Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations
Abstract
1. Introduction
2. Regulatory Mechanisms of LncRNAs in Cancer Progression
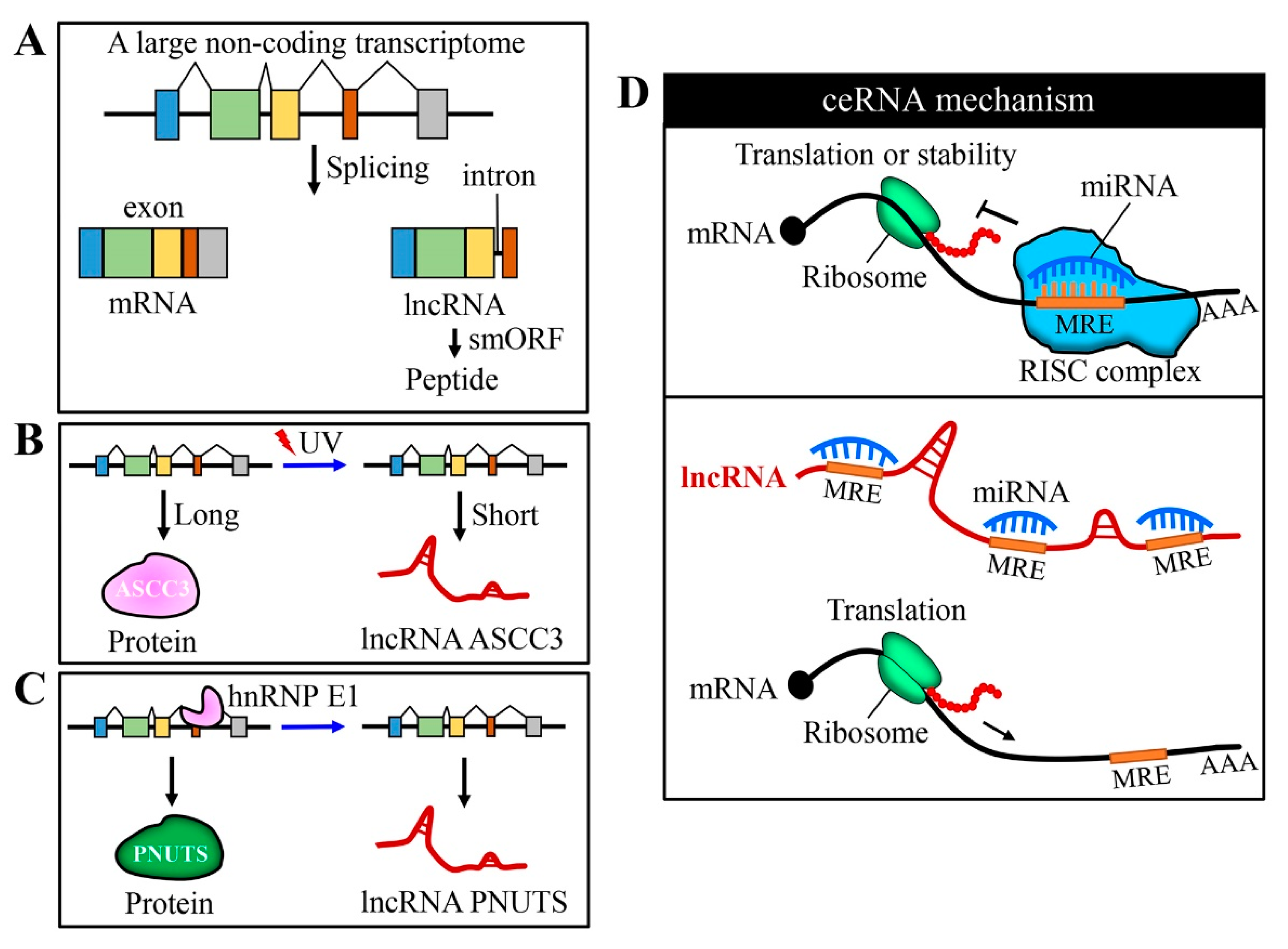
2.1. Competing Endogenous RNA (ceRNA) Mechanism
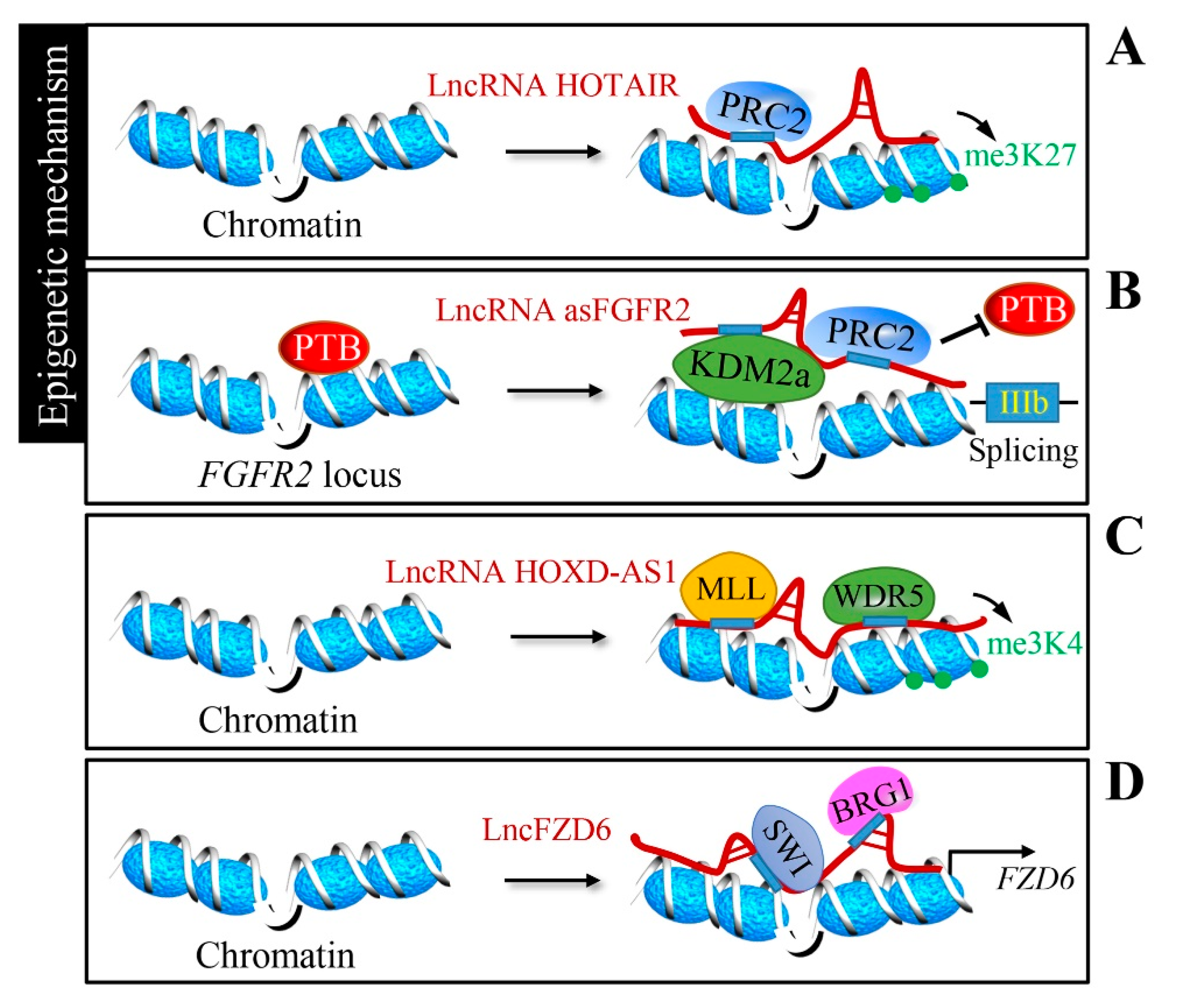
2.2. Epigenetic Regulation by lncRNAs
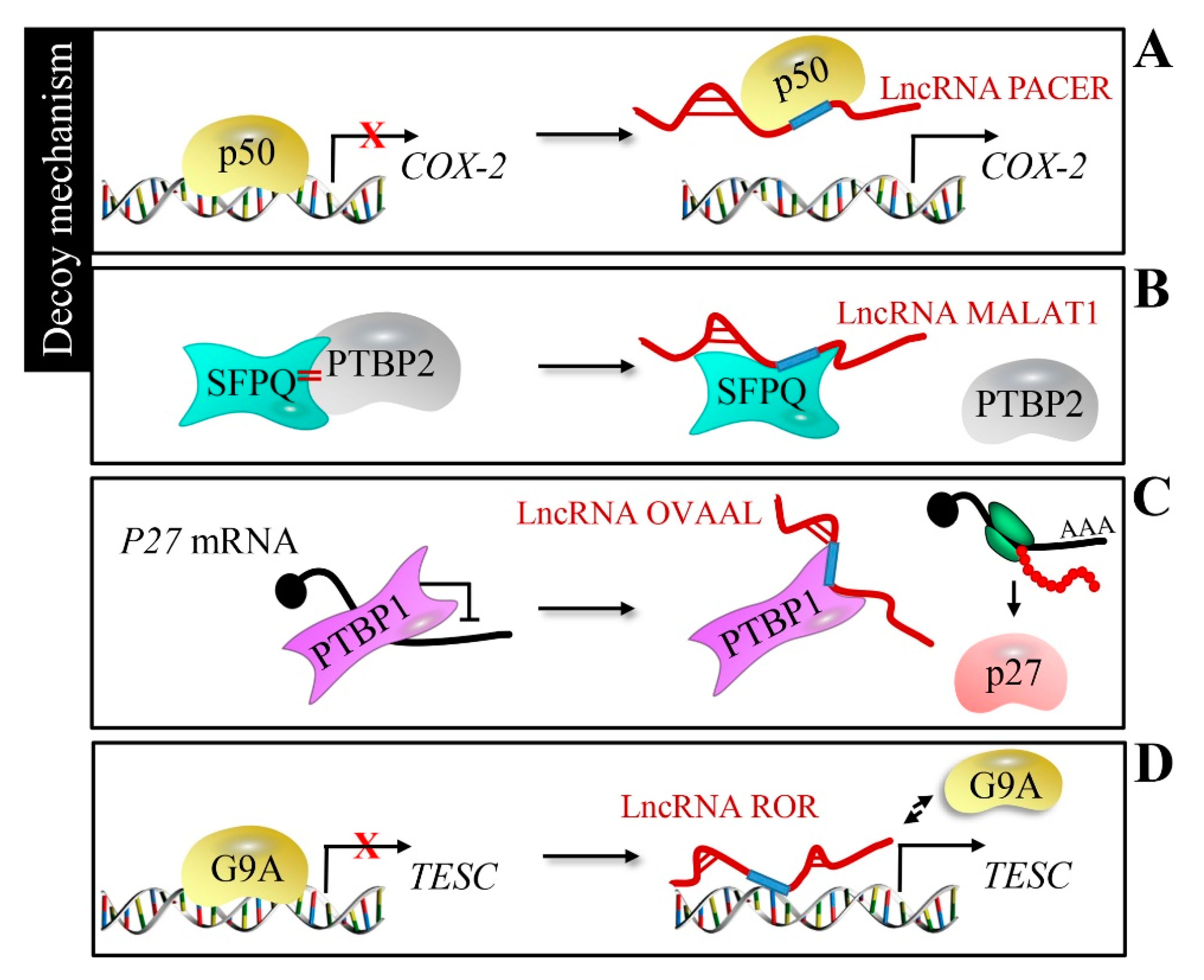
2.3. Decoy Mechanism
2.4. LncRNA Control of mRNA and Protein Stability
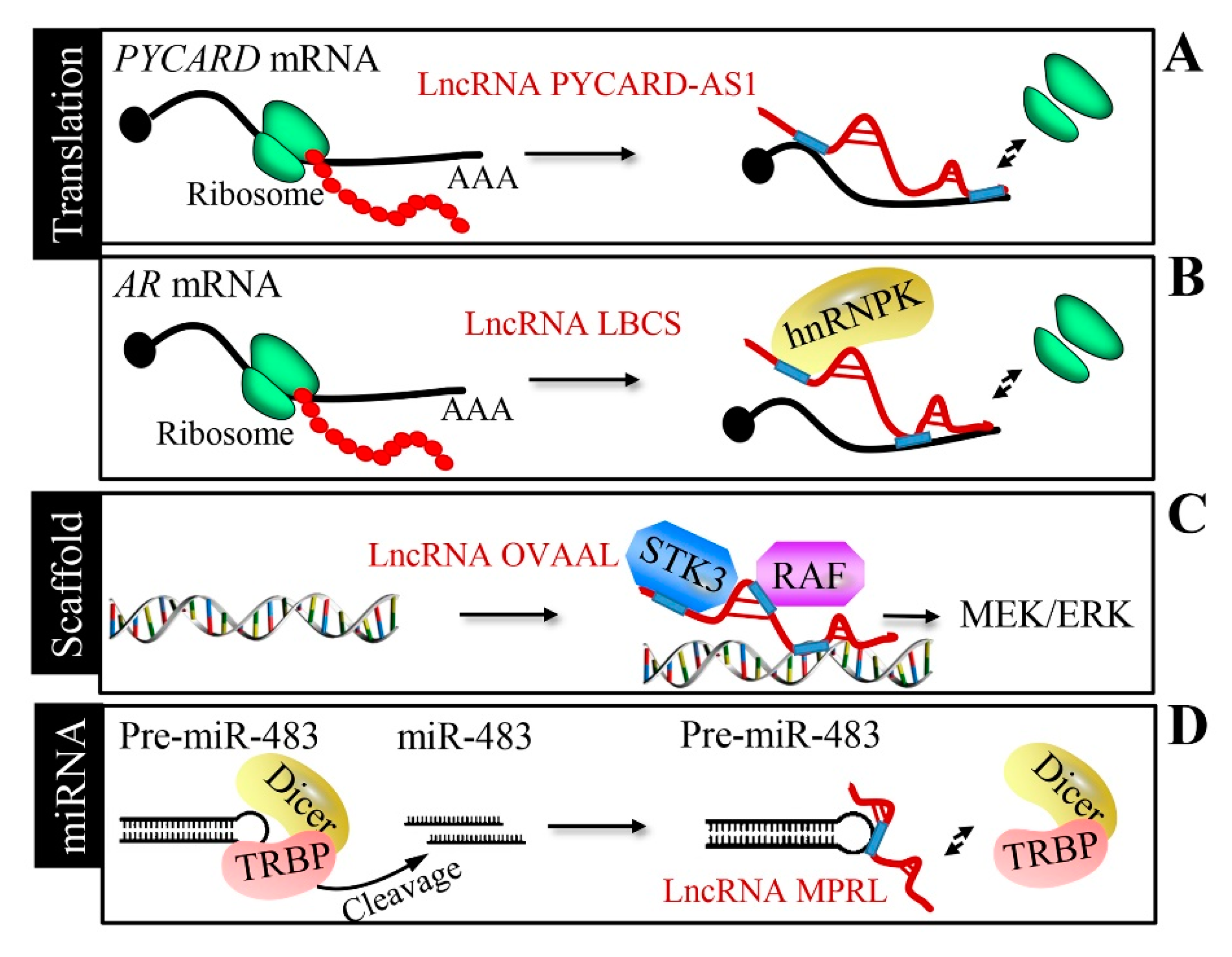
2.5. Transcriptional and Translational Regulation by LncRNAs
2.6. Scaffold of Transcription Factors by LncRNAs
2.7. miRNA Processing Mechanism
2.8. Architectural Role of LncRNAs
2.9. Other Mechanisms of LncRNA Function
3. Secondary Structure of LncRNAs
4. Dynamic Regulation of LncRNAs in Cancer Biology
4.1. Regulators of LncRNA Expression
4.2. LncRNA Turnover
4.3. Regulation of LncRNAs by Epitranscriptomics
5. Diverse Mechanisms of LncRNAs: An Example from Autophagy in Cancer
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Correction Statement
References
- Gottlieb, G. Normally occurring environmental and behavioral influences on gene activity: from central dogma to probabilistic epigenesis. Psychol. Rev. 1998, 105, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. Rna Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Song, Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013, 339, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Kondo, T.; Plaza, S.; Zanet, J.; Benrabah, E.; Valenti, P.; Hashimoto, Y.; Kobayashi, S.; Payre, F.; Kageyama, Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 2010, 329, 336–339. [Google Scholar] [CrossRef]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.A.; Alba, M.M. Long non-coding RNAs as a source of new peptides. eLife 2014, 3, e03523. [Google Scholar] [CrossRef]
- Huang, J.Z.; Chen, M.; Chen, D.; Gao, X.C.; Zhu, S.; Huang, H.; Hu, M.; Zhu, H.; Yan, G.R. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell 2017, 68, 171–184.e6. [Google Scholar] [CrossRef]
- Williamson, L.; Saponaro, M.; Boeing, S.; East, P.; Mitter, R.; Kantidakis, T.; Kelly, G.P.; Lobley, A.; Walker, J.; Spencer-Dene, B.; et al. UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell 2017, 168, 843–855.e813. [Google Scholar] [CrossRef] [PubMed]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Ramus, S.J.; Tyrer, J.; Bolton, K.L.; Gentry-Maharaj, A.; Wozniak, E.; Anton-Culver, H.; Chang-Claude, J.; Cramer, D.W.; DiCioccio, R.; et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat. Genet. 2009, 41, 996–1000. [Google Scholar] [CrossRef] [PubMed]
- Bolton, K.L.; Tyrer, J.; Song, H.; Ramus, S.J.; Notaridou, M.; Jones, C.; Sher, T.; Gentry-Maharaj, A.; Wozniak, E.; Tsai, Y.Y.; et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet. 2010, 42, 880–884. [Google Scholar] [CrossRef]
- Pharoah, P.D.; Tsai, Y.Y.; Ramus, S.J.; Phelan, C.M.; Goode, E.L.; Lawrenson, K.; Buckley, M.; Fridley, B.L.; Tyrer, J.P.; Shen, H.; et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 2013, 45, 362–370. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; Ramus, S.J.; Tyrer, J.; Lee, A.; Shen, H.C.; Beesley, J.; Lawrenson, K.; McGuffog, L.; Healey, S.; Lee, J.M.; et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet. 2015, 47, 164–171. [Google Scholar] [CrossRef]
- Kelemen, L.E.; Lawrenson, K.; Tyrer, J.; Li, Q.; Lee, J.M.; Seo, J.H.; Phelan, C.M.; Beesley, J.; Chen, X.; Spindler, T.J.; et al. Genome-wide significant risk associations for mucinous ovarian carcinoma. Nat. Genet. 2015, 47, 888–897. [Google Scholar] [CrossRef]
- Phelan, C.M.; Kuchenbaecker, K.B.; Tyrer, J.P.; Kar, S.P.; Lawrenson, K.; Winham, S.J.; Dennis, J.; Pirie, A.; Riggan, M.J.; Chornokur, G.; et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017, 49, 680–691. [Google Scholar] [CrossRef]
- Lu, Y.; Beeghly-Fadiel, A.; Wu, L.; Guo, X.; Li, B.; Schildkraut, J.M.; Im, H.K.; Chen, Y.A.; Permuth, J.B.; Reid, B.M.; et al. A transcriptome-wide association study among 97,898 women to identify candidate susceptibility genes for epithelial ovarian cancer risk. Cancer Res. 2018, 78, 5419–5430. [Google Scholar] [CrossRef]
- Guo, H.; Ahmed, M.; Zhang, F.; Yao, C.Q.; Li, S.; Liang, Y.; Hua, J.; Soares, F.; Sun, Y.; Langstein, J.; et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 2016, 48, 1142–1150. [Google Scholar] [CrossRef]
- Betts, J.A.; Moradi Marjaneh, M.; Al-Ejeh, F.; Lim, Y.C.; Shi, W.; Sivakumaran, H.; Tropee, R.; Patch, A.M.; Clark, M.B.; Bartonicek, N.; et al. Long noncoding RNAs CUPID1 and CUPID2 mediate breast cancer risk at 11q13 by modulating the response to DNA damage. Am. J. Hum. Genet. 2017, 101, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Akhade, V.S.; Kosalai, S.T.; Subhash, S.; Statello, L.; Meryet-Figuiere, M.; Abrahamsson, J.; Mondal, T.; Kanduri, C. PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nat. Commun. 2018, 9, 883. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Volinia, S.; Papaioannou, D.; Nicolet, D.; Kohlschmidt, J.; Yan, P.S.; Mrozek, K.; Bucci, D.; Carroll, A.J.; Baer, M.R.; et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2014, 111, 18679–18684. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z.; Xu, J.; Li, J.; Li, S.; Zhang, M.; Yang, D. Systematic identification of non-coding pharmacogenomic landscape in cancer. Nat. Commun. 2018, 9, 3192. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef]
- Lu, W.; Zhang, H.; Niu, Y.; Wu, Y.; Sun, W.; Li, H.; Kong, J.; Ding, K.; Shen, H.M.; Wu, H.; et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol. Cancer 2017, 16, 118. [Google Scholar] [CrossRef]
- Zhu, K.P.; Ma, X.L.; Zhang, C.L. LncRNA ODRUL Contributes to Osteosarcoma Progression through the miR-3182/MMP2 Axis. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 2383–2393. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Z.; Yu, S.; Nie, F.; Yan, S.; Ma, P.; Chen, Q.; Wei, C.; Fu, H.; Xu, T.; et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 2002–2014. [Google Scholar] [CrossRef]
- Zhang, G.; Li, S.; Lu, J.; Ge, Y.; Wang, Q.; Ma, G.; Zhao, Q.; Wu, D.; Gong, W.; Du, M.; et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol. Cancer 2018, 17, 87. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Chen, J.J.; Lv, Q.; Qin, J.; Huang, Y.Z.; Yu, M.H.; Zhong, M. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/beta-catenin signaling pathway. Cancer Lett. 2019, 440–441, 11–22. [Google Scholar] [CrossRef]
- Li, W.; Ma, S.; Bai, X.; Pan, W.; Ai, L.; Tan, W. Long noncoding RNA WDFY3-AS2 suppresses tumor progression by acting as a competing endogenous RNA of microRNA-18a in ovarian cancer. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zanet, J.; Benrabah, E.; Li, T.; Pelissier-Monier, A.; Chanut-Delalande, H.; Ronsin, B.; Bellen, H.J.; Payre, F.; Plaza, S. Pri sORF peptides induce selective proteasome-mediated protein processing. Science 2015, 349, 1356–1358. [Google Scholar] [CrossRef] [PubMed]
- Bazin, J.; Baerenfaller, K.; Gosai, S.J.; Gregory, B.D.; Crespi, M.; Bailey-Serres, J. Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc. Natl. Acad. Sci. USA 2017, 114, E10018–E10027. [Google Scholar] [CrossRef]
- Pan, J.; Meng, X.; Jiang, N.; Jin, X.; Zhou, C.; Xu, D.; Gong, Z. Insights into the Noncoding RNA-encoded Peptides. Protein Pept. Lett. 2018, 25, 720–727. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, S.; Meng, N.; He, Y.; Lu, R.; Yan, G.R. ncRNA-Encoded Peptides or Proteins and Cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2019, 27, 1718–1725. [Google Scholar] [CrossRef]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Yuan, Y.; Liu, B.; Xie, P.; Zhang, M.Q.; Li, Y.D.; Xie, Z.; Wang, X.W. Model-guided quantitative analysis of microRNA-mediated regulation on competing endogenous RNAs using a synthetic gene circuit. Proc. Natl. Acad. Sci. USA 2015, 112, 3158–3163. [Google Scholar] [CrossRef]
- Aguilo, F.; Zhou, M.M.; Walsh, M.J. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011, 71, 5365–5369. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef]
- Yap, K.L.; Li, S.; Munoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Kong, R.; Zhang, E.B.; Yin, D.D.; You, L.H.; Xu, T.P.; Chen, W.M.; Xia, R.; Wan, L.; Sun, M.; Wang, Z.X.; et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol. Cancer 2015, 14, 82. [Google Scholar] [CrossRef]
- Marin-Bejar, O.; Mas, A.M.; Gonzalez, J.; Martinez, D.; Athie, A.; Morales, X.; Galduroz, M.; Raimondi, I.; Grossi, E.; Guo, S.; et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017, 18, 202. [Google Scholar] [CrossRef]
- Gonzalez, I.; Munita, R.; Agirre, E.; Dittmer, T.A.; Gysling, K.; Misteli, T.; Luco, R.F. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 2015, 22, 370–376. [Google Scholar] [CrossRef]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef]
- Gu, P.; Chen, X.; Xie, R.; Han, J.; Xie, W.; Wang, B.; Dong, W.; Chen, C.; Yang, M.; Jiang, J.; et al. lncRNA HOXD-AS1 regulates proliferation and chemo-resistance of castration-resistant prostate cancer via recruiting WDR5. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 1959–1973. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Zhang, Y.; Weng, M.; Hu, Y.; Xuan, Y.; Hu, Y.; Lv, K. lncRNA GCAWKR promotes gastric cancer development by scaffolding the chromatin modification factors WDR5 and KAT2A. Mol. Ther. J. Am. Soc. Gene Ther. 2018, 26, 2658–2668. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhang, N.; Liu, W.; Liu, J.; Zhou, L.; Liu, Y.; Yang, M. The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J. Biol. Chem. 2018, 293, 17154–17165. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, Y.; Yao, L.; Liu, Y.; Huang, L.; Yan, Z.; Zhao, W.; Zhu, P.; Weng, H. LncFZD6 initiates Wnt/beta-catenin and liver TIC self-renewal through BRG1-mediated FZD6 transcriptional activation. Oncogene 2018, 37, 3098–3112. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Jiang, D.M.; Hu, S.S.; Zhao, L.; Wang, L.; Yang, M.H.; Ai, M.L.; Jiang, H.J.; Han, Y.; Ding, Y.Q.; et al. SATB2-AS1 suppresses colorectal carcinoma aggressiveness by inhibiting SATB2-dependent Snail transcription and epithelial-mesenchymal transition. Cancer Res. 2019, 79, 3542–3556. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, P.; Luo, J.; Wang, J.; Liu, Z.; Wu, W.; Du, Y.; Ye, B.; Wang, D.; He, L.; et al. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019, 38, e101110. [Google Scholar] [CrossRef]
- Shao, M.; Yang, Q.; Zhu, W.; Jin, H.; Wang, J.; Song, J.; Kong, Y.; Lv, X. LncHOXA10 drives liver TICs self-renewal and tumorigenesis via HOXA10 transcription activation. Mol. Cancer 2018, 17, 173. [Google Scholar] [CrossRef]
- Miao, H.; Wang, L.; Zhan, H.; Dai, J.; Chang, Y.; Wu, F.; Liu, T.; Liu, Z.; Gao, C.; Li, L.; et al. A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 2019, 15, e1008144. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Yang, Y.W.; Flynn, R.A.; Chen, Y.; Qu, K.; Wan, B.; Wang, K.C.; Lei, M.; Chang, H.Y. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife 2014, 3, e02046. [Google Scholar] [CrossRef]
- Krawczyk, M.; Emerson, B.M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. eLife 2014, 3, e01776. [Google Scholar] [CrossRef]
- Ji, Q.; Zhang, L.; Liu, X.; Zhou, L.; Wang, W.; Han, Z.; Sui, H.; Tang, Y.; Wang, Y.; Liu, N.; et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br. J. Cancer 2014, 111, 736–748. [Google Scholar] [CrossRef]
- Sang, B.; Zhang, Y.Y.; Guo, S.T.; Kong, L.F.; Cheng, Q.; Liu, G.Z.; Thorne, R.F.; Zhang, X.D.; Jin, L.; Wu, M. Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc. Natl. Acad. Sci. USA 2018, 115, E11661–E11670. [Google Scholar] [CrossRef]
- Fan, J.; Xing, Y.; Wen, X.; Jia, R.; Ni, H.; He, J.; Ding, X.; Pan, H.; Qian, G.; Ge, S.; et al. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015, 16, 139. [Google Scholar] [CrossRef]
- Cheadle, C.; Fan, J.; Cho-Chung, Y.S.; Werner, T.; Ray, J.; Do, L.; Gorospe, M.; Becker, K.G. Stability regulation of mRNA and the control of gene expression. Ann. New York Acad. Sci. 2005, 1058, 196–204. [Google Scholar] [CrossRef]
- Kumar, P.P.; Emechebe, U.; Smith, R.; Franklin, S.; Moore, B.; Yandell, M.; Lessnick, S.L.; Moon, A.M. Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. eLife 2014, 3. [Google Scholar] [CrossRef]
- Jadaliha, M.; Gholamalamdari, O.; Tang, W.; Zhang, Y.; Petracovici, A.; Hao, Q.; Tariq, A.; Kim, T.G.; Holton, S.E.; Singh, D.K.; et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 2018, 14, e1007802. [Google Scholar] [CrossRef]
- Tseng, Y.Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef]
- Lin, A.; Li, C.; Xing, Z.; Hu, Q.; Liang, K.; Han, L.; Wang, C.; Hawke, D.H.; Wang, S.; Zhang, Y.; et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat. Cell Biol. 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Li, Z.; Hou, P.; Fan, D.; Dong, M.; Ma, M.; Li, H.; Yao, R.; Li, Y.; Wang, G.; Geng, P.; et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017, 24, 59–71. [Google Scholar] [CrossRef]
- Ni, W.; Yao, S.; Zhou, Y.; Liu, Y.; Huang, P.; Zhou, A.; Liu, J.; Che, L.; Li, J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer 2019, 18, 143. [Google Scholar] [CrossRef]
- Gu, P.; Chen, X.; Xie, R.; Xie, W.; Huang, L.; Dong, W.; Han, J.; Liu, X.; Shen, J.; Huang, J.; et al. A novel AR translational regulator lncRNA LBCS inhibits castration resistance of prostate cancer. Mol. Cancer 2019, 18, 109. [Google Scholar] [CrossRef]
- Saayman, S.M.; Ackley, A.; Burdach, J.; Clemson, M.; Gruenert, D.C.; Tachikawa, K.; Chivukula, P.; Weinberg, M.S.; Morris, K.V. Long non-coding RNA BGas regulates the cystic fibrosis transmembrane conductance regulator. Mol. Ther. J. Am. Soc. Gene Ther. 2016, 24, 1351–1357. [Google Scholar] [CrossRef]
- Di Cecilia, S.; Zhang, F.; Sancho, A.; Li, S.; Aguilo, F.; Sun, Y.; Rengasamy, M.; Zhang, W.; Del Vecchio, L.; Salvatore, F.; et al. RBM5-AS1 is critical for self-renewal of colon cancer stem-like cells. Cancer Res. 2016, 76, 5615–5627. [Google Scholar] [CrossRef]
- Lingadahalli, S.; Jadhao, S.; Sung, Y.Y.; Chen, M.; Hu, L.; Chen, X.; Cheung, E. Novel lncRNA LINC00844 regulates prostate cancer cell migration and invasion through AR signaling. Mol. Cancer Res. Mcr 2018, 16, 1865–1878. [Google Scholar] [CrossRef]
- Wang, K.; Sun, T.; Li, N.; Wang, Y.; Wang, J.X.; Zhou, L.Y.; Long, B.; Liu, C.Y.; Liu, F.; Li, P.F. MDRL lncRNA regulates the processing of miR-484 primary transcript by targeting miR-361. PLoS Genet. 2014, 10, e1004467. [Google Scholar] [CrossRef]
- Tian, T.; Lv, X.; Pan, G.; Lu, Y.; Chen, W.; He, W.; Lei, X.; Zhang, H.; Liu, M.; Sun, S.; et al. Long Noncoding RNA MPRL promotes mitochondrial fission and cisplatin chemosensitivity via disruption of pre-miRNA processing. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 3673–3688. [Google Scholar] [CrossRef]
- Chen, L.L.; Carmichael, G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 2009, 35, 467–478. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Sasaki, Y.T.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef]
- Choudhry, H.; Albukhari, A.; Morotti, M.; Haider, S.; Moralli, D.; Smythies, J.; Schodel, J.; Green, C.M.; Camps, C.; Buffa, F.; et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 2015, 34, 4482–4490. [Google Scholar] [CrossRef]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.; Radaelli, E.; Vermi, W.; et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, Q.; Wang, Q.; Shen, Q.; Chen, X.; Li, Z.; Zhou, Y.; Hou, J.; Xu, B.; Li, N.; et al. NEAT1 paraspeckle promotes human hepatocellular carcinoma progression by strengthening IL-6/STAT3 signaling. Oncoimmunology 2018, 7, e1503913. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, S.B.; Wang, M.R.; Yao, R.W.; Wu, D.; Yang, L.; Chen, L.L. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 2018, 20, 1145–1158. [Google Scholar] [CrossRef]
- Cho, S.W.; Xu, J.; Sun, R.; Mumbach, M.R.; Carter, A.C.; Chen, Y.G.; Yost, K.E.; Kim, J.; He, J.; Nevins, S.A.; et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 2018, 173, 1398–1412.e22. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.S.; Sullivan, M.A.; Shah, R.N.; Nadadur, R.D.; Grzybowski, A.T.; Galat, V.; Moskowitz, I.P.; Ruthenburg, A.J. Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat. Struct. Mol. Biol. 2017, 24, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.A.; Babbs, B.; Cochrane, D.R.; Bitler, B.G.; Richer, J.K. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol. Carcinog. 2019, 58, 196–205. [Google Scholar] [CrossRef]
- Prensner, J.R.; Chen, W.; Iyer, M.K.; Cao, Q.; Ma, T.; Han, S.; Sahu, A.; Malik, R.; Wilder-Romans, K.; Navone, N.; et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014, 74, 1651–1660. [Google Scholar] [CrossRef]
- Somarowthu, S.; Legiewicz, M.; Chillón, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 2015, 58, 353–361. [Google Scholar] [CrossRef]
- Fang, R.; Moss, W.N.; Rutenberg-Schoenberg, M.; Simon, M.D. Probing Xist RNA structure in cells using targeted structure-seq. PLoS Genet. 2015, 11, e100566858. [Google Scholar] [CrossRef]
- Sztuba-Solinska, J.; Rausch, J.W.; Smith, R.; Miller, J.T.; Whitby, D.; Le Grice, S.F.J. Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA: a structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res. 2017, 45, 6805–6821. [Google Scholar] [CrossRef] [PubMed]
- McCown, P.J.; Wang, M.C.; Jaeger, L.; Brown, J.A. Secondary structural model of human MALAT1 reveals multiple structure-function relationships. Int. J. Mol. Sci. 2019, 20, 5610. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Somarowthu, S.; Pyle, A.M. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 2017, 13, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Zapien, D.; Legrand, P.; McEwen, A.G.; Proux, F.; Cragnolini, T.; Pasquali, S.; Dock-Bregeon, A.C. The crystal structure of the 5 functional domain of the transcription riboregulator 7SK. Nucleic Acids Res. 2017, 45, 3568–3579. [Google Scholar] [CrossRef]
- Eichhorn, C.D.; Yang, Y.; Repeta, L.; Feigon, J. Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7. Proc. Natl. Acad. Sci. USA 2018, 115, E6457–E6466. [Google Scholar] [CrossRef] [PubMed]
- Sas-Chen, A.; Aure, M.R.; Leibovich, L.; Carvalho, S.; Enuka, Y.; Korner, C.; Polycarpou-Schwarz, M.; Lavi, S.; Nevo, N.; Kuznetsov, Y.; et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO Mol. Med. 2016, 8, 1052–1064. [Google Scholar] [CrossRef]
- Bierhoff, H.; Dammert, M.A.; Brocks, D.; Dambacher, S.; Schotta, G.; Grummt, I. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell 2014, 54, 675–682. [Google Scholar] [CrossRef]
- Takayama, K.; Horie-Inoue, K.; Katayama, S.; Suzuki, T.; Tsutsumi, S.; Ikeda, K.; Urano, T.; Fujimura, T.; Takagi, K.; Takahashi, S.; et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013, 32, 1665–1680. [Google Scholar] [CrossRef]
- Ding, J.; Yeh, C.R.; Sun, Y.; Lin, C.; Chou, J.; Ou, Z.; Chang, C.; Qi, J.; Yeh, S. Estrogen receptor beta promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene 2018, 37, 5037–5053. [Google Scholar] [CrossRef]
- Niknafs, Y.S.; Han, S.; Ma, T.; Speers, C.; Zhang, C.; Wilder-Romans, K.; Iyer, M.K.; Pitchiaya, S.; Malik, R.; Hosono, Y.; et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat. Commun. 2016, 7, 12791. [Google Scholar] [CrossRef]
- Michelini, F.; Pitchiaya, S.; Vitelli, V.; Sharma, S.; Gioia, U.; Pessina, F.; Cabrini, M.; Wang, Y.; Capozzo, I.; Iannelli, F.; et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017, 19, 1400–1411. [Google Scholar] [CrossRef]
- Kim, T.; Jeon, Y.J.; Cui, R.; Lee, J.H.; Peng, Y.; Kim, S.H.; Tili, E.; Alder, H.; Croce, C.M. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef]
- Zhang, E.; He, X.; Zhang, C.; Su, J.; Lu, X.; Si, X.; Chen, J.; Yin, D.; Han, L.; De, W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018, 19, 154. [Google Scholar] [CrossRef]
- Geisler, S.; Lojek, L.; Khalil, A.M.; Baker, K.E.; Coller, J. Decapping of long noncoding RNAs regulates inducible genes. Mol. Cell 2012, 45, 279–291. [Google Scholar] [CrossRef]
- Ramaiah, M.; Shum, E.Y.; Wilkinson, M.F. How to activate a gene: decap its associated noncoding RNA. Mol. Cell 2012, 45, 271–273. [Google Scholar] [CrossRef]
- Beaulieu, Y.B.; Kleinman, C.L.; Landry-Voyer, A.M.; Majewski, J.; Bachand, F. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 2012, 8, e1003078. [Google Scholar] [CrossRef]
- Shafik, A.; Schumann, U.; Evers, M.; Sibbritt, T.; Preiss, T. The emerging epitranscriptomics of long noncoding RNAs. Biochim. Et Biophys. Acta 2016, 1859, 59–70. [Google Scholar] [CrossRef]
- Amort, T.; Souliere, M.F.; Wille, A.; Jia, X.Y.; Fiegl, H.; Worle, H.; Micura, R.; Lusser, A. Long non-coding RNAs as targets for cytosine methylation. Rna Biol. 2013, 10, 1003–1008. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Meng, J.; Rao, M.K.; Chen, Y.; Huang, Y. HEPeak: an HMM-based exome peak-finding package for RNA epigenome sequencing data. Bmc Genom. 2015, 16 (Suppl. S4), S2. [Google Scholar] [CrossRef]
- Liu, N.; Parisien, M.; Dai, Q.; Zheng, G.; He, C.; Pan, T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 2013, 19, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Dhir, A.; Dhir, S.; Proudfoot, N.J.; Jopling, C.L. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat. Struct. Mol. Biol. 2015, 22, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Peng, Z.; Cheng, Y.; Tan, B.C.; Kang, L.; Tian, Z.; Zhu, Y.; Zhang, W.; Liang, Y.; Hu, X.; Tan, X.; et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012, 30, 253–260. [Google Scholar] [CrossRef]
- Ramaswami, G.; Zhang, R.; Piskol, R.; Keegan, L.P.; Deng, P.; O’Connell, M.A.; Li, J.B. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 2013, 10, 128–132. [Google Scholar] [CrossRef]
- Sakurai, M.; Ueda, H.; Yano, T.; Okada, S.; Terajima, H.; Mitsuyama, T.; Toyoda, A.; Fujiyama, A.; Kawabata, H.; Suzuki, T. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 2014, 24, 522–534. [Google Scholar] [CrossRef]
- Salameh, A.; Lee, A.K.; Cardo-Vila, M.; Nunes, D.N.; Efstathiou, E.; Staquicini, F.I.; Dobroff, A.S.; Marchio, S.; Navone, N.M.; Hosoya, H.; et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl. Acad. Sci. USA 2015, 112, 8403–8408. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Cescon, M.; Bonaldo, P. Autophagy-mediated regulation of macrophages and its applications for cancer. Autophagy 2014, 10, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.F.; Wang, X.Y.; Fu, Z.Q.; Peng, Q.H.; Zhang, J.Y.; Ye, F.; Fu, Y.F.; Zhou, C.Y.; Lu, W.G.; Cheng, X.D.; et al. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy 2015, 11, 225–238. [Google Scholar] [CrossRef]
- YiRen, H.; YingCong, Y.; Sunwu, Y.; Keqin, L.; Xiaochun, T.; Senrui, C.; Ende, C.; XiZhou, L.; Yanfan, C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer 2017, 16, 174. [Google Scholar] [CrossRef]
- Huang, F.; Chen, W.; Peng, J.; Li, Y.; Zhuang, Y.; Zhu, Z.; Shao, C.; Yang, W.; Yao, H.; Zhang, S. LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol. Cancer 2018, 17, 98. [Google Scholar] [CrossRef]
- Xiu, Y.L.; Sun, K.X.; Chen, X.; Chen, S.; Zhao, Y.; Guo, Q.G.; Zong, Z.H. Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget 2017, 8, 31714–31725. [Google Scholar] [CrossRef]
- Xiong, H.; Ni, Z.; He, J.; Jiang, S.; Li, X.; He, J.; Gong, W.; Zheng, L.; Chen, S.; Li, B.; et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017, 36, 3528–3540. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, N.; Liu, Y.; Su, P.; Liang, Y.; Li, Y.; Wang, X.; Chen, T.; Song, X.; Sang, Y.; et al. Epigenetic regulation of NAMPT by NAMPT-AS drives metastatic progression in triple-negative breast cancer. Cancer Res. 2019, 79, 3347–3359. [Google Scholar] [CrossRef]
- Liu, C.; Fu, H.; Liu, X.; Lei, Q.; Zhang, Y.; She, X.; Liu, Q.; Liu, Q.; Sun, Y.; Li, G.; et al. LINC00470 coordinates the epigenetic regulation of ELFN2 to distract GBM cell autophagy. Mol. Ther. J. Am. Soc. Gene Ther. 2018, 26, 2267–2281. [Google Scholar] [CrossRef]
- Li, L.; Chen, H.; Gao, Y.; Wang, Y.W.; Zhang, G.Q.; Pan, S.H.; Ji, L.; Kong, R.; Wang, G.; Jia, Y.H.; et al. Long Noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol. Cancer Ther. 2016, 15, 2232–2243. [Google Scholar] [CrossRef]
- Xu, S.; Wang, P.; Zhang, J.; Wu, H.; Sui, S.; Zhang, J.; Wang, Q.; Qiao, K.; Yang, W.; Xu, H.; et al. Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol. Cancer 2019, 18, 89. [Google Scholar] [CrossRef] [PubMed]
- Bester, A.C.; Lee, J.D.; Chavez, A.; Lee, Y.R.; Nachmani, D.; Vora, S.; Victor, J.; Sauvageau, M.; Monteleone, E.; Rinn, J.L.; et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell 2018, 173, 649–664 e620. [Google Scholar] [CrossRef] [PubMed]

| LncRNA | miRNA | mRNA | Cancer Type | Ref |
|---|---|---|---|---|
| PTENP1 | PTEN-targeting miRNAs | PTEN | Various tumor cells | [27] |
| LINC00673 | miR-150-5p | ZEB1 | Lung cancer | [28] |
| ODRUL | miR-3182 | MMP2 | Osteosarcoma | [29] |
| LINC01234 | miR-204-5p | CBFB | Gastric cancer | [30,31] |
| NEAT1 | miR-34a | SIRT1 | Colorectal cancer | [32] |
| WDFY3-AS2 | miR-18a | RORA | Ovarian cancer | [33] |
| LncRNA | Chromatin Remodeling Complexes | Locus | Cancer Type | Ref |
|---|---|---|---|---|
| ANRIL | PRC1 (CBX7) | INK4b-ARF-INK4a | Prostate cancer | [45] |
| HOTAIR | PRC2 | HOXD cluster | Breast cancer | [46] |
| PTV1 | PRC2 (EZH2) | p15, p16 | Gastric cancer | [47] |
| LINK-PINT | PRC2 | Pro-invasive genes | Lung/colon cancer | [48] |
| FGFR2-AS | PRC2/KDM2a | FGFR2 | Epithelial cell | [49,50] |
| HOXD-AS1 | WDR5 | Target genes | Prostate cancer | [51] |
| GCAWKR | WDR5/KAT2A | Target genes | Gastric cancer | [52] |
| GAS8-AS1 | MLL1/WDR5 | GAS8 | Liver cancer | [53] |
| LncFZD6 | SWI/SNF (BRG1) | FZD6 | Liver cancer | [54] |
| SATB2-AS1 | p300 | SATB2 | Colorectal cancer | [55] |
| HAND2-AS1 | INO80 | BMPR1A | Liver cancer | [56] |
| LncHOXA10 | NURF | HOXA10 | Liver cancer | [57] |
| PYCARD-AS1 | DNMT1/G9a | PYCARD | Breast cancer | [58] |
| LncRNA | LncRNA Interactor | mRNA/Protein Stability | Cancer Type | Ref |
|---|---|---|---|---|
| UCA1 | hnRNPA1 | CDKN2A-p16 | - | [66] |
| PDCD4-AS1 | HuR | PDCD4 | Breast cancer | [67] |
| PVT1 | - | MYC | 8q24-amplified cancer cells | [68] |
| LINK-A | BRK, LRRK2 | HIF1α | Breast cancer | [69] |
| ANCR | - | EZH2 | Breast cancer | [70] |
| GAS5 | - | YAP | Colorectal cancer | [71] |
| Regulator | LncRNA | Regulatory Mechanism | Cancer Type | Ref |
|---|---|---|---|---|
| EGF | LIMT | Histone deacetylation of promoter | Breast cancer | [96] |
| Growth factors | Quiescence-induced lncRNAs | - | - | [97] |
| Androgen | CTBP1-AS | AR-binding sites | Prostate cancer | [98] |
| Estrogen receptor β | HOTAIR | Estrogen receptor β binding | Renal cell carcinoma | [99] |
| Oestrogen | DSCAM-AS1 | Oestrogen receptor binding | Breast cancer | [100] |
| DNA damage stress | Damage-inducing lncRNAs | MRE11-RAD50-NBS1 complex recruitment | - | [101] |
| MYC | MYCLos | Transcription regulation | Colorectal cancer | [102] |
| Histone modification | HOXC-AS3 | Promotor modification | Gastric cancer | [103] |
| LncRNA | Target | Mechanism | Cancer Type | Ref |
|---|---|---|---|---|
| MALAT1 | ATG12, miR-23b-3p | ceRNA | Gastric cancer | [125] |
| PVT1 | ULK1, miR-20a-5p | ceRNA | Pancreatic cancer | [126] |
| MEG3 | ATG3 | Protein degradation | Ovarian cancer | [127] |
| HULC | SIRT1 | Protein degradation | Liver cancer | [128] |
| NAMPT-AS | POU2F2, mTOR | Scaffold of transcription factor | Breast cancer | [129] |
| LINC00470 | AurkA, eIF2α | Epigenetic regulation | Glioblastoma | [130] |
| MALAT1 | HuR, TIA-1 | mRNA stability | Pancreatic cancer | [131] |
| EGOT | ITPR1 | Alternative splicing | Breast cancer | [132] |
| Identifier | Purpose | Application | Cancer Type |
|---|---|---|---|
| NCT03830619 | Evaluate the sensitivity/specificity of serum exosome lncRNA as a diagnosis biomarker | Diagnosis | Lung cancer |
| NCT04269746 | Evaluate the diagnostic value of lncRNA CCAT1 expression | Diagnosis | Colorectal cancer |
| NCT03469544 | Evaluate the value of lncRNA HOTAIR and midkine as biomarkers | Diagnosis | Thyroid cancer |
| NCT03738319 | Screen the candidate lncRNAs as biomarkers for the prognosis | Diagnosis | Epithelia Ovarian Cancer |
| NCT03057171 | Evaluate the expression of lncRNA THRIL and PACER by helicobacter pylori infection | Diagnosis | Stomach cancer |
| NCT03000764 | Seek a molecular signature (e.g., lncRNA) of pathological radiation-induced fibrosis | Diagnosis | Breast cancer |
| NCT04288739 | Detect the prognostic role of lncRNA XIST in acute myeloid leukemia | Diagnosis | Hematologic cancer |
| NCT03742869 | Detect the expression of lncRNAs in uterine cervical adenocarcinoma patients with and without HPV integration | Diagnosis | Uterine cervical adenocarcinoma |
| NCT04269746 | Validate the mRNA-lncRNA signature to predict the efficacy/recurrence risk after a combination of chemotherapy drugs | Prognosis | Triple-negative breast cancer |
| NCT03742856 | Predict the invasiveness and tumorigenesis of cancer cells with different FIGO stages and subtypes | Prognosis | Epithelial ovarian cancer |
| NCT04010487 | Predict the pathogenesis of the malignant transformation of adenomyosis | Prognosis | Endometrial cancer |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.-Z.; Liu, H.; Chen, S.-R. Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations. Cancers 2020, 12, 1245. https://doi.org/10.3390/cancers12051245
Zhang X-Z, Liu H, Chen S-R. Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations. Cancers. 2020; 12(5):1245. https://doi.org/10.3390/cancers12051245
Chicago/Turabian StyleZhang, Xiao-Zhen, Hao Liu, and Su-Ren Chen. 2020. "Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations" Cancers 12, no. 5: 1245. https://doi.org/10.3390/cancers12051245
APA StyleZhang, X.-Z., Liu, H., & Chen, S.-R. (2020). Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations. Cancers, 12(5), 1245. https://doi.org/10.3390/cancers12051245






