Role of Collagen Fiber Morphology on Ovarian Cancer Cell Migration Using Image-Based Models of the Extracellular Matrix
Abstract
1. Introduction
2. Methods and Materials
2.1. Microscope and Photochemistry
2.2. Sample Preparation
2.3. Cell Seeding and Time-Lapse Imaging
2.4. Cell Tracking
2.5. F-Actin, Focal Adhesion, and Cadherin Staining
2.6. Statistical Analysis
3. Results
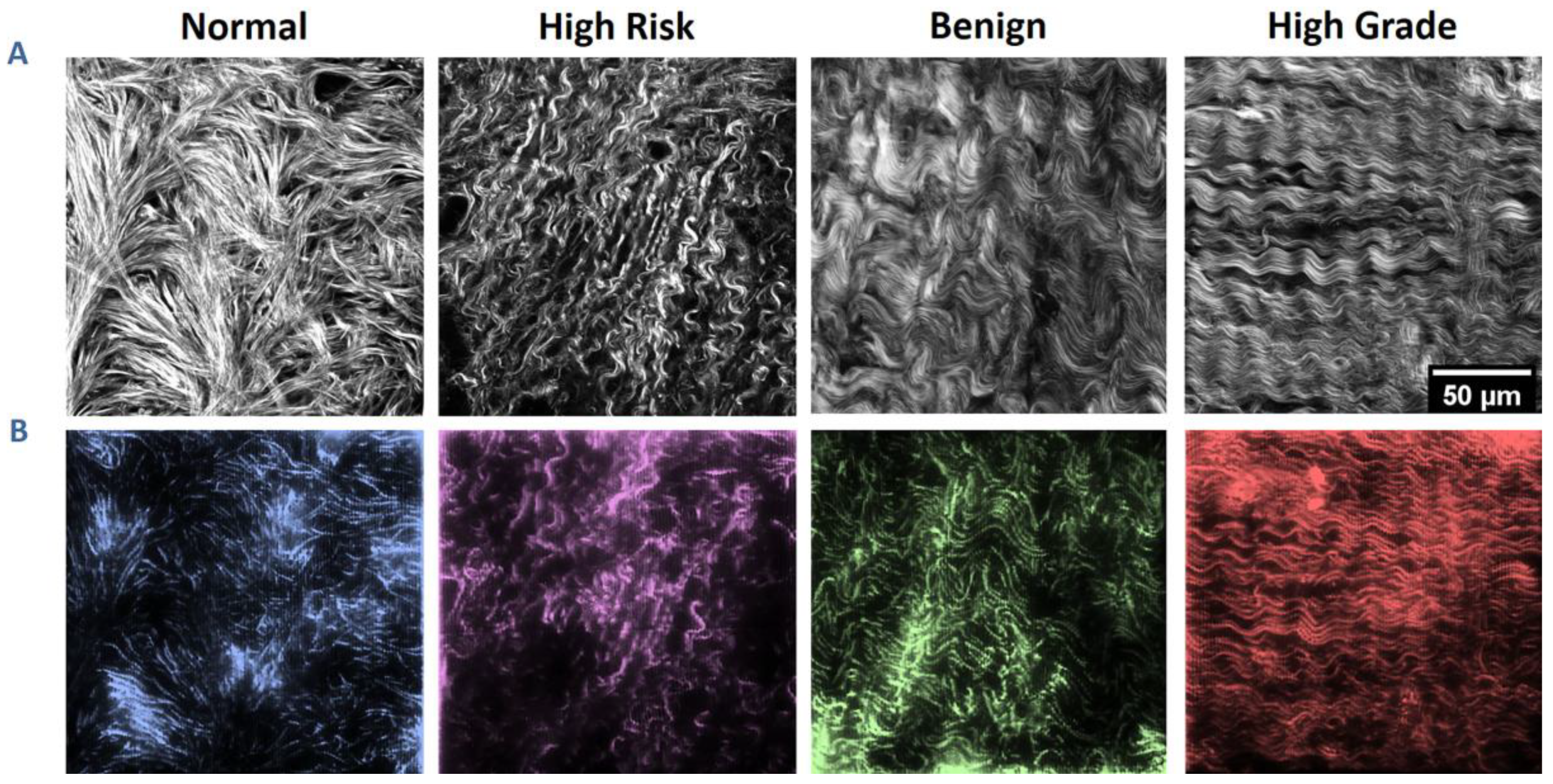
3.1. SHG Image-Based Blueprints for Fabrication
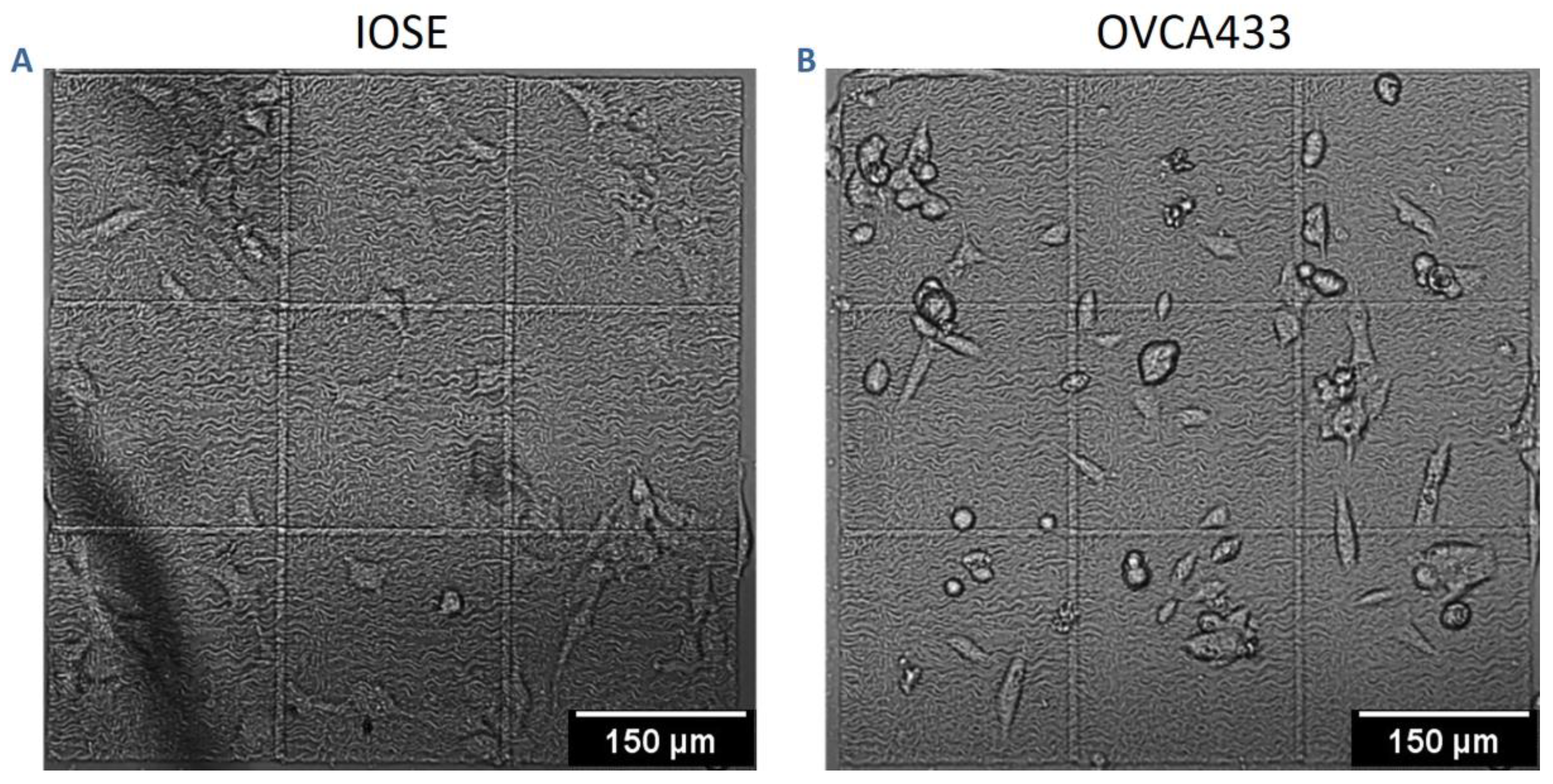
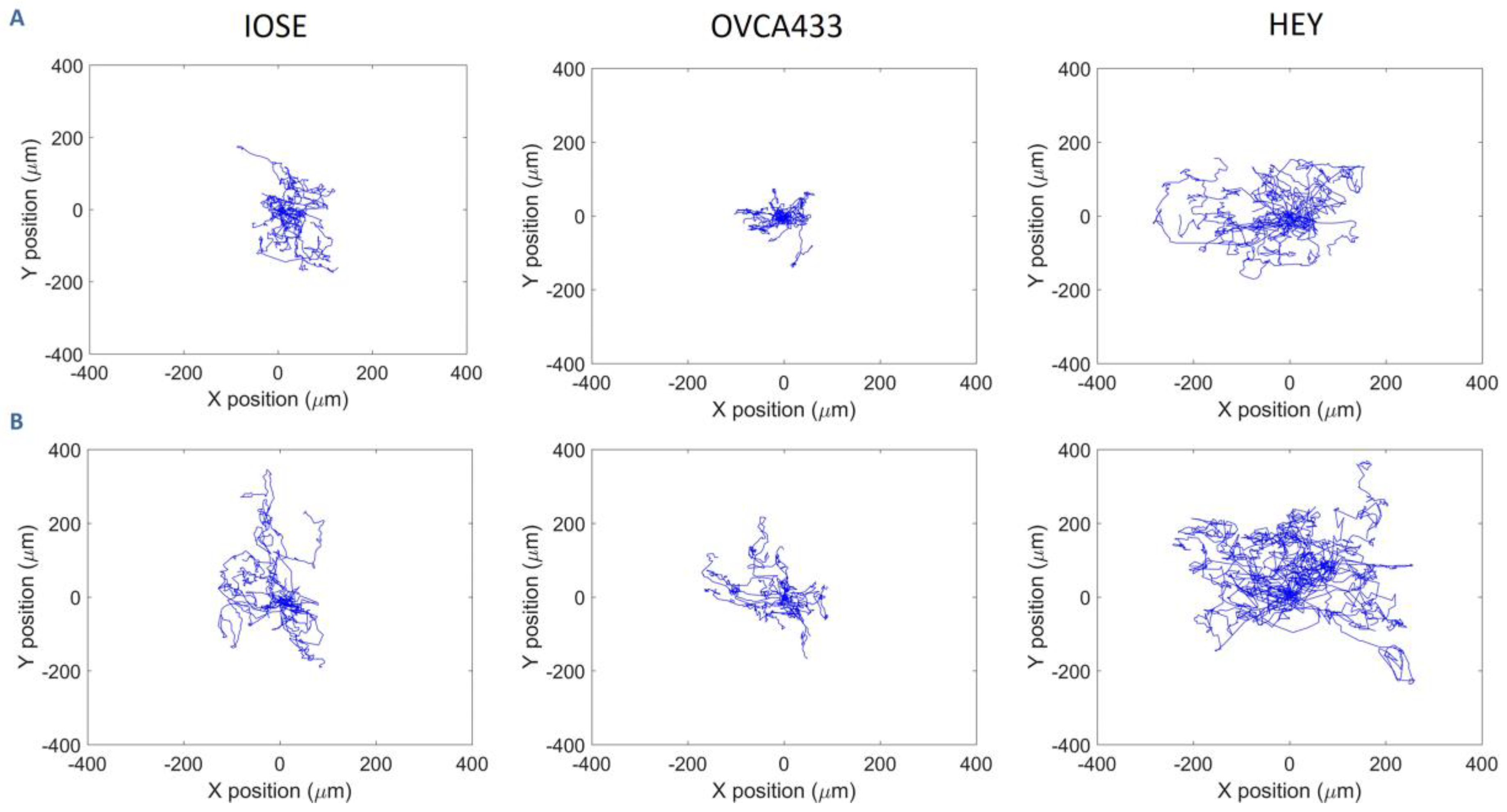
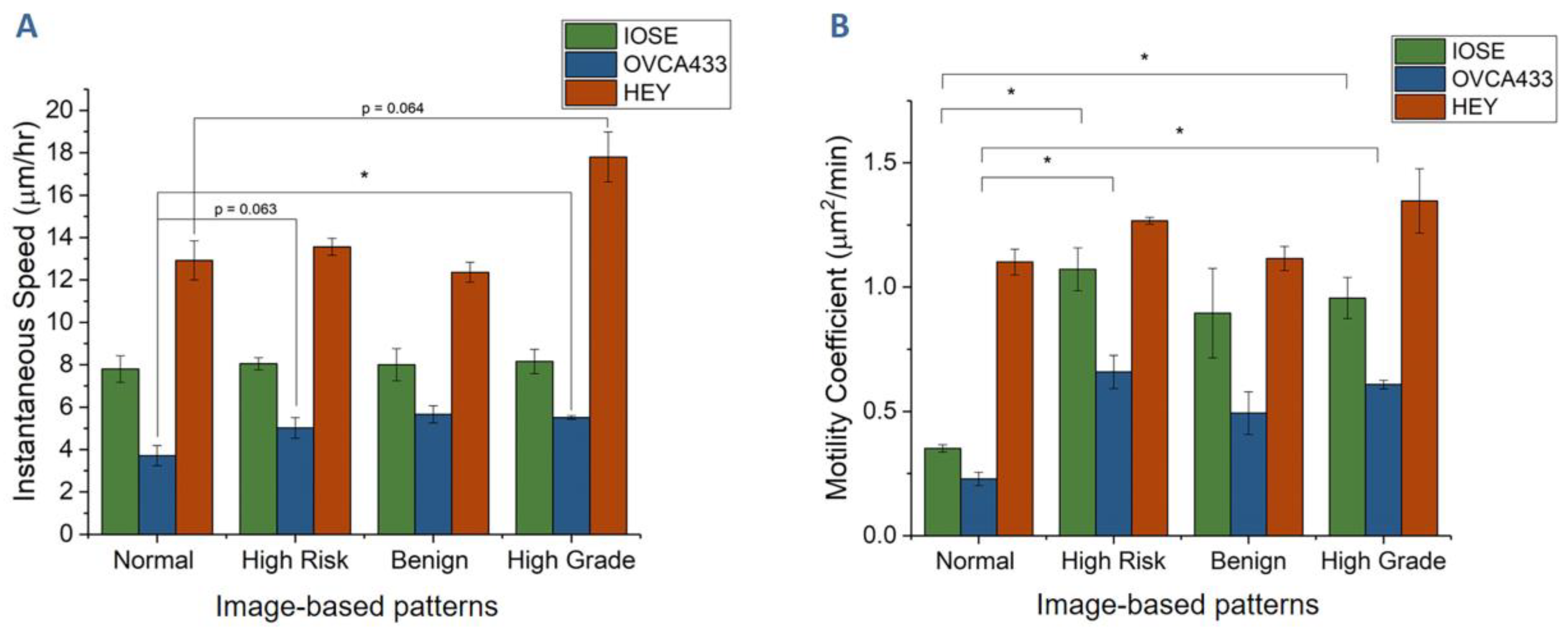
3.2. Cell Migration on Image-Based Scaffolds
3.3. Collagen Fiber Alignment Drives Cell Morphology
3.4. Fiber Alignment Governs Cytoskeletal Expression and Alignment
3.5. Matrix Morphology Determines Cadherin Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cancer Facts & Figures 2018; American Cancer Society: Atlanta, GA, USA, 2018.
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Karlan, B.Y. The status of ultrasound and color Doppler imaging for the early detection of ovarian carcinoma. Cancer Invest. 1997, 15, 265–269. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, M.W.; Drescher, C.; Karlan, B.; Scholler, N.; Urban, N.; Hellstrom, K.E.; Hellstrom, I. Combining CA 125 and SMR serum markers for diagnosis and early detection of ovarian carcinoma. Gynecol. Oncol. 2004, 95, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Petricoin, E.F.; Ardekani, A.M.; Hitt, B.A.; Levine, P.J.; Fusaro, V.A.; Steinberg, S.M.; Mills, G.B.; Simone, C.; Fishman, D.A.; Kohn, E.C.; et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002, 359, 572–577. [Google Scholar] [CrossRef]
- Qayyum, A.; Coakley, F.V.; Westphalen, A.C.; Hricak, H.; Okuno, W.T.; Powell, B. Role of CT and MR imaging in predicting optimal cytoreduction of newly diagnosed primary epithelial ovarian cancer. Gynecol. Oncol. 2005, 96, 301–306. [Google Scholar] [CrossRef]
- Bristow, R.E.; Giuntoli, R.L., II; Pannu, H.K.; Schulick, R.D.; Fishman, E.K.; Wahl, R.L. Combined PET/CT for detecting recurrent ovarian cancer limited to retroperitoneal lymph nodes. Gynecol. Oncol. 2005, 99, 294–300. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Landen, C.N., Jr.; Birrer, M.J.; Sood, A.K. Early events in the pathogenesis of epithelial ovarian cancer. J. Clin. Oncol. 2008, 26, 995–1005. [Google Scholar] [CrossRef]
- Doubeni, C.A.; Doubeni, A.R.; Myers, A.E. Diagnosis and Management of Ovarian Cancer. Am. Fam. Physician 2016, 93, 937–944. [Google Scholar]
- Fritsche, H.A.; Bast, R.C. CA 125 in ovarian cancer: Advances and controversy. Clin. Chem. 1998, 44, 1379–1380. [Google Scholar] [CrossRef]
- Liotta, L.A.; Kohn, E.C. The microenvironment of the tumour-host interface. Nature 2001, 411, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Ngalame, Y.; Panelli, M.C.; Nguyen-Jackson, H.; Deavers, M.; Mueller, P.; Hu, W.; Savary, C.A.; Kobayashi, R.; Freedman, R.S.; et al. Peritoneal and Subperitoneal Stroma May Facilitate Regional Spread of Ovarian Cancer. Clin. Cancer Res. 2005, 11, 113–122. [Google Scholar] [PubMed]
- Mueller, M.M.; Fusenig, N.E. Friends or foes—bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer 2004, 4, 839. [Google Scholar] [CrossRef] [PubMed]
- Mullany, L.K.; Richards, J.S. Minireview: Animal Models and Mechanisms of Ovarian Cancer Development. Endocrinology 2012, 153, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Jin, V.; Bergsten, T.M.; Austin, J.R.; Lantvit, D.D.; Russo, A.; Burdette, J.E. Loss of PTEN in Fallopian Tube Epithelium Results in Multicellular Tumor Spheroid Formation and Metastasis to the Ovary. Cancers 2019, 11, 884. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Czarnecki, A.A.; Dean, M.; Modi, D.A.; Lantvit, D.D.; Hardy, L.; Baligod, S.; Davis, D.A.; Wei, J.J.; Burdette, J.E. PTEN loss in the fallopian tube induces hyperplasia and ovarian tumor formation. Oncogene 2018, 37, 1976–1990. [Google Scholar] [CrossRef]
- Cho, A.; Howell, V.M.; Colvin, E.K. The extracellular matrix in epithelial ovarian cancer—A piece of a puzzle. Front. Oncol. 2015, 5, 245. [Google Scholar] [CrossRef]
- Ricciardelli, C.; Rodgers, R.J. Extracellular matrix of ovarian tumors. Semin. Reprod. Med. 2006, 24, 270–282. [Google Scholar] [CrossRef]
- Kauppila, S.; Bode, M.K.; Stenbäck, F.; Risteli, L.; Risteli, J. Cross-linked telopeptides of type I and III collagens in malignant ovarian tumours in vivo. Br. J. Cancer 1999, 81, 654–661. [Google Scholar] [CrossRef]
- Chen, X.; Nadiarynkh, O.; Plotnikov, S.; Campagnola, P.J. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc. 2012, 7, 654–669. [Google Scholar] [CrossRef]
- Tilbury, K.; Lien, C.H.; Chen, S.J.; Campagnola, P.J. Differentiation of Col I and Col III Isoforms in Stromal Models of Ovarian Cancer by Analysis of Second Harmonic Generation Polarization and Emission Directionality. Biophys. J. 2014, 106, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Tilbury, K.B.; Campbell, K.R.; Eliceiri, K.W.; Salih, S.M.; Patankar, M.; Campagnola, P.J. Stromal alterations in ovarian cancers via wavelength dependent Second Harmonic Generation microscopy and optical scattering. BMC Cancer 2017, 17, 102. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.L.; Brewer, M.A.; Nadiarnykh, O.; Hocker, J.; Singh, V.; Mackie, T.R.; Campagnola, P.J. Texture analysis applied to second harmonic generation image data for ovarian cancer classification. J. Biomed. Opt. 2014, 19, 096007. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Campbell, K.R.; Tilbury, K.; Nadiarnykh, O.; Brewer, M.A.; Patankar, M.; Singh, V.; Eliceiri, K.W.; Campagnola, P.J. 3D texture analysis for classification of second harmonic generation images of human ovarian cancer. Sci. Rep.-Uk 2016, 6, 35734. [Google Scholar] [CrossRef] [PubMed]
- Nadiarnykh, O.; Lacomb, R.B.; Brewer, M.A.; Campagnola, P.J. Alterations of the extracellular matrix in ovarian cancer studied by Second Harmonic Generation imaging microscopy. BMC Cancer 2010, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Capo-Chichi, C.D.; Smith, E.R.; Yang, D.H.; Roland, I.H.; Vanderveer, L.; Cohen, C.; Hamilton, T.C.; Godwin, A.K.; Xu, X.X. Dynamic alterations of the extracellular environment of ovarian surface epithelial cells in premalignant transformation, tumorigenicity, and metastasis. Cancer 2002, 95, 1802–1815. [Google Scholar] [CrossRef]
- Bast, R.C.; Boyer, C.M.; Jacobs, I.; Xu, F.J.; Wu, S.; Wiener, J.; Kohler, M.; Berchuck, A. Cell growth regulation in epithelial ovarian cancer. Cancer 1993, 71, 1597–1601. [Google Scholar] [CrossRef]
- Kruk, P.A.; Uitto, V.J.; Firth, J.D.; Dedhar, S.; Auersperg, N. Reciprocal interactions between human ovarian surface epithelial cells and adjacent extracellular matrix. Exp. Cell Res. 1994, 215, 97–108. [Google Scholar] [CrossRef]
- Carlson, J.W.; Jarboe, E.A.; Kindelberger, D.; Nucci, M.R.; Hirsch, M.S.; Crum, C.P. Serous tubal intraepithelial carcinoma: Diagnostic reproducibility and its implications. Int. J. Gynecol. Pathol. 2010, 29, 310–314. [Google Scholar] [CrossRef]
- Riching, K.M.; Cox, B.L.; Salick, M.R.; Pehlke, C.; Riching, A.S.; Ponik, S.M.; Bass, B.R.; Crone, W.C.; Jiang, Y.; Weaver, A.M.; et al. 3D Collagen Alignment Limits Protrusions to Enhance Breast Cancer Cell Persistence. Biophys. J. 2014, 107, 2546–2558. [Google Scholar] [CrossRef]
- Ajeti, V.; Nadiarnykh, O.; Ponik, S.M.; Keely, P.J.; Eliceiri, K.W.; Campagnola, P.J. Structural changes in mixed Col I/Col V collagen gels probed by SHG microscopy: Implications for probing stromal alterations in human breast cancer. Biomed. Opt. Express 2011, 2, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Casey, R.C.; Oegema, T.R., Jr.; Skubitz, K.M.; Pambuccian, S.E.; Grindle, S.M.; Skubitz, A.P. Cell membrane glycosylation mediates the adhesion, migration, and invasion of ovarian carcinoma cells. Clin. Exp. Metastasis 2003, 20, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Casey, R.C.; Skubitz, A.P. CD44 and beta1 integrins mediate ovarian carcinoma cell migration toward extracellular matrix proteins. Clin. Exp. Metastasis 2000, 18, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Bersini, S.; Miermont, A.; Pavesi, A.; Kamm, R.D.; Thiery, J.P.; Moretti, M.; Adriani, G. A combined microfluidic-transcriptomic approach to characterize the extravasation potential of cancer cells. Oncotarget 2018, 9, 36110–36125. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.K.; Pan, Y.; Nyberg, K.; Marra, M.A.; Lim, E.L.; Jones, S.J.; Maar, D.; Gibb, E.A.; Gunaratne, P.H.; Robertson, A.G.; et al. Tumour-suppressor microRNAs regulate ovarian cancer cell physical properties and invasive behaviour. Open Biol. 2016, 6, 160275. [Google Scholar] [CrossRef]
- Nakamura, O.; Kawata, S.; Maruo, S. Three-dimensional microfabrication with two-photon-absorbed photopolymerization. Opt. Lett. 1997, 22, 132–134. [Google Scholar] [CrossRef]
- Cumpston, B.H.; Ananthavel, S.P.; Barlow, S.; Dyer, D.L.; Ehrlich, J.E.; Erskine, L.L.; Heikal, A.A.; Kuebler, S.M.; Lee, I.-Y.S.; McCord-Maughon, D.; et al. Two-photon polymerization initiators for three-dimensional optical data storage and microfabrication. Nature 1999, 398, 51. [Google Scholar] [CrossRef]
- Pitts, J.D.; Campagnola, P.J.; Epling, G.A.; Goodman, S.L. Reaction efficiencies for sub-micron multi-photon freeform fabrications of proteins and polymers with applications in sustained release. Macromolecules 2000, 33, 1514–1523. [Google Scholar] [CrossRef]
- Maruo, S.; Fourkas, J.T. Recent progress in multiphoton microfabrication. Laser Photonics Rev. 2008, 2, 100–111. [Google Scholar] [CrossRef]
- Culver, J.C.; Hoffmann, J.C.; Poche, R.A.; Slater, J.H.; West, J.L.; Dickinson, M.E. Three-Dimensional Biomimetic Patterning in Hydrogels to Guide Cellular Organization. Adv. Mater. 2012, 24, 2344–2348. [Google Scholar] [CrossRef]
- Sridhar, M.; Basu, S.; Scranton, V.L.; Campagnola, P.J. Construction of a laser scanning microscope for multiphoton excited optical fabrication. Rev. Sci. Instrum. 2003, 74, 3474–3477. [Google Scholar] [CrossRef]
- Pins, G.D.; Bush, K.A.; Cunningham, L.P.; Campagnola, P.J. Multiphoton Excited Fabricated Nano and MicroPatterned Extracellular Matrix Proteins Direct Cellular Morphology. J. Biomed. Mater. Res. 2006, 78, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Su, Y.-D.; Ajeti, V.; Chen, S.J.; Campagnola, P.J. Cell adhesion on micro-structured fibronectin gradients fabricated by multiphoton excited photochemistry. Cell. Mol. Bioeng. 2012, 5, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Ajeti, V.; Lara-Santiago, J.; Alkmin, S.; Campagnola, P.J. Ovarian and Breast Cancer Migration Dynamics on Laminin and Fibronectin Bidirectional Gradient Fibers Fabricated via Multiphoton Excited Photochemistry. Cell. Mol. Bioeng. 2017, 10, 295–311. [Google Scholar] [CrossRef]
- Alkmin, S.; Brodziski, R.; Simon, H.; Hinton, D.; Goldsmith, R.H.; Patankar, M.; Campagnola, P.J. Migration dynamics of ovarian epithelial cells on micro-fabricated image-based models of normal and malignant stroma. Acta Biomater. 2019, 100, 92–104. [Google Scholar] [CrossRef]
- Ajeti, V.; Lien, C.H.; Chen, S.J.; Su, P.J.; Squirrell, J.M.; Molinarolo, K.H.; Lyons, G.E.; Eliceiri, K.W.; Ogle, B.M.; Campagnola, P.J. Image-inspired 3D multiphoton excited fabrication of extracellular matrix structures by modulated raster scanning. Opt. Express 2013, 21, 25346–25355. [Google Scholar] [CrossRef]
- Kojima, K.; Ito, M.; Morishita, H.; Hayashi, N. A novel water-soluble photoinitiator for the acrylic photopolymerization type resist system. Chem. Mater. 1998, 10, 3429–3433. [Google Scholar] [CrossRef]
- Allen, N.S. Photoinitiators for UV and visible curing of coatings: Mechanisms and properties. J. Photochem. Photobiol. A Chem. 1996, 100, 101–107. [Google Scholar] [CrossRef]
- Nichol, J.W.; Koshy, S.T.; Bae, H.; Hwang, C.M.; Yamanlar, S.; Khademhosseini, A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials 2010, 31, 5536–5544. [Google Scholar] [CrossRef]
- Klotz, B.J.; Gawlitta, D.; Rosenberg, A.; Malda, J.; Melchels, F.P.W. Gelatin-Methacryloyl Hydrogels: Towards Biofabrication-Based Tissue Repair. Trends Biotechnol. 2016, 34, 394–407. [Google Scholar] [CrossRef]
- Engelhardt, S.; Hoch, E.; Borchers, K.; Meyer, W.; Krüger, H.; Tovar, G.E.M.; Gillner, A. Fabrication of 2D protein microstructures and 3D polymer–protein hybrid microstructures by two-photon polymerization—IOPscience. Biofabrication 2011, 3, 025003. [Google Scholar] [CrossRef] [PubMed]
- Brewer, M.; Wharton, J.T.; Wang, J.; McWatters, A.; Auersperg, N.; Gershenson, D.; Bast, R.; Zou, C. In vitro model of normal, immortalized ovarian surface epithelial and ovarian cancer cells for chemoprevention of ovarian cancer. Gynecol. Oncol. 2005, 98, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Robinson, J.B.; Deguzman, A.; Bucana, C.D.; Fidler, I.J. Blockade of nuclear factor-kappaB signaling inhibits angiogenesis and tumorigenicity of human ovarian cancer cells by suppressing expression of vascular endothelial growth factor and interleukin 8. Cancer Res. 2000, 60, 5334–5339. [Google Scholar] [PubMed]
- Dickinson, R.B.; Tranquillo, R.T. A stochastic model for adhesion-mediated cell random motility and haptotaxis. J. Math. Biol. 1993, 31, 563–600. [Google Scholar] [CrossRef]
- Bredfeldt, J.S.; Liu, Y.; Conklin, M.W.; Keely, P.J.; Mackie, T.R.; Eliceiri, K.W. Automated quantification of aligned collagen for human breast carcinoma prognosis. J. Pathol. Inform. 2014, 5, 28. [Google Scholar] [CrossRef]
- He, R.-Y.; Ajeti, V.; Chen, S.-J.; Brewer, M.A.; Campagnola, P.J. Ovarian Cancer Cell Adhesion/Migration Dynamics on Micro-Structured Laminin Gradients Fabricated by Multiphoton Excited Photochemistry. Bioengineering 2015, 2, 139–159. [Google Scholar] [CrossRef]
- Chen, X.; Brewer, M.A.; Zou, C.; Campagnola, P.J. Adhesion and migration of ovarian cancer cells on crosslinked laminin fibers nanofabricated by multiphoton excited photochemistry. Integr. Biol. 2009, 1, 469–476. [Google Scholar] [CrossRef]
- Humphries, J.D.; Wang, P.; Streuli, C.; Geiger, B.; Humphries, M.J.; Ballestrem, C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell. Biol. 2007, 179, 1043–1057. [Google Scholar] [CrossRef]
- Atherton, P.; Stutchbury, B.; Jethwa, D.; Ballestrem, C. Mechanosensitive components of integrin adhesions: Role of vinculin. Exp. Cell Res. 2016, 343, 21–27. [Google Scholar] [CrossRef]
- Tisler, M.; Alkmin, S.; Chang, H.Y.; Leet, J.; Bernau, K.; Sandbo, N.; Campagnola, P.J. Analysis of fibroblast migration dynamics in idiopathic pulmonary fibrosis using image-based scaffolds of the lung extracellular matrix. Am. J. Physiol. Lung Cell. Mol. Physiol. 2019, 318, L276–L286. [Google Scholar] [CrossRef]
- Barbolina, M.V.; Moss, N.M.; Westfall, S.D.; Liu, Y.; Burkhalter, R.J.; Marga, F.; Forgacs, G.; Hudson, L.G.; Stack, M.S. Microenvironmental regulation of ovarian cancer metastasis. Cancer Treat. Res. 2009, 149, 319–334. [Google Scholar] [CrossRef]
- Patel, I.S.; Madan, P.; Getsios, S.; Bertrand, M.A.; MacCalman, C.D. Cadherin switching in ovarian cancer progression. Int. J. Cancer 2003, 106, 172–177. [Google Scholar] [CrossRef]
- Wu, C.; Cipollone, J.; Maines-Bandiera, S.; Tan, C.; Karsan, A.; Auersperg, N.; Roskelley, C.D. The morphogenic function of E-cadherin-mediated adherens junctions in epithelial ovarian carcinoma formation and progression. Differentiation 2008, 76, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef] [PubMed]
- Petrie, R.J.; Doyle, A.D.; Yamada, K.M. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Knittel, J.G.; Yan, L.; Rueden, C.T.; White, J.G.; Keely, P.J. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Sung, K.E.; Su, G.; Pehlke, C.; Trier, S.M.; Eliceiri, K.W.; Keely, P.J.; Friedl, A.; Beebe, D.J. Control of 3-dimensional collagen matrix polymerization for reproducible human mammary fibroblast cell culture in microfluidic devices. Biomaterials 2009, 30, 4833–4841. [Google Scholar] [CrossRef] [PubMed]
- Sung, K.E.; Yang, N.; Pehlke, C.; Keely, P.J.; Eliceiri, K.W.; Friedl, A.; Beebe, D.J. Transition to invasion in breast cancer: A microfluidic in vitro model enables examination of spatial and temporal effects. Integr. Biol. (Camb.) 2011, 3, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Carlson, J.W.; Miron, A.; Jarboe, E.A.; Parast, M.M.; Hirsch, M.S.; Lee, Y.; Muto, M.G.; Kindelberger, D.; Crum, C.P. Serous tubal intraepithelial carcinoma: Its potential role in primary peritoneal serous carcinoma and serous cancer prevention. J. Clin. Oncol. 2008, 26, 4160–4165. [Google Scholar] [CrossRef]
- Doyle, A.D.; Wang, F.W.; Matsumoto, K.; Yamada, K.M. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 2009, 184, 481–490. [Google Scholar] [CrossRef]
- Kubow, K.E.; Shuklis, V.D.; Sales, D.J.; Horwitz, A.R. Contact guidance persists under myosin inhibition due to the local alignment of adhesions and individual protrusions. Sci. Rep. 2017, 7, 14380. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Trier, S.M.; Keely, P.J. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys. J. 2008, 95, 5374–5384. [Google Scholar] [CrossRef]
- Chang, F.; Zhang, Y.; Mi, J.; Zhou, Q.; Bai, F.; Xu, X.; Fisher, D.E.; Sun, Q.; Wu, X. ROCK inhibitor enhances the growth and migration of BRAF-mutant skin melanoma cells. Cancer Sci. 2018, 109, 3428–3437. [Google Scholar] [CrossRef]
- Rodriguez-Hernandez, I.; Cantelli, G.; Bruce, F.; Sanz-Moreno, V. Rho, ROCK and actomyosin contractility in metastasis as drug targets. F1000Res 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.S.; Maines-Bandiera, S.L.; Rosen, B.; Wheelock, M.J.; Johnson, K.R.; Leung, P.C.; Roskelley, C.D.; Auersperg, N. Constitutive and conditional cadherin expression in cultured human ovarian surface epithelium: Influence of family history of ovarian cancer. Int. J. Cancer 1999, 81, 180–188. [Google Scholar] [CrossRef]
- Wheelock, M.J.; Shintani, Y.; Maeda, M.; Fukumoto, Y.; Johnson, K.R. Cadherin switching. J. Cell Sci. 2008, 121, 727–735. [Google Scholar] [CrossRef]
- Rosso, M.; Majem, B.; Devis, L.; Lapyckyj, L.; Besso, M.J.; Llaurado, M.; Abascal, M.F.; Matos, M.L.; Lanau, L.; Castellvi, J.; et al. E-cadherin: A determinant molecule associated with ovarian cancer progression, dissemination and aggressiveness. PLoS ONE 2017, 12, e0184439. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.B.; Phillips, G.R.; Qiao, R.F.; Norton, L.; Aaronson, S.A. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J. Cell Biol. 2000, 148, 779–790. [Google Scholar] [CrossRef]
- Islam, S.; Carey, T.E.; Wolf, G.T.; Wheelock, M.J.; Johnson, K.R. Expression of N-cadherin by human squamous carcinoma cells induces a scattered fibroblastic phenotype with disrupted cell-cell adhesion. J. Cell Biol. 1996, 135, 1643–1654. [Google Scholar] [CrossRef]
- Nieman, M.T.; Prudoff, R.S.; Johnson, K.R.; Wheelock, M.J. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J. Cell Biol. 1999, 147, 631–644. [Google Scholar] [CrossRef]
- Kim, D.-H.; Provenzano, P.P.; Smith, C.L.; Levchenko, A. Matrix nanotopography as a regulator of cell function. J. Cell Biol. 2012, 197, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Keely, P.J. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and Rho GTPase signaling. J. Cell Sci. 2011, 124, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Keely, P.J. The role of focal adhesion kinase in tumor initiation and progression. Cell Adhes. Migr. 2009, 3, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Wyckoff, J.; Philippar, U.; Segall, J.E.; Gertler, F.; Condeelis, J. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnol. 2005, 5, 14. [Google Scholar] [CrossRef]
- Sidani, M.; Wyckoff, J.; Xue, C.; Segall, J.E.; Condeelis, J. Probing the microenvironment of mammary tumors using multiphoton microscopy. J. Mammary Gland Biol. Neoplasia 2006, 11, 151–163. [Google Scholar] [CrossRef]
- Ray, A.; Morford, R.K.; Ghaderi, N.; Odde, D.J.; Provenzano, P.P. Dynamics of 3D carcinoma cell invasion into aligned collagen. Integr. Biol. (Camb.) 2018, 10, 100–112. [Google Scholar] [CrossRef]
- Ray, A.; Slama, Z.M.; Morford, R.K.; Madden, S.A.; Provenzano, P.P. Enhanced Directional Migration of Cancer Stem Cells in 3D Aligned Collagen Matrices. Biophys. J. 2017, 112, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Fraley, S.I.; Wu, P.H.; He, L.; Feng, Y.; Krisnamurthy, R.; Longmore, G.D.; Wirtz, D. Three-dimensional matrix fiber alignment modulates cell migration and MT1-MMP utility by spatially and temporally directing protrusions. Sci. Rep. 2015, 5, 14580. [Google Scholar] [CrossRef]
- Erdogan, B.; Ao, M.; White, L.M.; Means, A.L.; Brewer, B.M.; Yang, L.; Washington, M.K.; Shi, C.; Franco, O.E.; Weaver, A.M.; et al. Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J. Cell Biol. 2017, 216, 3799–3816. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Eliceiri, K.W.; Campbell, J.M.; Inman, D.R.; White, J.G.; Keely, P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006, 4, 38. [Google Scholar] [CrossRef]
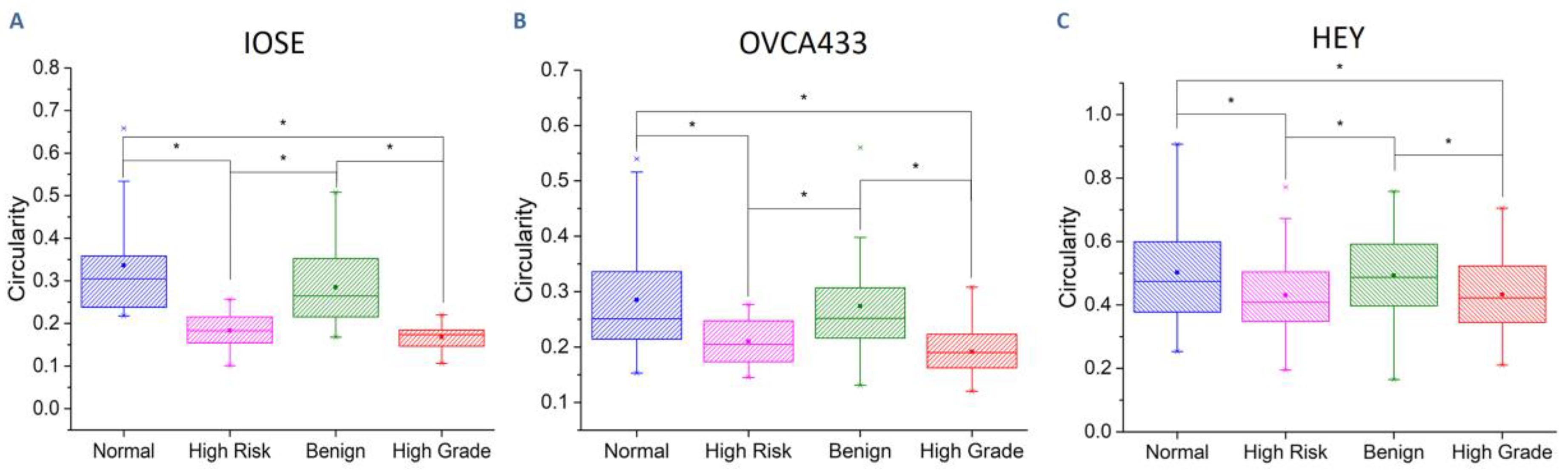

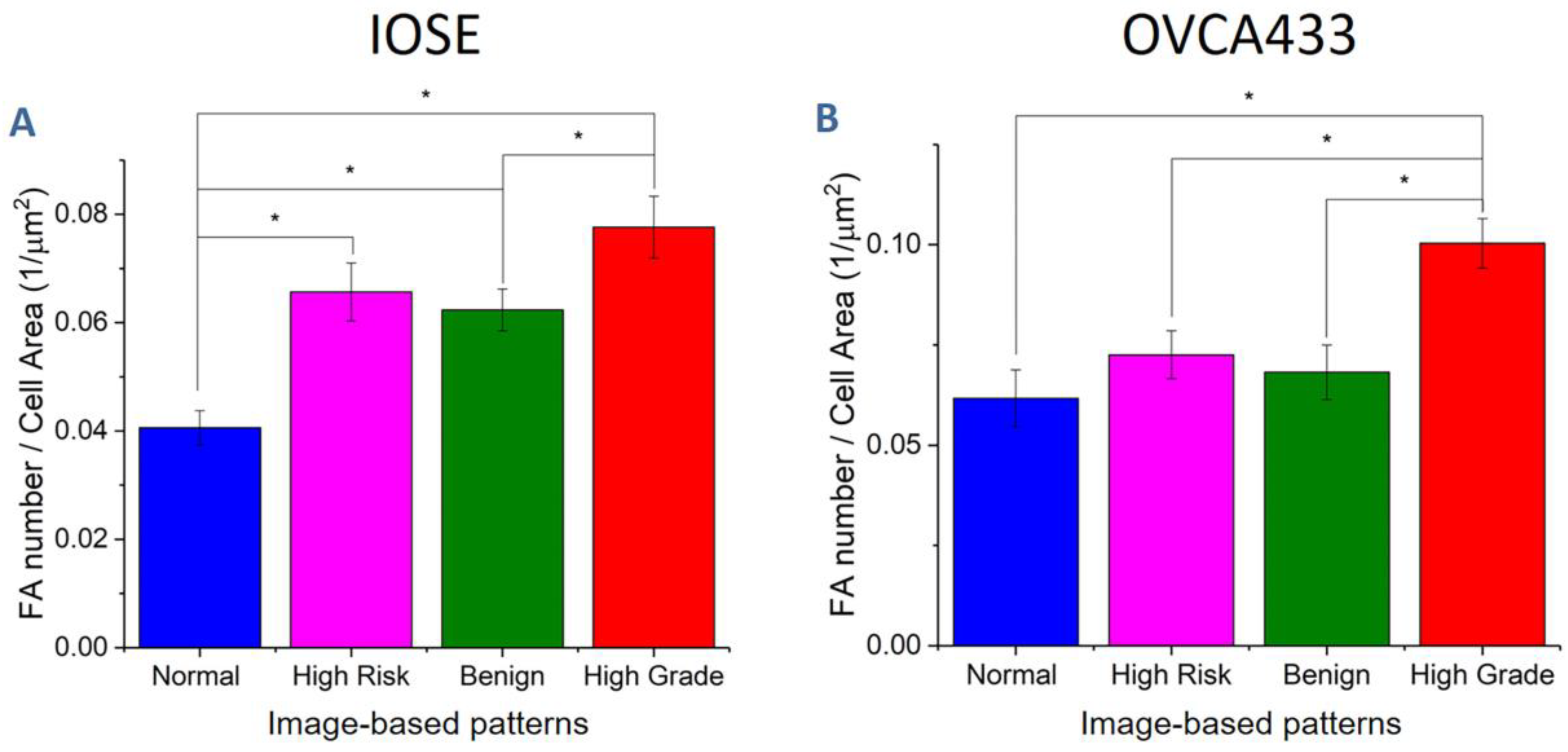
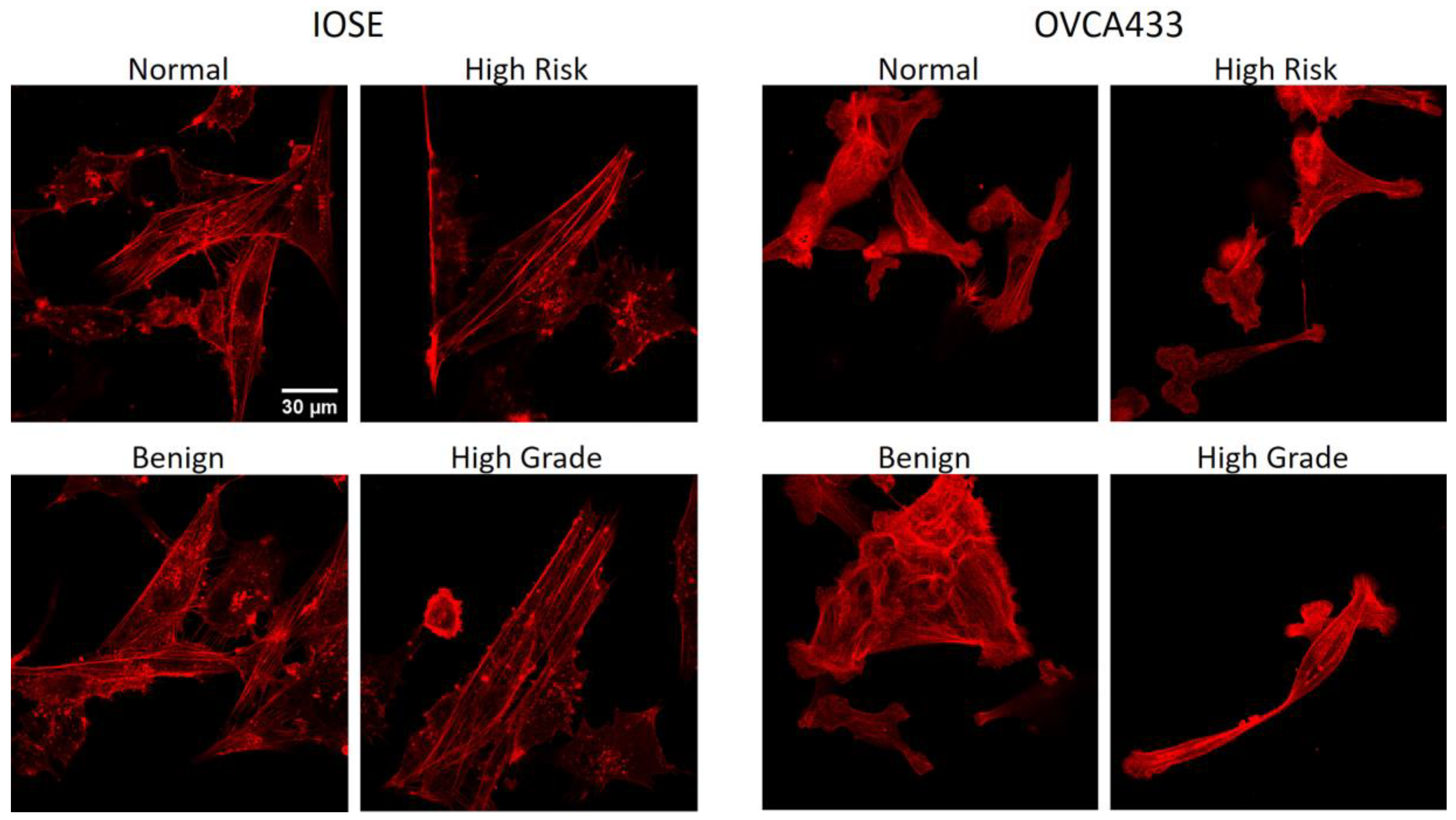
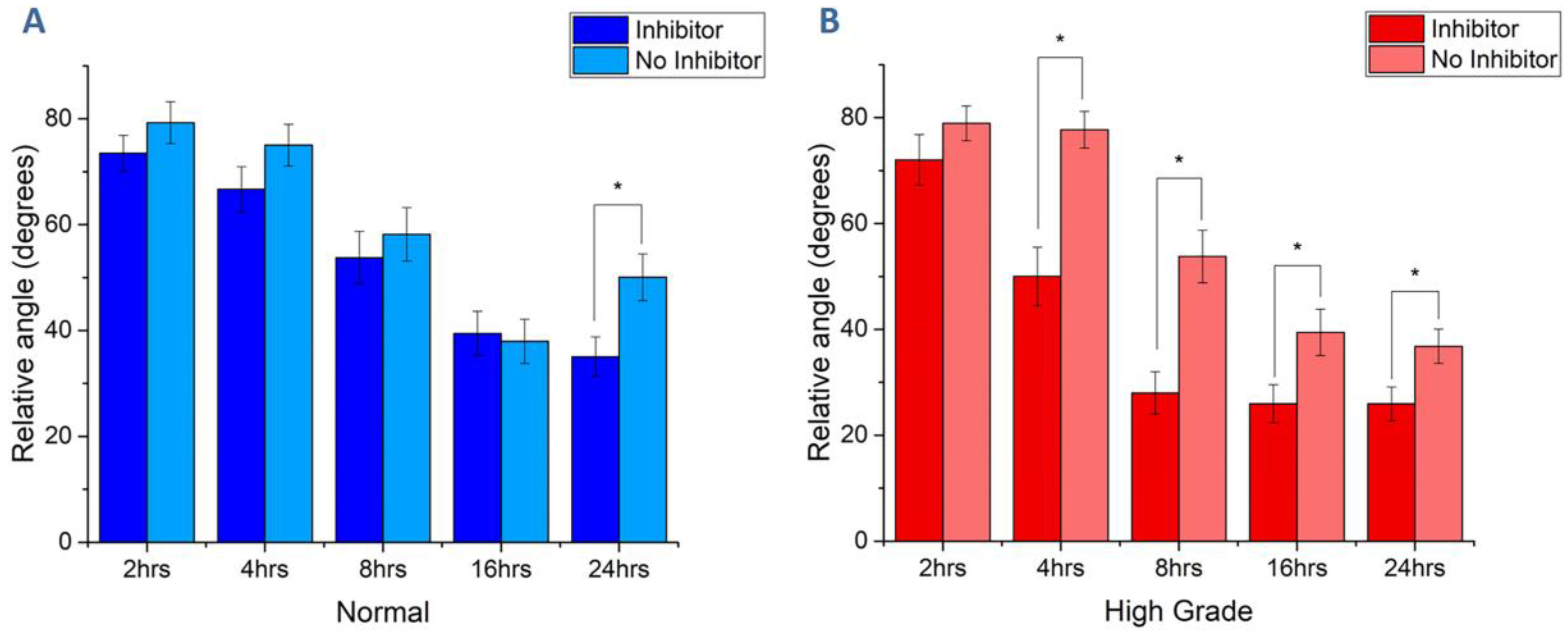
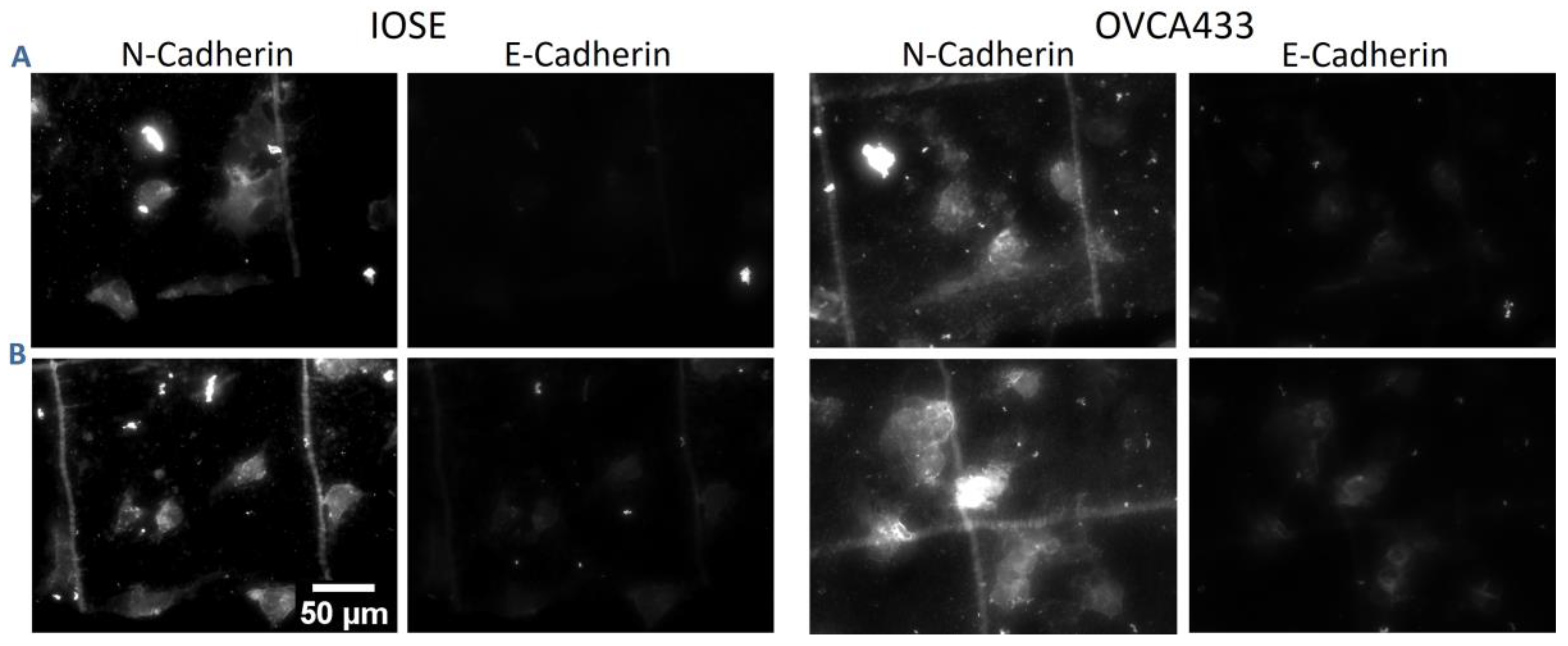
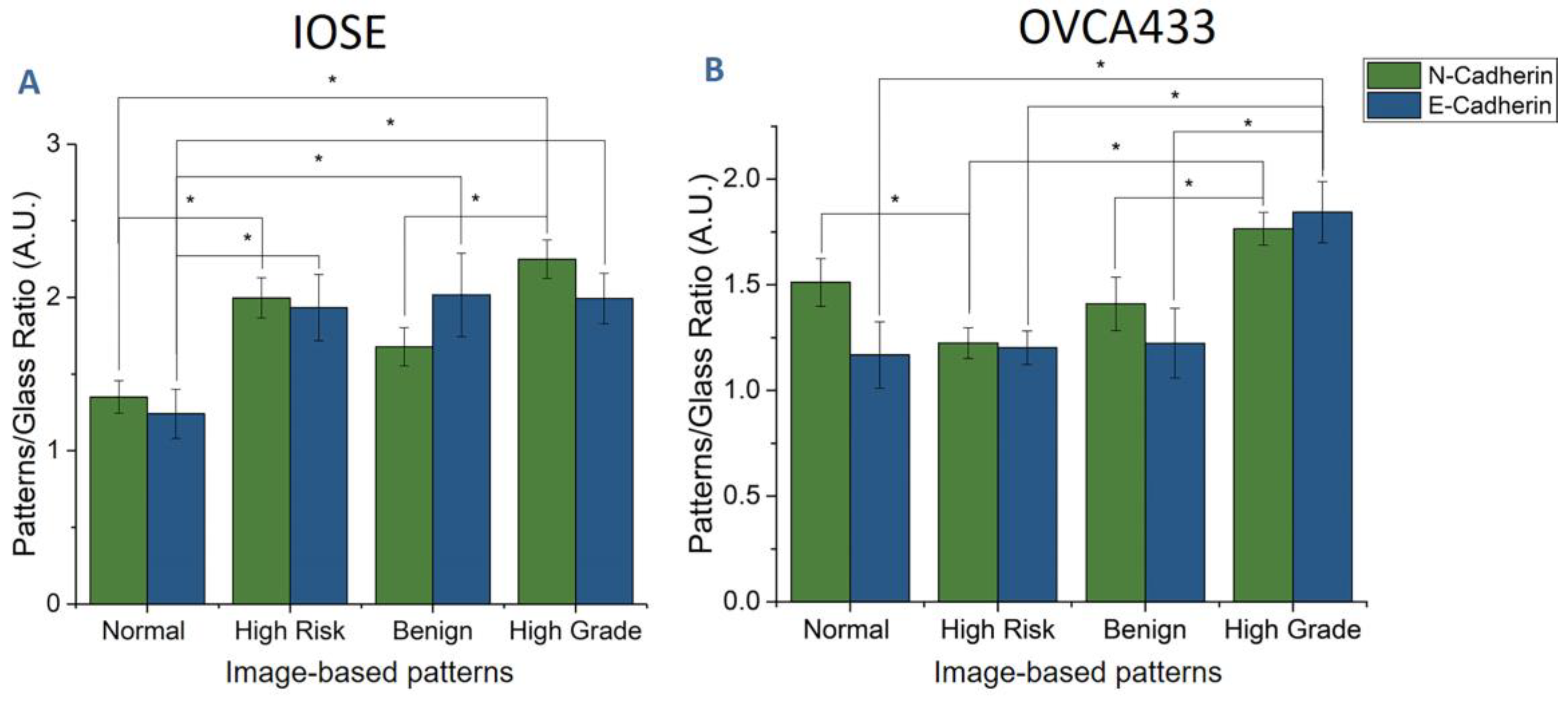
| Cell Line | Normal | High Risk | Benign | High Grade |
|---|---|---|---|---|
| IOSE | 0.37 | 0.83 | 0.32 | 0.97 |
| OVCA433 | 0.59 | 0.76 | 0.16 | 0.78 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkmin, S.; Brodziski, R.; Simon, H.; Hinton, D.; Goldsmith, R.H.; Patankar, M.; Campagnola, P.J. Role of Collagen Fiber Morphology on Ovarian Cancer Cell Migration Using Image-Based Models of the Extracellular Matrix. Cancers 2020, 12, 1390. https://doi.org/10.3390/cancers12061390
Alkmin S, Brodziski R, Simon H, Hinton D, Goldsmith RH, Patankar M, Campagnola PJ. Role of Collagen Fiber Morphology on Ovarian Cancer Cell Migration Using Image-Based Models of the Extracellular Matrix. Cancers. 2020; 12(6):1390. https://doi.org/10.3390/cancers12061390
Chicago/Turabian StyleAlkmin, Samuel, Rebecca Brodziski, Haleigh Simon, Daniel Hinton, Randall H. Goldsmith, Manish Patankar, and Paul J. Campagnola. 2020. "Role of Collagen Fiber Morphology on Ovarian Cancer Cell Migration Using Image-Based Models of the Extracellular Matrix" Cancers 12, no. 6: 1390. https://doi.org/10.3390/cancers12061390
APA StyleAlkmin, S., Brodziski, R., Simon, H., Hinton, D., Goldsmith, R. H., Patankar, M., & Campagnola, P. J. (2020). Role of Collagen Fiber Morphology on Ovarian Cancer Cell Migration Using Image-Based Models of the Extracellular Matrix. Cancers, 12(6), 1390. https://doi.org/10.3390/cancers12061390






