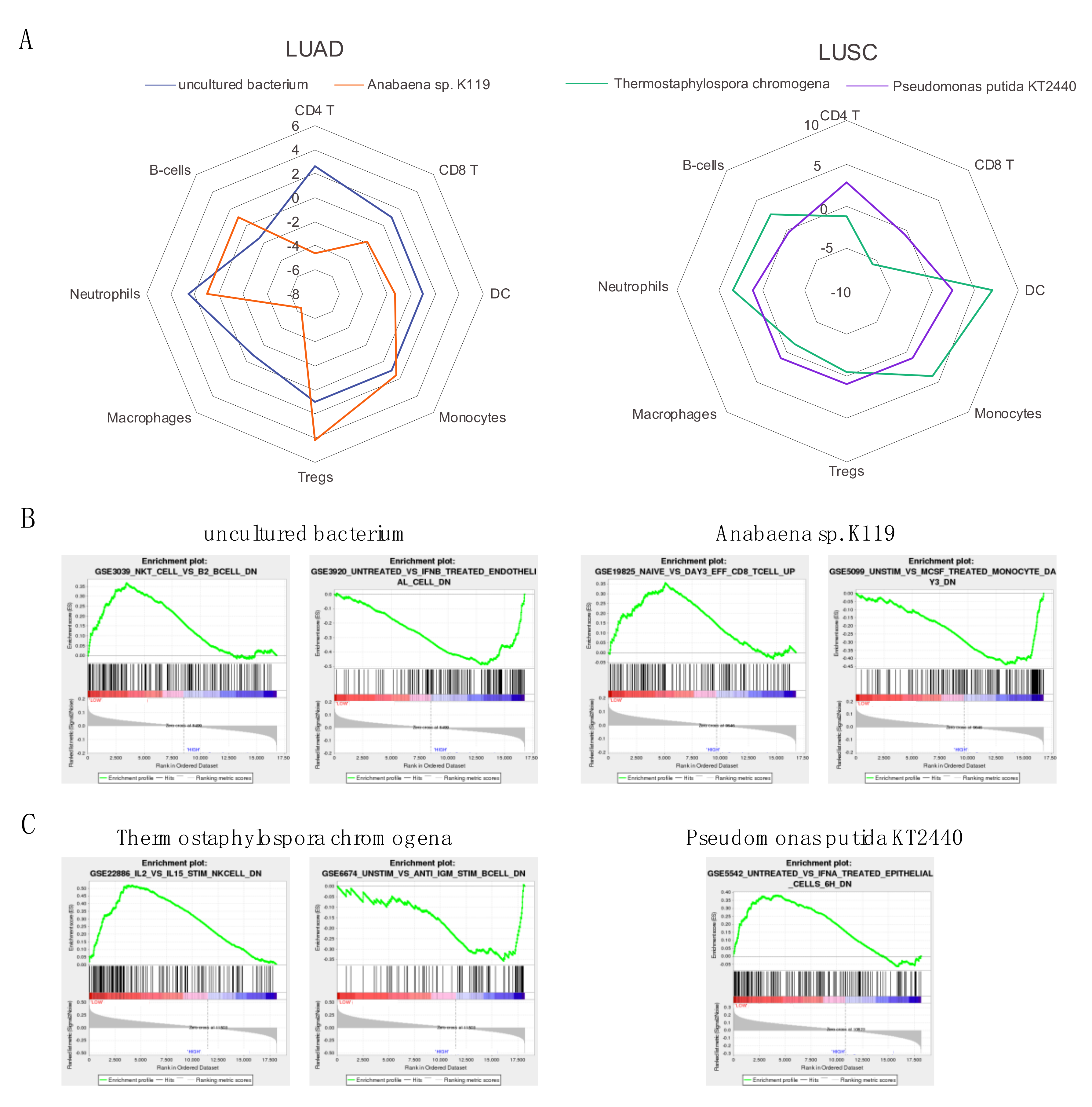
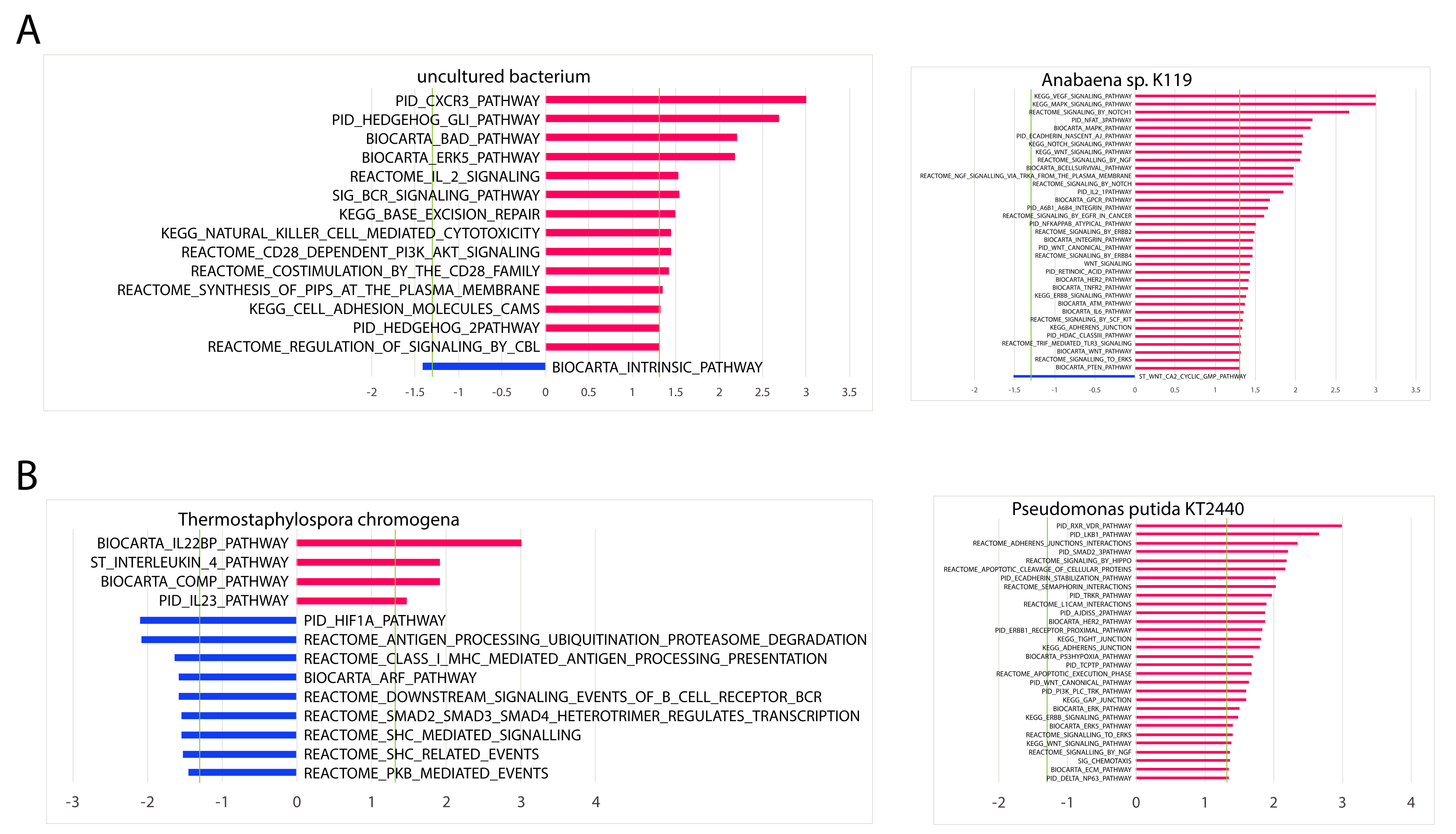
In contrast to the analyses that correlated microbial abundance to patient survival and immune cell populations, there were a large number of significant microbial associations with immune signatures and immune and cancer pathways. For the two most significant microbes for each cancer, which included uncultured bacterium and Anabaena sp. K119 for LUAD and T. chromogena and P. putida str. KT2440 for LUSC, we found that the abundance of these microbes is significantly associated with several immune signatures that are related to specific immune cell types. All four microbes were found to negatively associate with CD8 T cells and macrophages and positively associate with neutrophils and monocytes. Other immune cell types, including regulatory T cells and dendritic cells were positively correlated with at least one LUAD-associated microbe and negatively associated with at least one LUSC-associated microbe. We also selected examples of immune signatures that were significantly dysregulated as a result of low or high abundance of these four microbes. The abundance of the four microbes was also correlated to immune and cancer pathways. Uncultured bacterium and T. chromogena had the fewest associated cancer and immune pathways. Although three of the microbes had more positive associations than negative associations with immune and cancer pathways (P. putida str. KT2440 had all positive associations), T. chromogena exhibited the opposite proportions of positive to negative associations of immune and cancer pathways. Finally, we performed Repeated Evaluation of Variables conditionAL Entropy and Redundancy (REVEALER) to correlate the significant microbes following the gender and age bin comparisons to genomic alterations. Only E. coli str. K-12 substr. W3110, which is an LUAD-associated microbe, was significantly correlated with genomic alterations, which consisted of one amplification loci and mainly deletion and mutation loci.
Out of the microbes that were significantly correlated to at least one analyses, only
E. coli str. K-12 substr. W3110,
S. aureus,
R. dentocariosa str. ATCC 17931, and
P. putida str. KT2440 were found in humans [
23,
24,
25,
26]. Because the majority of bacteria is unculturable, only a small fraction of the bacteria is represented in the data. For the microbe that appeared most frequently in our analyses, which was the LUAD-associated microbe
E. coli str. K-12 substr. W3110, no other studies have found this microbe, or microbes closely related to it, to be implicated in the human microbiome and cancer.
E. coli str. K-12 is commonly used as a model organism, and the substrain W3110 is used as a wild-type strain in experiments globally [
26]. On the other hand, the LUAD-associated microbe
S. aureus, which is a known pathogen that colonizes nasal areas, has been found to increase the risk of patients dying from cancer [
25,
27]. It has been discovered that the colonization of
S. aureus occurs more frequently in men than women [
28]. Furthermore, LUSC-associated microbes
R. dentocariosa str. ATCC 17931, which is normally found in the oral cavity, was discovered to cause lung infections and pneumonia [
24]. Finally,
P. putida str. KT2440 was isolated from blood samples in cancer patients, including one lung cancer patient [
23]. In conclusion, we have discovered novel associations for several bacteria known to be present in humans but never before correlated to gender and age in lung cancer. Our results could be critical to efforts to diagnose, treat, or prevent lung cancer using the microbiome composition. Lastly, gender and age difference in microbe levels may also contribute to biological differences in the mechanism of lung cancer.
4. Materials and Methods
4.1. Acquisition of TCGA RNA-Sequencing Datasets
RNA-sequencing tumor tissue data for 497 LUAD and 433 LUSC patients along with adjacent solid tissue normal data for 49 LUSC patients and 59 LUAD patients was obtained from The Cancer Genome Atlas (
https://portal.gdc.cancer.gov/legacy-archive/search/f) on 5 Aug 2018. Clinical information for each patient was downloaded from the Broad GDAC Firehose (
https://gdac.broadinstitute.org/). Data related to genomic alterations for each patient were downloaded from the Broad Institute TCGA Genome Data Analysis Center’s (
http://gdac.broadinstitute.org/runs/analyses__latest/reports/) analysis report (2016). Pathoscope 2.0 [
14] was used to filter RNA-sequencing data for bacterial reads using direct alignment through Bowtie2. The NCBI’s nucleotide database was accessed for bacterial sequences. Pathoscope’s best hit output data, the absolute count of each species in the data, was used to measure the amount of bacterial species present in a sample.
4.2. Differential Microbial Abundance between Cancer and Normal Samples
Differential abundance analysis was performed to compare microbe abundance (percent abundance) in cancer tissues to microbe abundance in normal tissues of the same body site. Microbes that are present in less than 10 patients of the same cancer were excluded. The Kruskal–Wallis analysis test was then applied to determine differential abundance (p < 0.05).
4.3. Differential Microbe Abundance Based on Patient Age and Gender
Kruskal–Wallis testing was performed on microbial abundance for male cancer patients and female cancer patients in order to determine the association between abundance and gender. Patients were also divided into four age bins. The boundaries of the age bins were defined by the minimum age, maximum age, or one of the three quartiles when including ages of all patients of the same cancer. Kruskal–Wallis testing was performed on the microbe abundances vs. age bins to determine the association between abundance and age. Kruskal–Wallis tests were also performed on microbe abundances between 8 cancer patient groups, divided by age and gender. These included four age bins of female cancer patients and four age bins of male cancer patients. For both female and male patients in LUAD, age bins 1, 2, 3, and 4 correspond to age ranges of 33–58, 59–65, 66–72, 73–88, respectively. For both female and male patients in LUSC, age bins 1, 2, 3, and 4 correspond to age ranges of 45–61, 62–67, 68–72, 73–85, respectively. The number of patients in each patient group is listed in
Table S3. The distribution of tumor stages for each age bin is listed in
Table S4. For all groups, comparisons were between microbe abundances of the group and microbe abundances in normal tissue.
Associations found to be significant (p < 0.05) were separated based on whether microbe abundance was higher in a particular patient group or in normal tissue. Venn diagrams were made to show unique microbes and microbes in common between different combinations of groups. Diagrams included microbes that were differentially abundant in the same direction for both groups. Venn diagram comparisons were made between male patients and female patients in the same age bin with the same cancer, between patients in different age bins with the same gender and the same cancer, and between patients with the same age bin, the same gender and different cancers.
4.4. Visualization of Microbial Population Distribution Using PCoA
We first determined that principle component analysis (PCA) and correspondence analysis (CA) were not ideal because of undesirable gradient and plot shape, respectively. The Bray–Curtis dissimilarity measure was then calculated for all cancer samples for LUAD and LUSC, and then PCoA was performed using the dissimilarity matrix as the input. Analyses were performed using R and the R package vegan.
4.5. Identification of Smoking Associated Microbes in LUSC and LUAD
We identified smoking associated microbes by comparing the microbe abundance in cancer samples from smokers at time of diagnosis vs. that in cancer samples from life-long nonsmokers (χ2 test, p < 0.05).
4.6. Logistic Regression to Correlate Clinical Variables with Microbe Abundance
To identify confounding variables, we correlated age and gender-associated microbes to other clinical variables using logistic regression. The variables included were ethnicity, pathologic stage, vital status, number of pack-years smoked, pathologic TNM stages, tobacco smoking history, and race.
4.7. Correlation of Microbial Abundance to Patient Survival
Survival analyses were performed using the Kaplan–Meier model, with microbe expression designated as a binary variable based on the presence or absence of a microbe in tumor samples. Univariate Cox regression analysis was used to identify candidates that were significantly associated with patient survival (p < 0.05).
4.8. Correlation of Microbial Abundance to Immune Infiltration
Estimated relative immune cell infiltration levels for 22 cell types were computed using the software Cibersortx [
29]. Microbe abundance was then correlated with immune cell infiltration levels for each microbe using the Kruskal–Wallis test (
p < 0.05). Microbe abundance was modeled as a binary variable of presence and absence. The immune cell types examined include naïve B-cells, memory B-cells, plasma cells, CD8 T-cells, CD4 naïve T-cells, CD4 memory resting T-cells, CD4 memory activated T-cells, follicular helper T-cells, regulatory T-cells, gamma-delta T-cells, resting NK cells, activated NK cells, monocytes, M0-M2 macrophages, resting dendritic cells, activated dendritic cells, resting mast cells, activated mast cells, eosinophils, and neutrophils.
4.9. Immune Pathway Association with Microbial Expression Using GSEA
GSEA was utilized to identify microbes associated with the dysregulation of biological pathways and signatures, which are obtained from the Molecular Signature Database (MSigDB) [
30]. Specifically, canonical pathways (C2) and immunologic signatures (C7) were examined. The abundance data for each microbe were inputted as a categorical variable (presence or absence) in the phenotype file. The gene expression dataset consisted of the expression values of all genes in counts per million (CPM). Using Pearson’s correlation for the continuous phenotypes and signal-to-noise ratio for categorical phenotypes, the microbe abundance is correlated to the above gene sets to generate enrichment scores. Higher enrichment scores indicate stronger correlation between microbe abundance and expression of genes within a gene set.
4.10. REVEALER Association of Microbe Abundance with Genomic Alterations
The Repeated Evaluation of Variables conditionAL Entropy and Redundancy (REVEALER) program was used to identify a statistically significant association of genomic alterations (amplifications, deletions, or mutations) with the abundance of individual microbes. We define an association as significant if the absolute value of its Conditional Information Coefficient (CIC) value was greater than 0.2 and if the p-value was less than 0.05.
4.11. Evaluation of Possible Contamination Using Plates and Date of Sequencing
The abundance values of microbes were associated with the plates on which the samples were stored prior to sequencing using the Kruskal–Wallis test and visual examination of abundance differences between different plates using a boxplot. For visual examination, the microbe abundance for each sample was plotted in order of date sequenced on a dot plot. If the microbe is a contaminant, samples sequenced near the same date would have a similar overexpression of specific bacterial species. Therefore, we applied a heuristic algorithm based on divisive clustering using the DIANA R package to extract the sample ranges where this overexpression occurs, which allowed us to determine potential contaminants’ relationship with the sequencing date.